Abstract
Parkinson’s disease (PD), the most common movement disorder, is characterized by age-dependent degeneration of dopaminergic neurons in the substantia nigra of the mid-brain. Non-motor symptoms of PD, however, precede the motor features caused by dysfunction of the dopaminergic system, suggesting that PD is a systemic disorder. Mitochondrial dysfunction has long been observed in PD patients and animal models, but the mechanistic link between mitochondrial dysfunction and PD pathogenesis is not well understood. Recent studies have revealed that genes associated with autosomal recessive forms of PD such as PINK1 and Parkin are directly involved in regulating mitochondrial morphology and maintenance, abnormality of which is also observed in the more common, sporadic forms of PD, although the autosomal recessive PDs lack Lewy-body pathology that is characteristic of sporadic PD. These latest findings suggest that at least some forms of PD can be characterized as a mitochondrial disorder. Whether mitochondrial dysfunction represents a unifying pathogenic mechanism of all PD cases remains a major unresolved question.
Introduction
Mitochondrial dysfunction has long been implicated in the etiology of PD. The discovery of the Parkinsonism-inducing neurotoxin 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP), which is a selective inhibitor of mitochondrial complex I, directed researchers’ attention to pathological roles of mitochondria in PD and raised the possibility that environmental toxins affecting mitochondria might cause PD. Other mitochondrial toxins characterized as parkinsonism-inducing reagents include 6-Hydroxy-Dopamine (6-OHDA), rotenone and paraquat. Studies of animal models of PD induced with these toxins suggest that mitochondrial dysfunction and oxidative stress are important pathogenic mechanisms [1]. In humans, reduced complex I activity has been reported in both post-mortem brain samples and platelets of sporadic PD cases [2-4], and mutations or polymorphisms in mitochondrial DNA can confer genetic risk for PD [5]. Genetic evidence has also come from studies of familial forms of PD (FPD). The identification and characterization of FPD genes have provided an unprecedented opportunity to understand pathogenic mechanisms underlying dopaminergic neurodegeneration. Studies of FPD have revealed two distinct but potentially inter-connected disease pathways: the autosomal dominant genes represented by α-Synuclein that lead to Lewy-body pathology, and the autosomal recessive genes Parkin, PINK1, DJ-1, and HtrA2/Omi that have been linked to regulation of mitochondria. In this review, we focus on recent findings from molecular genetic and cell biological studies that reveal the roles of the autosomal recessive FPD genes in governing mitochondrial functions and discuss how loss of function of these genes may lead to neurodegeneration. It is anticipated that studies of these autosomal recessive FPD genes will also help understand the pathogenesis of sporadic and the autosomal dominant FPD cases, which also feature mitochondrial pathology.
Regulation of mitochondrial dynamics by PINK1 and Parkin
Mutations of the Parkin gene cause an autosomal recessive juvenile form of PD (AR-JP). The gene product contains a ubiquitin-like (Ubl) domain at the N-terminus and two RING fingers flanking a cysteine-rich domain, termed In Between RING fingers (IBR), which confer E3 ubiquitin-ligase activity. To study Parkin function, several Parkin-deficient mice have been generated. However, most of them do not fully recapitulate dopaminergic neurodegeneration, which has hindered elucidation of the pathological mechanisms of AR-JP. The discovery of a genetic interaction between Parkin and PINK1 in Drosophila has shed light on Parkin function in vivo [6-8]. The PINK1 gene, mutations of which also cause juvenile PD, encodes a serine-threonine kinase with a mitochondria-targeting signal at the N-terminus. Loss of PINK1 or Parkin genes in Drosophila results in mitochondrial aggregation and cellular degeneration in dopaminergic neurons muscle and sperm, leading to motor impairment and decreased fertility [6-8]. Overexpression of wild-type Parkin can rescue the phenotypes caused by PINK1 deficiency, but not the other way around [6-8]. These studies suggest that Parkin is epistatic to PINK1 and that it affects mitochondrial function. Parkin protein is mainly localized to the cytosol, and the molecular mechanism by which it regulates mitochondrial function is an open question.
In contrast to the textbook view of kidney bean-shaped organelles, mitochondria exhibit dynamic morphological changes in vivo associated with changes in distribution and function. These morphological changes are regulated by a delicate balance between the opposing processes of mitochondrial fusion and fission. Increased fission leads to mitochondrial fragmentation, while increased fusion leads to mitochondrial elongation or aggregation. One remarkable feature of the PINK1-deficient fly is the presence of highly aggregated mitochondria in dopaminergic neurons [8,9]. A similar mitochondrial morphological abnormality is observed in the flight muscle of PINK1- and Parkin-deficient flies, in which swollen mitochondria, often with disintegrated cristae, are observed [10,11]. Interestingly, PINK1 and Parkin mutant phenotypes are partly rescued by increased activity of Drp1, which is a major component of the mitochondrial fission machinery, or by reduced activity of Mitofusin (Mfn) or OPA1, which together control mitochondrial fusion [9,11,12]. Abnormal mitochondrial morphology and dynamics are also observed in mammalian cultured cells and hippocampal and dopaminergic neurons [9,13]. These findings suggest that PINK1 and Parkin may have conserved roles in the regulation of neuronal mitochondrial morphology and function. This represents a breakthrough in PD research.
Regulation of mitophagy by PINK1 and Parkin
Another breakthrough in our understanding of PINK1/Parkin function came from a series of elegant cell biological studies. When the mitochondrial membrane potential is disrupted by mitochondria-damaging reagents such as carbonyl cyanide m-chlorophenylhydrazone (CCCP) in mammalian [14-18] or Drosophila cultured cells [19], Parkin translocates to mitochondria with low membrane potential, where it promotes LC3-mediated autophagic elimination of the damaged mitochondria in a process called mitophagy (Figure 1) [20]. After Parkin translocation, mitochondrial accumulation of poly-ubiquitinated proteins, consisting mainly of Lys63-linked poly-ubiquitin and a small portion of Lys48-linkages [21,22], recruits the ubiquitin- and LC3-binding adaptor protein p62/SQSTM1 [16,23,24] and the ubiquitin-binding deacetylase HDAC6 [21]. Although important details are still unresolved, Lys63-linked poly-ubiquitination may contribute to proteasomal degradation of mitochondrial proteins [25] and HDAC6- and/or p62-mediated sequestration of mitochondria [21,22]. Mitochondria depolarized by CCCP or paraquat accumulate in the perinuclear compartment in a p62/SQSTM1-dependent manner [16,23,24]. This is followed by engulfment of the damaged mitochondria by autophagosomes and subsequent lysosomal degradation [20]. The clustering of ubiquitinated mitochondria by p62 and HDAC6 is reminiscent of their sequestration of ubiquitinated proteins into aggresomes [26,27].
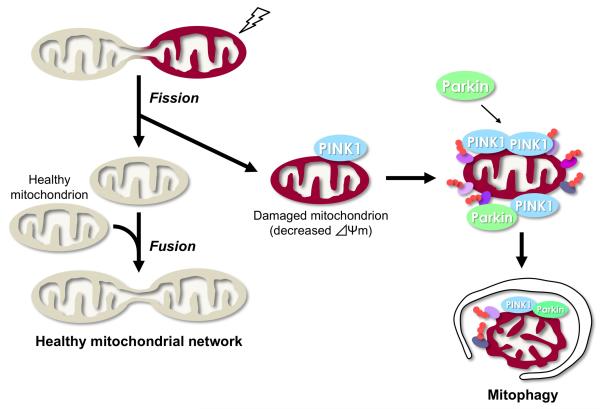
Figure 1.
Mitochondrial fusion and fission events are required for the maintenance of a healthy mitochondrial population (beige). Mitochondrial fusion is thought to facilitate the interchange of internal components such as copies of the mitochondrial genome, respiratory proteins and metabolic products. Mitochondrial fission may play a role in the removal of dysfunctional mitochondria (dark red) with reduced mitochondrial membrane potential (Δψm), through an autophagy-lysosomal pathway named “mitophagy”. PINK1 and Parkin are likely to be involved in this process. PINK1 normally has a short half-life in healthy mitochondria. Upon reduction of the Δψm, PINK1 is stabilized on the OM. Accumulation of PINK1 induces the translocation of Parkin from the cytosol to the mitochondria, leading to Parkin-dependent ubiquitination and degradation of the mitochondrial proteins, and subsequent activation of the autophagy machinery. Ubiquitnated proteins of the mitochondria are shown as ovals with small orange circles.
The translocation of Parkin from the cytosol to the mitochondria, which requires intact PINK1 with kinase activity, is an essential step for mitophagy [17,18]. Through the ubiquitin-proteasome pathway, Parkin ubiquitinates and degrades several proteins localized at the mitochondrial outer membrane, including Mfn [28-30], Drp1 [31], voltage-dependent anion channel 1 (VDAC1) [16,30] and Bcl-2 [32]. The degradation of the mitochondrial fusion factor Mfn by Parkin was also observed in Drosophila cultured cells [19,33]. This may contribute to the fragmentation of mitochondria and facilitate mitophagy. This finding is consistent with the in vivo observations that loss of PINK1 or Parkin leads to mitochondrial elongation, which is rescued by a reduction of Mfn activity. However, the elimination of Mfn by Parkin and the perinuclear aggregation of mitochondria by p62/SQSTM1 appear to be dispensable for mitophagy in mammalian cells [23,24,30], although the requirement of p62 is controversial [16]. Mfn degradation and mitochondrial perinuclear clustering may prevent the re-fusion of depolarized mitochondria with healthy ones, or compromise the axonal transport of damaged mitochondria [23,28]. In addition, these events may facilitate the isolation of mitochondria by the autophagosomes [28].
How is the autophagy machinery targeted to mitochondria? In yeast, an outer mitochondrial protein ATG32 is reported to recruit the autophagy machinery [34,35]. Although there is no homologue of ATG32 in higher animals, mammalian BNIP3 (BCL2 and adenovirus E1B 19 kDa-interacting protein 3) and NIX/BNIP3-like (BNIP3L), which belong to the BH3-only mitochondrial protein family, induce both cell death and mitophagy. NIX is involved in the programmed mitochondrial clearance by mitophagy during reticulocyte maturation [36,37], and is reported to be required for Parkin translocation to depolarized mitochondria treated with CCCP [38]. But it is unclear whether NIX functions to prime the recruitment of the autophagy machinery in this context as ATG32 does in yeast, or whether it acts as a regulator of PINK1.
Regulation of PINK1 and Parkin
Although endogenous PINK1 is difficult to detect under normal conditions, PINK1 rapidly accumulates in depolarized mitochondria [14,17,18]. This suggests that PINK1 protein is regulated by a post-translational degradation mechanism (Figure 2). Several studies indicate that the rhomboid family protease presenilin-associated rhomboid-like protein (PARL), which is localized to the mitochondrial inner membrane, processes PINK1 in a mitochondrial membrane potential-dependent manner [39-43]. Newly synthesized PINK1 in the cytosol is imported and inserted into the mitochondrial inner membrane (IM), and is cleaved in its putative transmembrane domain by PARL to generate the 52-kD form of PINK1, which is rapidly removed by a proteasome-dependent pathway, likely after its release into the cytosol from the mitochondrial intermembrane space (IMS) [40-42]. Upon depolarization of the mitochondrial membrane potential, the IM insertion and the subsequent processing of PINK1 by PARL may be inhibited, leading to full-length PINK1 accumulating in the mitochondrial outer membrane (OM), probably facing the cytosol [41,42,44]. However, there is discrepancy as to whether the processing of PINK1 by PARL is required for Parkin recruitment upon mitochondria depolarization, and further studies are necessary to completely resolve the changes in topology of the processed forms of PINK1 [41,43]. The accumulation of PINK1 with kinase activity is sufficient for Parkin recruitment to the mitochondrial surface, where Parkin’s E3 activity appears to be stimulated, although the phosphorylation target(s) of PINK1 remains unknown [17]. Conformational change of Parkin on the mitochondria may serve to activate its E3 activity, as the Ubl domain of Parkin normally inhibits its E3 activity intramolecularly [45]. Parkin is also upregulated by ATF4, a transcription factor of the unfolded protein response (UPR) [46]. Mitochondrial damage may induce the activation of the UPR, leading to the upregulation of Parkin expression [46].
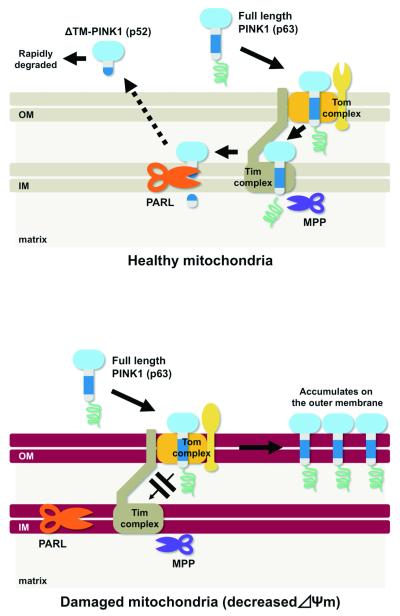
Figure 2.
Proposed model of post-translational processing of PINK1. (Upper) Newly synthesized PINK1 (p63) is targeted to the IM via the Tom and Tim complexes. PINK1 p63 may be processed by mitochondrial processing protease (MPP), which cleaves the mitochondrial targeting sequence to generate a 60-kD PINK1. PINK1 is then cleaved to a 52-kD form within the IM by PARL. The 52-kD PINK1 is released into the cytosol and is degraded by proteasome activity. (Lower) Upon reduction of the Δψm, PINK1 is accumulated at the OM, probably due to inhibition of the translocation through the Tim complex.
Loss of mitochondrial control and possible disease relevance
Although the relevance of mitophagy observed with mitochondria-damaging reagents to PD etiology remains debatable, accumulating evidence of mitochondrial abnormality in animal models and PD patients has increased our understanding of disease pathogenesis. The ubiquitination and elimination of Mfn following oxidative stress induction as well as mitochondrial depolarization are reproduced in human fibroblasts derived from PINK1- or Parkin patients [47]. Recent studies of Parkin- or PINK1-deficient mice have reported morphological and functional alterations of mitochondria in both neurons [48,49] and astrocytes [50]. A missense mutation in PARL found in PD cases abolishes its PINK1-processing activity and the ensuing Parkin-mediated mitophagy [43]. Like the muscle degeneration in Drosophila, the function of cardiac muscle, in which mitochondria are abundant, is also impaired by increased oxidative stress in PINK1-null mice [51]. Ischemic preconditioning has cardioprotective effects in heart failure models, where the mitochondrial translocation of Parkin is induced. Parkin deletion abolishes this effect [52]. Although the roles of PINK1 and Parkin in human cardiac function are unknown, it is worth noting that the prevalence of heart failure in elderly PD patients is double that of non-PD controls [53].
Contribution of DJ-1 and HtrA2 to mitochondrial regulation
DJ-1, which can exert neuroprotective effect by scavenging hydrogen peroxide through self-oxidation, has been reported to be involved in mitochondrial maintenance. Recent data suggest that DJ-1 acts in parallel to the PINK1-Parkin pathway to control mitochondrial polarization and morphology in cultured cells [54,55] and mitochondrial coupling and ATP production in Drosophila [56] in certain contexts, but surprisingly DJ-1/PINK1/Parkin triple knockout mice do not exhibit degeneration in the nigrostriatal system [57]. Although the linkage of the HtrA2 gene to PD pathogenesis is under debate [58], loss of the HtrA2 gene, which encodes a mitochondrial serine protease, leads to selective loss of striatal neurons in mice [59]. Genetic studies in Drosophila showed that HtrA2 mutants do not exhibit mitochondrial morphological defects and there is no genetic interaction that supports HtrA2 functioning in the same genetic pathway as Pink1 in terms of regulation of mitochondrial integrity and dynamics [60]. Furthermore, HtrA2-associated neurodegeneration was not rescued by a Parkin transgene in mice [61]. Together, these results suggest that HtrA2 may not be functioning in the PINK1-Parkin pathway.
Concluding remarks
Prominent pathological features of PD include mitochondrial dysfunction and the accumulation of protein inclusions into Lewy-bodies. These disease phenotypes could arise from impairments in the cellular quality control systems for mitochondria and cytoplasmic proteins involving mitochondrial fission/fusion dynamics, the ubiquitin-proteasome system, and the autophagy pathway. These cellular quality control systems do not work in isolation but rather are inter-connected. This could explain why mutations in the autosomal recessive and autosomal dominant FPD genes, which impair the mitochondrial quality control and cytoplasmic protein quality control, respectively, lead to distinct pathological hallmarks but similar clinical outcomes. Impairment of the ubiquitin-proteasome pathway can induce the accumulation of reactive oxygen species in mitochondria [62], with the affected mitochondria later removed by the autophagy pathway [62]. In addition to impaired mitophagy, decreased mitochondrial biogenesis, which may be closely linked to the TOR-mediated protein translation pathway [64], is also implicated in PD pathogenesis [63,65,66]. Thus, pathways for protein synthesis, quality control, mitochondrial maintenance, and mitochondrial dynamics are mechanistically inter-connected in the pathogenesis of PD, and represent novel targets for disease prevention and treatment.
Box 1. Physiological meaning of mitophagy.
The elimination of damaged mitochondria by the PINK1-Parkin pathway appears to be divided into two phases. The first step may be characterized as Parkin- and proteasome-dependent protein degradation of a broad range of the mitochondrial OM proteins, including Mfn1, Mfn2, Tom70 and Tom20 [30,67]. The second step involves Parkin-dependent mitophagy, with which the proteasomal activity may be functionally coupled [30]. Alternatively, proteasomal activity in the second step may be required only for destruction of the mitochondrial OM [67]. The degradation of a wide range of OM proteins by a proteasome- and a AAA+ family ATPase p97-dependent pathway raises the possibility that Parkin performs quality control of the OM proteins even under steady state, which bears some resemblance to ER-associated degradation (ERAD). ERAD is an important cellular event needed to eliminate aberrant membrane and secretory proteins at the ER, which also involves the proteasome and p97 activities.
Highlights.
PINK1 and Parkin are involved in regulating mitochondrial fission and fusion dynamics.
PINK1 and Parkin are implicated in a process of autophagic removal of dysfunctional mitochondria called mitophagy.
Mitochondrial dynamics and Mitophagy are thought to be required for the maintenance of a healthy mitochondrial network.
The pathological relevance of mitophagy in PD etiology awaits further investigation.
Acknowledgments
We apologize to those authors whose work was not cited due to space limitation. We thank T. Sawada for providing artworks for the figures. Supported by Brain Science Foundation Research Grant, Suzuken Memorial Foundation Research Grant, Astellas Foundation for Research on Metabolic Disorders, Grant-in-Aid for Young Scientists (B) from MEXT in Japan (YI), and the NIH (R01AR054926 and R01MH080378) (BL)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
- 1.Hisahara S, Shimohama S. Toxin-induced and genetic animal models of Parkinson’s disease. Parkinsons Dis. 2010;2011:951709. doi: 10.4061/2011/951709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker WD, Jr., Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol. 1989;26:719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- 3.Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 4.Keeney PM, Xie J, Capaldi RA, Bennett JP., Jr. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 8.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- *9.Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. This study and [11*,12*] first showed that PINK1 and Parkin genes genetically interact with genes that directly control mitochondrial fission and fusion events in Drosophila. Genetic manipulations that increase mitochondrial fission activity or decrease fusion activity partially suppressed mutant phenotypes in PINK1- and Parkin-deficient flies.
- 10.Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6:e1001229. doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *11.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. See annotation to Ref. [9*].
- *12.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. See annotation to Ref. [9*].
- 13.Yu W, Sun Y, Guo S, Lu B. The PINK1/Parkin pathway regulates mitochondrial dynamics and function in mammalian hippocampal and dopaminergic neurons. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr235. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- **17.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. This study determined in detail how pathogenic mutations in PINK1 and Parkin affect the mitophagy process and biochemically examined the molecular mechanisms regulating PINK1 processing and accumulation.
- **18.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. This study characterized the molecular mechanism of PINK1 protein turnover, which involves both processing by protease(s) and proteasome-dependent degradation. Mitochondrial accumulation of PINK1 following the depolarization of mitochondria was characterized as an initial step of mitophagy.
- 19.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **20.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. This paper is the first report describing the translocation of Parkin to depolarized mitochondria and the involvement of Parkin in the mitophagy pathway.
- 21.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 27.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- *28.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. The study showed Parkin-mediated ubiquitination of Mfn upon mitochondrial depolarization. The finding is closely related to the results of Drosophila genetic studies showing the genetic relationships between Parkin and the mitochondrial fission/fusion pathway [9*][11*][12*]. Involvement of the proteasome and p97 in mitophagy was also described.
- 29.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. This study and [67*] revealed that Parkin-mediated mitophagy consist of two steps: The first step involves an ubiquitin proteasome-dependent process of OM protein degradation. The second one recruits the autophagy machinery to remove damaged mitochondria. Moreover, this study conducted exhaustive proteomic identification of proteins with increased or decreased abundance in mitochondria when mitophagy is induced.
- 31.Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, Wang G. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 36.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. discussion 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. This study and [40,42,43*] demonstrated that PINK1 protein inserted into OM is cleaved by the mitochondrial protease PARL.
- 42.Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- *43.Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, Ross OA, Farrer M, McQuibban GA, Bulman DE. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum Mol Genet. 2011;20:1966–1974. doi: 10.1093/hmg/ddr077. The study demonstrated processing of PINK1 by PARL and is the first to report that a mutation of PARL lacking PINK1 procesing activity is found in PD patients.
- 44.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011 doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouman L, Schlierf A, Lutz AK, Shan J, Deinlein A, Kast J, Galehdar Z, Palmisano V, Patenge N, Berg D, et al. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress. Cell Death Differ. 2011;18:769–782. doi: 10.1038/cdd.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. Using fibroblasts derived from PD patients with PINK1- and Parkin-linked mutations, this study provided evidence that impairment of the mitophagy pathway is involved in PD etiology. Elimination of Mfn upon mitochondrial depolarization was abolished in patient samples. In addition, oxidative stress by peroxide treatment was shown to induce mitophagy.
- 48.Gispert S, Ricciardi F, Kurz A, Azizov M, Hoepken HH, Becker D, Voos W, Leuner K, Muller WE, Kudin AP, et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt S, Linnartz B, Mendritzki S, Sczepan T, Lubbert M, Stichel CC, Lubbert H. Genetic mouse models for Parkinson’s disease display severe pathology in glial cell mitochondria. Hum Mol Genet. 2011;20:1197–1211. doi: 10.1093/hmg/ddq564. [DOI] [PubMed] [Google Scholar]
- 51.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning Involves Selective Mitophagy Mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zesiewicz TA, Strom JA, Borenstein AR, Hauser RA, Cimino CR, Fontanet HL, Cintron GB, Staffetti JF, Dunne PB, Sullivan KL. Heart failure in Parkinson’s disease: analysis of the United States medicare current beneficiary survey. Parkinsonism Relat Disord. 2004;10:417–420. doi: 10.1016/j.parkreldis.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Irrcher I, Aleyasin H, Seifert EL, Hewitt SJ, Chhabra S, Phillips M, Lutz AK, Rousseaux MW, Bevilacqua L, Jahani-Asl A, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 55.Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci U S A. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitada T, Tong Y, Gautier CA, Shen J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J Neurochem. 2009;111:696–702. doi: 10.1111/j.1471-4159.2009.06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon-Sanchez J, Singleton AB. Sequencing analysis of OMI/HTRA2 shows previously reported pathogenic mutations in neurologically normal controls. Hum Mol Genet. 2008;17:1988–1993. doi: 10.1093/hmg/ddn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- 60.Yun J, Cao JH, Dodson MW, Clark IE, Kapahi P, Chowdhury RB, Guo M. Loss-of-function analysis suggests that Omi/HtrA2 is not an essential component of the PINK1/PARKIN pathway in vivo. J Neurosci. 2008;28:14500–14510. doi: 10.1523/JNEUROSCI.5141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshida T, Mizuta T, Shimizu S. Neurodegeneration in mnd2 mutant mice is not prevented by parkin transgene. Biochem Biophys Res Commun. 2010;402:676–679. doi: 10.1016/j.bbrc.2010.10.083. [DOI] [PubMed] [Google Scholar]
- 62.Takeda K, Yoshida T, Kikuchi S, Nagao K, Kokubu A, Pluskal T, Villar-Briones A, Nakamura T, Yanagida M. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Natl Acad Sci U S A. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *65.Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet. 2010;6:e1001237. doi: 10.1371/journal.pgen.1001237. This paper showed that activation of autophagy is protective in fly PINK1 mutant. Surprisingly, autophagy did not appear to be required for the rescue of PINK1 mutant by Parkin, suggesting that in vivo, Parkin may act through multiple pathways. It also showed that increasing protein synthesis was detrimental in PINK1 mutant, whereas decreasing protein synthesis was protective. Moreover, increaded autophagy and decreased translation are already induced in PINK1 mutant, suggesting that they represent compensatory responses.
- 66.Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *67.Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin Mediates Proteasome-dependent Protein Degradation and Rupture of the Outer Mitochondrial Membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. See annotation to Ref. [30**].