Abstract
The most primitive hematopoietic stem cell (HSC)/progenitor cell (PC) population reported to date is characterized as being Lin−CD34+CD38−CD90+CD45R. We have a long-standing interest in comparing the characteristics of hematopoietic progenitor cell populations enriched from normal subjects and patients with chronic myelogenous leukemia (CML). In order to investigate further purification of HSCs and for potential targetable differences between the very primitive normal and CML stem/PCs, we have phenotypically compared the normal and CML Lin−CD34+CD38−CD90+CD45RA− HSC/PC populations. The additional antigens analyzed were HLA-DR, the receptor tyrosine kinases c-kit and Tie2, the interleukin-3 cytokine receptor, CD33 and the activation antigen CD69, the latter of which was recently reported to be selectively elevated in cell lines expressing the Bcr-Abl tyrosine kinase. Notably, we found a strikingly low percentage of cells from the HSC/PC sub-population isolated from CML patients that were found to express the c-kit receptor (<1%) compared with the percentages of HSC/PCs expressing the c-kitR isolated from umbilical cord blood (50%) and mobilized peripheral blood (10%). Surprisingly, Tie2 receptor expression within the HSC/PC subset was extremely low from both normal and CML samples. Using in vivo transplantation studies, we provide evidence that HLA-DR, c-kitR, Tie2 and IL-3R may not be suitable markers for further partitioning of HSCs from the Lin−CD34+CD38−CD90+CD45RA− sub-population.
Keywords: stem cells, phenotype, normal, myeloid, leukemic
Introduction
Hematopoietic stem cells (HSCs) are a very primitive and rare population of cells that reside in the bone marrow and give rise to all the cells of the blood including erythrocytes, granulocytes, monocytes, platelets, natural killer cells and lymphocytes of B and T lineage. Because of the unique ability of stem cells to both self-renew and differentiate, the continued replenishment of these various specialized blood cells (the process of homeostasis) is sustained for the entire lifespan of the species.1
Largely because of the relative ease with which one can access hematopoietic tissue and the development of functional assays to measure their activity, the HSC is perhaps the most well-characterized stem cell on both the biological and molecular levels. However, despite much progress, HSC studies remain quite difficult because of their rarity within the total heterogeneous population of hematopoietic cells (<1 in 50 000).2 Having a homogenous population of HSCs would immensely enhance directly identifying and examining the biological and molecular programs controlling normal stem cell function and, importantly, determine how these programs may be altered in leukemic stem cells (LSCs). In addition, purified HSCs ultimately may prove to be extremely valuable for therapeutic purposes as well.3 Over the past 40 years or so, the enrichment of HSCs on the basis of their physical (for example, cell size, density), cell surface antigen, and biochemical characteristics (for example, elevated levels of aldehyde dehydrogenase) has been intensely pursued by a multitude of investigators including ourselves.4, 5, 6, 7, 8, 9, 10, 11, 12 Whereas such efforts have culminated in the ability to purify the HSC to near homogeneity from mouse bone marrow and fetal liver,13, 14 the same cannot be said for human HSCs. However, Irving Weissman and colleagues15, who have made major contributions in the area of HSC purification and characterization over the years, have moved us ever closer toward that goal. Recently, they reported the enrichment of human HSCs from umbilical cord blood (CB; also known to be a very rich source of HSCs16) from an estimated starting frequency of <1 in 50 000 cells to a frequency of 1 in 10 cells.17 Thus, they were able to demonstrate long-term multilineage engraftment (post 12-week transplantation) in newborn nonobese diabetes/severe combined immunodeficiency (NOD/SCID) IL-2R gammanull (NOG) mice upon facial injection of as little as 10 cells from their purified HSC fractions. Mature and very primitive hematopoietic cell populations express somewhat different cell surface antigens.15 Fluorophore-conjugated monoclonal antibodies directed against these antigens were used with multicolor flow cytometry to isolate phenotypically defined populations simultaneously. Their highly enriched HSC fraction was identified as being lineage (Lin) negative(−) CD34 positive(+) CD38−CD90+CD45RA−. This HSC phenotypic profile indeed encompasses previous findings by Weissman and colleagues15 and those of numerous other investigators demonstrating that the most primitive human hematopoietic progenitor cell (PC) populations are lineage−,6, 18 CD34+,19 CD38−,20 CD90+18 and CD45RA−.21
Given our long-standing interest in comparing the biological, biochemical and molecular characteristics of primitive hematopoietic PC populations enriched from normal human subjects and from patients with chronic myelogenous leukemia (CML),22, 23 we decided to compare the expression of other well-known hematopoietic cell surface antigens by the Lin−CD34+CD38−CD90+CD45RA− HSC/PC population isolated from both these sources in order to determine (1) if HSCs could be further purified on the basis of another cell surface antigen(s) and (2) if there are any consistent phenotypic differences between highly enriched normal HSCs and CML LSCs. If consistent differences can be identified, they could have significant therapeutic potential such as in purging strategies for use in autologous transplantation or elimination of quiescent LSCs that survive current treatment regimens. CML is a clonal disease originating in a multipotent HSC or early PC because of fusion of portions of the Bcr and Abl genes, which results in constitutively increased Abl kinase activity that is thought to be necessary and sufficient for the initiation of CML.24, 25, 26 Currently, the front-line treatment of CML is the Bcr–Abl kinase inhibitor, imatinib mesylate.27, 28, 29, 30 Although this treatment has been remarkably successful, only a minority (∼25%) achieve major molecular remissions and a significant number develop Bcr–Abl kinase domain mutations or Bcr-Abl gene amplifications that confer resistance.31, 32, 33, 34 Also, there is now very strong evidence that quiescent LSCs (or early quiescent leukemic progenitors functioning as LSCs35) of the patients are refractory to imatinib mesylate as well as other newly designed more potent second-generation Bcr–Abl tyrosine kinase inhibitors such as nilotinib and dasatinib.36, 37, 38, 39 Because of this, patients must continuously take these tyrosine kinase inhibitors, as discontinuation results in the re-emergence of excessive numbers of myeloid cells. Thus, intensive efforts remain focused on identifying new molecular targets within or on the surface of Ph+ LSCs that can be exploited therapeutically.
The quiescent leukemic stem cell or early PCs functioning as LSCs are known to be concentrated in the most primitive cell compartments, because like normal cells, once stem or early PCs become committed to differentiation and maturation, they continue to proliferate.38
In the present studies, we have further phenotypically examined the Lin−CD34+CD38−CD90+CD45RA− HSC/PC population (hereafter, sometimes referred to as the CD90+CD45RA− HSC sub-population) purified from two normal tissue sources (CB and granulocyte colony-stimulating factor (G-CSF) mobilized peripheral blood (MPB)), as well as from peripheral blood or bone marrow samples from patients with CML. The cell surface antigens analyzed include HLA-DR, the receptor tyrosine kinases c-kitR (CD117) and Tie2 (CD202b), the interleukin-3 cytokine receptor (IL-3R, CD123) and CD33. Although all of these antigens have been well documented to be expressed by various classes of human hematopoietic PCs,40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57 the number of studies investigating their expression by human HSCs is sparse.58, 59, 60, 61, 62
In addition, we have looked at the expression of CD69, an immunoregulatory glycoprotein that is transiently expressed on the surface of all hematopoietic cells (except erythrocytes) following their activation.63 This antigen was of particular interest to us in light of a very recently published study64 demonstrating an upregulated expression of CD69 in the Ph+ cell line, K562, as well as in other hematopoietic cell lines retrovirally expressing the Bcr–Abl protein. Furthermore, they found that this upregulated expression was inhibited by the Bcr–Abl kinase inhibitors, nilotinib and dasatinib. Unfortunately, in that study, primary cells from CML patients were not examined. If CD69 is consistently elevated on the surface of LSCs from CML patients, it might serve as a therapeutic target.
In order to analyze the Lin−CD34+CD38−CD90+CD45RA− sub-population for their surface expression of HLA-DR, c-kitR, Tie2, CD33, IL-3 receptor and CD69, we have directly conjugated (or commercially obtained) the Alexa Fluor 700 dye to mouse monoclonal antibodies directed against these antigens. We chose this fluorophore as it displays high fluorescence intensity, is very photostable and exhibits essentially no spectral overlap with the other fluorescent dyes when used according to the multi-color cell sorting strategy described below. The only other fluorochrome used in this stain that spectrally overlaps with Alexa 700 (Invitrogen, Carlsbad, CA, USA) is allophycocyanin (APC) and this spillover is negated because APC-negative cells were analyzed.
This study is the first, to our knowledge, to examine the surface expression of these antigens by the very primitive Lin−CD34+CD38−CD90+CD45RA− HSC/PC population purified from UB, G-CSF MPB and peripheral blood and/or bone marrow samples obtained from CML patients. In addition, we have further partitioned this sub-population into HLA-DR + and −, c-kitR + and − and Tie2 + and − (and IL-3R+ and IL-3R− in a few instances) subsets by fluorescence-activated cell sorting and have transplanted cells from these fractions into sub-lethally irradiated NOG mice to measure their long-term marrow-repopulating ability. The results of these studies are the subject of the present report.
Materials and methods
Antibodies
A panel of mouse anti-human monoclonal antibodies (MoAbs) was used for analysis and sorting of HSC-enriched populations including: CD2, CD3, CD5, CD10, CD14, CD19, CD20, CD56 and My-8 (generously supplied by Coulter Immunology; now Beckman Coulter, Brea, CA, USA). CD235a (anti-Glycophorin A) was kindly supplied by Dr Paul Edwards (Ludwig Institute for Cancer Research). These MoAbs are directed against antigens expressed by B and T lymphocytes, monocytes, maturing granulocytes, erythroid cells and natural killer cells.65 All these antibodies were biotinylated in-house using the EZ-Link Micro Sulfo-NHS-LC-Biotinylation Kit (Pierce, Rockford, IL, USA), purified using a Zebra desalt spin column (Pierce), and subsequently tested and titrated (by flow cytometry) by staining established myeloid and lymphoid cell lines expressing these antigens. Biotinylated CD7 (cat. no. MHCD0715) was obtained from Invitrogen. In order to detect these biotinylated MoAbs, streptavidin PE-Cy5 (Biolegend, San Diego, CA, USA) was used as a secondary reagent. Fluorescein isothiocyanate (FITC) anti-CD34 (clone 581), PE-anti-CD90 (clone 5E10) and V450 anti-CD45RA (clone HI100) were obtained from BD Biosciences (San Jose, CA, USA). APC-anti-CD38 was purchased from eBioscience (San Diego, CA, USA). Alexa Fluor 700 anti-CD69 (clone FN50) was purchased from BioLegend. Anti-CD117, c-kitR (clone 104D2); CD123, α-chain of IL-3 receptor (clone 6H6); CD202b (Tie2; clone 33.1; Biolegend); CD33 (clone 906) and HLA-DR (clone H279; gifts from Coulter) were all labeled with Alexa Fluor 700 (cat no. A20010, Invitrogen) in-house according to the instructions accompanying Invitrogen's Alexa Fluor Monoclonal Antibody Labeling Kits. Conjugated antibodies were purified by passing the labeling reactions over a fluorescent dye removal spin column (Pierce) to remove free Alexa 700 dye. The specificity of these labeled antibodies was confirmed by staining established cell lines known to express these antigens.
Analysis of human engraftment was performed using anti-human FITC–CD45 (clone HI30) and FITC–IgG1 ê isotype control (BD Biosciences). In some instances, lineage analysis of human cells was performed using anti-human Abs: FITC–CD13 (clone SJ1D1) and FITC–CD19 (clone J4.119) (BD Biosciences). Mouse leukocytes were identified by staining with PE-CD45.1 (clone A20; Biolegend) and PE-IgG1 isotype control (BD Biosciences). Cell samples were incubated with mouse and human FcR blocking reagents (Miltenyi Biotec, Inc., Auburn, CA, USA) for 15 min at 4 °C before the addition of MoAbs.
Cell sources and CD34 cell enrichment
Fresh CB units were obtained from the New York Blood Center's National Cord Blood Program (Long Island City, NY, USA). The blood was diluted 1:1 with Iscove's modified Dulbecco's medium containing 5% fetal calf serum (FCS) and platelets were depleted by centrifugation over Percoll (1.050 g/cm3; GE Healthcare, Piscataway, NJ, USA) before obtaining the mononuclear cell (MNC) fraction by centrifugation over Ficoll-Hypaque as previously described.65, 66 In order to reduce the total number of MNCs while recovering essentially all the HSC/PCs, in some cases the MNCs were further subjected to a 1.071 g/cm3 Percoll density cut.65 Enriched CD34+ cells were then obtained by using the MACS CD34 cell isolation kit (Miltenyi Biotec).
Cryopreserved MPB samples were obtained from the Stem Cell Processing Laboratory at the Memorial Sloan-Kettering Cancer Center. Patients with solid tumors and no hematological involvement were mobilized with 10 μg/kg of G-CSF for 5 days. The hematopoietic progenitor cell-Apheresis products were volume reduced and mixed with an equal volume (50% v/v final) of cryoprotectant media containing 5% dimethyl sulfoxide and 5% human serum albumin. The cryoproducts were placed in a −80 °C freezer and frozen utilizing the ‘passive' cryopreservation method. The products were then transferred to a LN2 vapor storage device and maintained at −180 to −190 °C.
After appropriate Human Protection Committee Validation and informed consent, marrow aspirations or peripheral blood specimens were obtained from chronic-phase CML patients hospitalized at Memorial Sloan-Kettering during the period 1990–2006. Samples were also obtained from Dr Richard Silver (Emeritus Director, Clinical Oncology Chemotherapy Research, Division of Hematology and Medical Oncology, New York Presbyterian Cornell Medical Center, New York, NY, USA). Buffy coat cells were depleted of platelets and MNCs were obtained and cryopreserved as previously described by us in detail.65 In some instances, cryopreserved CML patient cell samples were kindly provided by Drs Tessa Holyoake (University of Glasgow, Scotland, UK) and Adrian Morales Maravilla (Puebla, Mexico). All patients had 100% Ph positivity in the bone marrow as assessed by either direct cytogenetic analysis or fluorescence in situ hybridization.
After thawing, mobilized blood cells or CML patient cells were suspended in Iscove's modified Dulbecco's medium containing 20% FCS and 80 units/ml of deoxyribonuclease (DNase; Worthington Biochemical Corp., Lakewood, NJ, USA). Dead cells were removed by Ficoll-Hypaque centrifugation and viable MNCs were recovered at the interface, washed and resuspended in Iscove's modified Dulbecco's medium+20% FCS+DNase and processed for CD34+ cell enrichment as described above. In some instances, two or three CML patient samples were pooled after thawing in order to obtain sufficient numbers of CD34+ cells for the present studies.
Isolation of HSC/PC sub-populations by multicolor flow cytometry
Enriched CD34+ cells were stained simultaneously with FITC–CD34, APC-CD38, PE-CD90, V450-CD45RA and the plethora of biotinylated mouse anti-human lineage-specific MoAbs. Following 30 min on ice, the cells were washed in cold sorting buffer (phosphate-buffered saline supplemented with 2% FCS and 1% bovine serum albumin) and resuspended in sorting buffer containing 0.5 μg/ml of streptavidin PE-Cy5. Cells were then immediately aliquoted and an optimal amount of one of the Alexa 700-conjugated antibodies described above was added to each aliquot. A control aliquot was always included in which no Alexa 700-conjugated MoAb was added. This control is referred to as the FMO (fluorescence minus one). After a 30-min incubation on ice, cells were washed and resuspended in cold sorting buffer containing 1 μg/ml of propidium iodide (for staining nonviable cells). Analyses and sorting were performed on a BD FACSAria IIu cytometer (BD Biosciences). For analyses, positivity was defined as fluorescence >99% of that in the absence of Alexa 700 (FMO). When sorting the desired subsets of Lin−CD34+CD38−CD90+CD45RA− cells, the fluorescence sorting gates were set such that ∼80% of the most negative cells and 80% of the most positive cells for a given cell surface antigen were selected. Sorted cells were collected into sterile 12 × 75 mm polypropylene round-bottom tubes containing 1 ml of the serum-free medium, QBSF-60 (Quality Biologicals, Gaithersburg, MD, USA) and kept in ice until transplantation or for other assays described below. Flow cytometric data were analyzed with FlowJo software version 9 (Tree Star, Ashland, OR, USA).
Mouse transplantation
Animal studies and procedures were approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee. NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice were either purchased from The Jackson Laboratory (Bar Harbor, ME, USA) or bred in-house, and were housed in specific pathogen-free facilities and fed a sulfatrim-containing diet. Mice, 6–8 weeks old, were irradiated (2.2 Gy) 3 h before transplantation and cells of interest were resuspended in 0.2 ml of QBSF and transplanted intravenously via the tail vein. Unless otherwise indicated, mice were killed 12 weeks after transplant, and the cells from both the tibiae and femurs of each mouse were collected into cold phosphate-buffered saline containing FCS. Red cells were lysed with ACK lysing buffer (Invitrogen) and residual white blood cells were processed for flow cytometry as described above. To serve as a control, bone marrow cells from a NOG mouse that was not transplanted was always included.
Colony-forming assays and detection of Bcr–Abl transcripts
For CML patient samples, purified HSC sub-populations were assayed for their colony-forming ability as previously described in detail,10 with some modifications. Briefly, cell aliquots were seeded in Iscove's modified Dulbeccco's medium containing 24% FCS, 1.3% methylcellulose, c-kit ligand (KL; 100 ng/ml), IL-3 (10 ng/ml), G-CSF (10 ng/ml), granulocyte-monocyte (GM)-CSF (10 ng/ml) (all gifts of the Kirin Brewing Co., Gunma, Japan), IL-6 (10 ng/ml; R&D Systems, Minneapolis, MN, USA) and erythropoietin (EPO; 1 unit/ml; Toyobo, New York, NY, USA) and incubated at 37 °C in a humidified atmosphere of 5% CO2 in air. After 12–14 days, triplicate cultures were scored for granulocytic/monocytic (CFU-GM) and erythroid (BFU-E) colonies.
Individual colonies were analyzed for the Bcr-Abl transcript by methods previously described by us in detail with some modifications.67 Briefly, colonies were removed from the methylcellulose with a sterile, RNase-free filtered micro-pipette tip, dispensed into 0.8 ml of Trizol reagent (Invitrogen) and RNA was extracted following standard Trizol protocol. Complementary DNA was then generated with Transcriptor Reverse Transcriptase (Roche, Indianapolis, IN, USA) using random hexamers and PCR amplification was carried out using FastStart Taq DNA Polymerase. Amplified products were run on a 2% agarose gel containing ethidium bromide and visualized by ultraviolet light transillumination. The expected products for Bcr-Abl were 394 base pairs (bp) and 319 bp, which correspond to the b3a2 and b2a2 fusion transcripts, respectively,67 whereas the expected PCR product generated from normal Abl (which served as a control transcript in the same reaction vessel) was 185 bp. Primers used to amplify normal Abl were published previously.68 In order to serve as a negative control for PCR, colonies were also generated from CD34+ cells obtained from CB samples and processed in the same manner.
Suspension cultures
For some CML samples, desired cell sub-populations were assayed in vitro for their growth potential in response to the early growth factors KL, thrombopoietin (TPO) and FMS-like tyrosine kinase 3 (FLT-3). Briefly, cells (500–750) were suspended in 1 ml of QBSF-60 containing KL (100 ng), TPO (20 ng) and FLT-3 (20 ng) (R&D Systems). Cultures were done in 48-well plates and maintained in a humidified atmosphere of 5% CO2 at 37 °C for 21 days and aliquots were removed at 14 and 21 days to assess viability by Trypan blue exclusion.
Results and discussion
Further phenotypic characterization of the Lin− CD34+ CD38− CD90+ CD45RA− HSC/PC sub-population purified from CB, G-CSF MPB and CML patient samples
Data compiled from numerous investigators have indicated that the most primitive hematopoietic cells including the HSCs reside in a lineage-negative sub-population with a CD34+ CD38− CD90+ CD45RA− phenotype. Very recently, using a multicolor sorting strategy to purify this subset from CB and subsequently transplanting them into NOG mice, Weissman's lab demonstrated that up to 10% of these cells were long-term SCID-mouse repopulating cells (LT-SRCs).17 This enrichment strategy is quite impressive given that the starting frequency of these LT-SRCs in unseparated CB cells is most likely <1 in 50 000. Using in-vitro colony-forming assays, they also found that ∼33% of the cells from this primitive sub-population were committed colony-forming cells, predominantly CFU-GM progenitors.
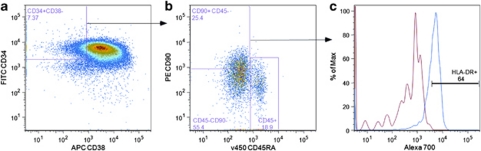
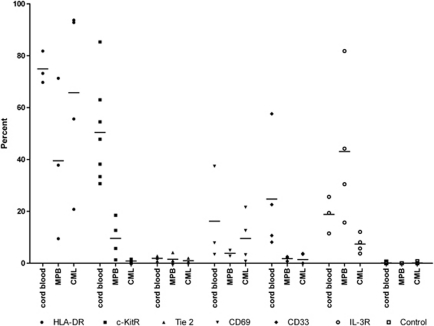
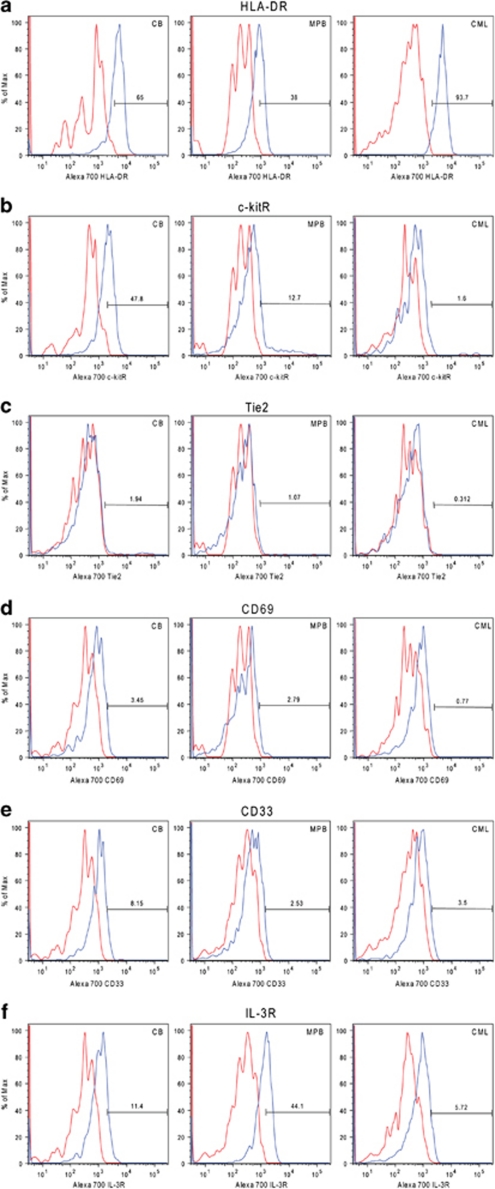
Employing a very similar multicolor sorting scheme, likewise, we were able to identify a Lin−CD34+CD38−CD90+ CD45RA− cell sub-population from CB (Figure 1). This sub-population comprised 16.7±7.1% (n=14) out of the Lin−CD34+CD38− cells, which is in the range of what Weissman's lab previously observed.17 As the expression of CD34 and CD38 antigens is not bi-modal (on/off), placement of gates defining positive and negative antigen-expressing cells is subjective, leading to some variability in published reports. Similar antigen expression profiles (Figures 1a and b) were also obtained when this strategy was used for sorting cells from MPB and CML patient samples (Supplementary Figure S1); however, the mean percentages of CD90+CD45RA− cells within the Lin−CD34+CD38− cell population were somewhat higher for MPB (28.2±16.4% s.d., n=7) and CML patients (32.3±20.5% s.d., n=10). We then examined the Lin−CD34+CD38− CD90+CD45RA− sub-population for their expression of HLA-DR, c-kitR, Tie2, CD33, IL-3R and CD69 antigens by using MoAbs (to these antigens) that were labeled with the fluorophore Alexa700. For illustrative purposes, Figure 1c shows an example of an HLA-DR expression profile that was obtained by gating on CB CD90+CD45RA− cells. Figure 2 shows the percentage of CD90+CD45RA− cells, in individual experiments, expressing the above-named antigens, whereas Figure 3 depicts representative Alexa 700 flow cytometry profiles (gated on the CD90+CD45RA− subset) obtained for each of these antigens. The percentage of CD90+CD45RA− cells expressing the HLA-DR antigen was consistently high from those obtained from CB (75±3.6% s.e.m.), whereas the percentage of CD90+CD45RA− cells purified from CML samples and MPB were lower and highly variable, 66±18% s.e.m. and 40±18% s.e.m., respectively (Figure 2). However, it can be seen in Figure 3 that the population of CD90+CD45RA− cells stained with anti-HLA-DR antibody showed an increased Alexa 700 fluorescence compared with the Alexa 700 unstained control; essentially the entire population shifted. Such a shift suggests that the cells classified as negative in our gating scheme were in fact expressing low amounts of HLA-DR antigen but the number of antigen molecules were not enough to cleanly separate the dim cells from autofluorescence.
Figure 1.
CB- enriched CD34-positive cells were analyzed for the expression of lineage markers CD34, CD38, CD90, CD45RA and HLA-DR by flow cytometry. The left panel (a) is gated on lineage-negative (Lin−) viable cells whereas the center panel (b) is gated on the Lin−CD34+CD38− cells. The right panel (c) is gated on the Lin−CD34+CD38−CD90+CD45RA− HSC/PC sub-population and shows the distribution of the HLA-DR antigen (blue histogram) within this primitive subset. The histogram in red is the fluorescence histogram obtained for the gated cells (CD90+CD45RA−) in the absence of staining with an Alexa 700-conjugated MoAb and is referred to as the FMO (Fluorescence Minus One) control. The number under each descriptive sub-population represents the percentage of positive cells within the total population of Lin−CD34+ enriched (a) or the CD34+CD38− (b) cells. Data shown represent one of three separate MPB experiments.
Figure 2.
CB-, MPB- and CML-enriched CD34+ cells were gated on the Lin−CD34+CD38−CD90+CD45RA− sub-population (as demonstrated in Figure 1) and analyzed for their expression of HLA-DR, c-kitR, Tie2, CD69, CD33 and the IL-3R by flow cytometry using Alexa Fluor 700-conjugated MoAbs to each of these antigens. Each data point in the figure represents the percentage of cells in an individual experiment that was positive for the antigen shown. Control samples represent percentage of positive cells obtained in the absence of any Alexa 700 MoAb (FMO). Crossbars represent the arithmetic means of the data points in each column.
Figure 3.
Representative expression profiles of six surface markers in the Lin− CD34+CD38−CD90+CD45RA− sub-population isolated from CB, MPB and CML patient samples. The six markers were (a) HLA-DR, (b) c-kitR, (c) Tie2, (d) CD69, (e) CD33 and (f) IL-3R. Red histograms are the control FMOs (see Figure 1) whereas those in blue are the expression profiles obtained by staining the cells with the Alexa 700-conjugated MoAbs.
With respect to the expression of the c-kit receptor, the mean percentage of CD90+CD45RA− cells classified as c-kitR+ was very disparate between the three tissue sources (Figure 2), 50±7% s.e.m., 10±4% s.e.m. and 0.9±0.3% s.e.m. for CB, MPB and CML, respectively. There is also a significant Alexa 700 fluorescence profile shift seen with anti-c-kitR antibody-stained CB cells (Figure 3), indicating that there could be a low expression of the c-kit receptor on the CD90+CD34RA− cell population classified as c-kitR negative. The Alexa 700 fluorescence profile shift on anti-c-kitR-stained MPB and CML was much less pronounced than that seen for CB; however, MPB and CML contained an additional population of cells expressing high amounts of c-kitR antigen, although at different frequencies. Most intriguing was that, relative to the percentages of CB and MPB CD90+CD45RA− cells that expressed c-kitR+, the number of CD90+CD45RA− cells isolated from CML patients that expressed the c-kit receptor was drastically lower. Although the significance of this novel finding is presently not clear, it is provocative that we previously reported10 that primitive GM progenitors isolated from CML patients have a greatly reduced requirement for KL in order to achieve optimal growth with the cytokines, G-CSF and GM-CSF. More recently,69 using the tyrosine kinase inhibitor PD173955, we provided pharmacological evidence that the Bcr–Abl kinase activity constitutively present in Ph+ primary primitive PCs disrupts the normal synergistic activity of KL with G-CSF+GM-CSF such that primitive CML GM progenitors have a greatly reduced requirement for KL in achieving optimal activation and growth in the presence of G-CSF+ GM-CSF. Whether or not these abnormal responses of CML primitive progenitors to KL are attributable to an abnormally low number of kit receptors on these cells must be investigated.
Other studies have shown that CML erythroid progenitors are also more sensitive than normal progenitors to growth stimulation with KL as well as to EPO and other cytokines known to act on stem/progenitor (S/P) cells.38, 70, 71 Once quiescent CML progenitors are stimulated to begin proliferating, they undergo further differentiation and maturation more rapidly than normal progenitors, but both granulopoiesis and erythropoiesis are less efficient than in normal hematopoiesis, as shown in cloning experiments in which the CML cells form many more small colonies and clusters but fewer large and extra-large CFU-GM and BFU-E than normal S/P cells.10, 12, 23, 38, 70, 71 Whereas normal CD34+ cells form almost entirely CFU-GM clusters and colonies in clonogenic experiments when stimulated by cytokines in the absence of EPO, CML G0 cells consistently form a mixture of GM and CFU-E and BFU-E without EPO; this spontaneous formation of erythroid colonies by CML G0 cells appears to be correlated with overexpression of numerous genes belonging to the megakaryocyte-erythroid lineages as well as to underexpression of Prominin-1 (CD133/2).72
Correlative biological and gene expression studies comparing highly enriched normal and CML quiescent (CD34 G0) and proliferating (CD34+ G1/S/G2/M) S/P cells have also shown that CML CD34+ G0 cells are in a more advanced stage of development and that gene expression is more similar to the proliferating (G1/S/G2/M) cells than normal G0 cells are to G1/S/G2/M cells.72 Numerous genes that are known to be overexpressed in normal hematopoietic S/P cells compared with more differentiated cells are downregulated in CML G0 cells compared with normal G0 cells, including six genes belonging to the polychrome repressive complex 1 (PRC1) and one (EPC2) belonging to the PRC2 group, providing additional evidence that CML G0 cells are more differentiated than normal G0 cells as both PRC complexes belong to groups of epigenetic regulators involved in maintenance of adult and embryonic stem cells. In keeping with their more advanced stage of differentiation and greater similarity to G1/S/G2/M cells, there is upregulation of genes involved in DNA replication and mitotic spindle machinery in CML G0 blasts compared with normal G0 blasts. The gene expression differences indicating that CML G0 cells are more poised to begin proliferating than normal quiescent S/P cells correlates nicely with the biological studies showing that CML G0 cells are more sensitive than normal G0 cells to growth stimulation by single cytokines (for example, KL, G-CSF, GM-CSF and EPO), or to combinations (KL+FL+TPO) known to act on early S/P cells.72 Although CML and normal G0 blasts are almost equally responsive to maximum stimulation by multiple cytokines, the CML cells are triggered into cycle more rapidly.
In examining the expression of Tie2, it can be seen in Figure 2 that the number of CD90+CD45RA− cells expressing this tyrosine kinase receptor was extremely low, regardless of whether they were purified from CB, MPB or CML (1.9±0.4% s.e.m., 1.5±0.9% s.e.m. and 1.0±0.5% s.e.m., respectively). Curiously, others have shown that the Tie2 receptor is present on HSCs in the quiescent state in adult murine bone marrow;52 yet, in some of the present experiments we have found that >96% of the primitive CD90+CD45RA− cells purified from CB are quiescent (as assessed in situ by the absence of the proliferation-associated Ki67 protein;73 data not shown). Moreover, our results appear to be at odds with other previously published studies50, 51 demonstrating that Tie2 is expressed by the majority of CD34+CD38− cells isolated from both human bone marrow and CB. However, it must be kept in mind that Lin−CD34+CD38−CD90+CD45RA− cells represent only a subset (∼8–30%) out of the total CD34+CD38−population. It should also be pointed out that the antibodies used to detect Tie2 in other studies (clones 10F11, 3C4, 7E8, 8G3 pooled, see Shackelford et al.40; cloneHT50, see Schlossman et al.41) were different than the one presently used (clone 33.1). In light of the findings of these other investigators, we will confirm the present observations by examining the CD90+CD45RA− HSC/PC subset using other Alexa700-labeled Tie2 antibodies. Also, it would be worthwhile to examine the expression of Tie2 by Lin−CD34+CD38−CD90+CD45RA− HSC/PCs purified from human bone marrow.
With regard to expression of the myeloid antigen CD33, we found that the mean percentages of MPB and CML CD90+CD45RA− cells that expressed the CD33 antigen was very low (<2% for both; Figure 2). In contrast, a significantly higher percentage (although highly variable) of CB CD90+CD45RA− cells expressed CD33 (mean= 25%±11 s.e.m.). Also, there was an Alexa 700 fluorescence profile shift (Figure 3) on the anti-CD33-stained cells, again indicating that there could be a low expression of CD33 on the population of CB CD90+CD45RA− cells classified as CD33 negative.
In looking at the expression of the IL-3 cytokine receptor, the CD90+CD45RA− sub-population isolated from MPB contained the highest percentage of IL-3R-positive cells (43±14% s.e.m.) followed by those isolated from CB (19±4% s.e.m.) and CML (7±2% s.e.m.) (Figure 1). However, IL-3R staining also exhibited a fluorescence profile shift for CB, MPB and CML (Figure 2), indicating that there could be low expression of the IL-3R on the populations of CD90+CD45RA− cells classified as IL-3R negative from all three tissue sources. Our finding of a relatively high percentage of IL-3R-positive cells within the CD90+CD45RA− subset isolated from MPB would be consistent with the fact that MPB is known to be enriched for committed myeloid PCs whose growth and maturation are well documented to be supported by the IL-3 cytokine.56
Last, in examining the CD69 molecule on the surface of the CD90+CD45RA− subsets, we found that this so-called activation antigen was expressed by a relatively minor percentage of these primitive cells from all three sources and, in addition, there was no salient increase in either the percentage of CD69+ cells (Figure 2) or its intensity of expression (Figure 3) within the CD90+CD45RA− HSC/PC sub-population isolated from patients with CML in the chronic phase versus those purified from CB and MPB (CB: 16±11% s.e.m., MPB: 4±1% s.e.m., CML: 10±5% s.e.m.). Nevertheless, it will be interesting to sort this CD69+ cell subset for further functional studies in order to determine what these primitive cells represent. In light of the recently reported finding of Hantschel et al.64 that antigen CD69 is increased on the surface of cells from cell lines expressing the Bcr–Abl protein, it remains possible that CD69 is elevated on HSCs isolated from CML patients in the accelerated and/or blastic phases of the disease; however, this warrants further examination.
LT- repopulating HSCs from CB are heterogeneous with respect to their HLA-DR, c-kitR or Tie2 cell surface expression
In an effort to systematically determine if the LT-SRCs residing within the Lin−CD34+CD38−CD90+CD45RA− sub-population can be further partitioned according to their surface antigen expression (or lack of), we sorted this primitive HSC/PC sub-population into HLA-DR+/−, c-kitR+/− and Tie2 +/− cell subsets, and injected these cells into NOG mice for detection of human marrow-repopulating cell activity. Because of their extremely high rates of engraftment of human HSCs,74 NOG mice currently serve as perhaps the best model for xenotransplantation studies. It should be noted that for the majority of these sorting experiments, a fluorescence gate separating cells as Alexa 700 negative and positive was set such that 80% of most negative and 80% of most positive cells were sorted. Our results to date are summarized in Table 1 (see Supplementary Table S1 for total engraftment data) show that long-term repopulating HSCs residing within the CB Lin−CD34+CD38−CD90+CD45RA− sub-population are heterogeneous with respect to their cell surface expression of either HLA-DR, c-kitR or Tie2. In the present studies, engraftment is defined as having detected at least 0.1% human CD45-positive cells in mouse bone marrow cell samples and chimerism is defined as the percentage of human CD45-positive cells within a mouse bone marrow cell sample. Thus, post 12-week transplantation, HLA-DR+ and HLA-DR− subsets exhibited comparable rates of engraftment (83% and 88%, respectively) and comparable levels of chimerism (mean of 19% CD45 human cells and 14% CD45 human cells, respectively). Likewise, c-kitR+ and c-kitR− subsets showed similar mean rates of engraftment (100% vs 80%) and mean percentages of CD45 cells (13% vs 18%, respectively). Although Tie2+ and Tie2− cell subsets exhibited the same rates of engraftment (100%), the mean level of human chimerism achieved with the former cell subset was much lower than with the latter (2.7% CD45+ vs 28% CD45+). Coupled with the fact that an average of 96% of the cells are recovered in the Tie2− subset indicates that the overwhelming majority of LT-HSCs residing within the Lin−CD34+CD38−CD90+CD45RA− sub-population from CB are Tie2−. Lineage specificity of the human CD45+ cells was analyzed on the bone marrow cells of some of the NOG mice engrafted with cells from the HLA-DR + and − and c-kitR + and − subsets. We found the human CD45+ cells to contain both myeloid (CD13) and B-lymphoid (CD19) cells, indicating that LT-HSCs in CB possessed myeloid and lymphoid differentiating capability (data not shown); a finding consistent with that of Majeti et al.17
Table 1. Summary of long-term (12 weeks) bone marrow engraftment of sub-populations of the human cord blood Lin−CD34+CD38−CD90+CD45RA− HSC/PC fraction injected into NOG mice.
| Subset | No. of CB samples | Median no. of cells injected (range) | % Engraftment | Avg % human chimerism (range) |
|---|---|---|---|---|
| HLA-DR (−) | 7 | 137 (50–194) | 83% (10/12) | 19% (0–55) |
| HLA-DR (+) | 6 | 128 (50–194) | 88% (7/8) | 14% (0–47) |
| c-kitR (−) | 4 | 150 (40–200) | 100% (5/5) | 13% (0.3–57) |
| c-kitR (+) | 4 | 150 (40–200) | 80% (4/5) | 18% (0.06–47) |
| Tie2 (−) | 4 | 100 (35–200) | 100% (4/4) | 28% (0.35–62) |
| Tie2 (+) | 4 | 90 (35–150) | 100% (4/4) | 2.7% (0.17–6.9) |
Abbreviations: CB, cord blood; HSC, hematopoietic stem cell; NOG, nonobese diabetes/severe combined immunodeficiency (NOD/SCID) IL-2R gammanull; PC, progenitor cell.
Based on these preliminary studies, it would appear, on first pass, that HLA-DR, c-kitR and Tie2 surface markers may not be useful for the further purification of LT-SRCs. However, previous studies by other investigators75, 76 have clearly demonstrated that HSCs are functionally heterogeneous and represent a hierarchy of cells with differing self-renewal, proliferative and repopulating abilities. In light of this heterogeneity, secondary and perhaps tertiary transplants using bone marrow cells harvested from primary transplanted NOG mice will be required to determine if there are differences in the self-renewal potential (and therefore primitiveness) of the HSCs residing within the HLA-DR+/−, c-kitR+/− and Tie2+/− subsets.
We have also sorted the Lin−CD34+CD38−CD90+CD45RA− HSC fraction purified from MPB into HLA-DR+/−, c-kitR+/− or IL-3R+/− subsets for xenotransplantation and the results are summarized in Table 2. Of note, we did not detect CD45+ human cells post 12 weeks after transplantation of HLA-DR+/− and c-kitR+/−subsets, although a similar number of cells were transplanted as with CB transplants. In two separate MPB experiments, IL-3R+/− subsets were sorted from the CD90+CD45RA− sub-population and both these subsets (150–2500 cells injected) elicited similar low levels of chimerism at 12 weeks post transplantation. This suggests that LT-HSCs at least in MPB are heterogeneous for their expression of the IL-3R. Clearly, more transplantation experiments must be performed using higher numbers of cells in order to quantitate LT-SRC activity from HLA-DR+/−, c-kitR+/− and IL-3R+/− subsets. However, our preliminary xenotransplantation studies indicate that there are fewer LT-SRCs within the Lin−CD34+CD38−CD90+CD45RA− HSC/PC population derived from MPB versus those purified from CB. This is consistent with the previous xenotransplantation studies of Wang et al.2 demonstrating a significantly higher frequency of LT-SRCs in CB compared with MPB. Rather, MPB has been shown to be enriched for committed myeloid progenitors including colony-forming cells.56
Table 2. Engraftment data of long-term bone marrow transplantation of sub-populations of human mobilized peripheral blood Lin−CD34+CD38−CD90+CD45RA− HSC/PC fraction into mice.
| Subset | Sample | No. of cells injected | % Chimerism |
|---|---|---|---|
| HLA-DR (−) | 1 | 100 | Na |
| 2 | 120 | N | |
| 3 | 150 | N | |
| HLA-DR (+) | 1 | 100 | N |
| 2 | 120 | N | |
| 3 | 150 | N | |
| c-kitR (−) | 1 | 100 | N |
| 2 | 120 | N | |
| 3 | 78 | N | |
| c-kitR (+) | 2 | 120 | N |
| 3 | 73 | N | |
| IL-3R (−) | 4 | 150 | 0.7 |
| 4 | 450 | 1.0 | |
| 5 | 1000 | Nb | |
| 5 | 2500 | 1.4b | |
| IL-3R (+) | 4 | 500 | 2.7 |
| 4 | 2000 | 6.0 | |
| 5 | 715 | 1.4b | |
| 5 | 2500 | 0.14b |
Abbreviations: CB, cord blood; HSC, hematopoietic stem cell; IL-3R, interleukin-3 cytokine receptor; NOG, nonobese diabetes/severe combined immunodeficiency (NOD/SCID) IL-2R gammanull; PC, progenitor cell.
N=no engraftment, that is, no human CD45-positive cells (or <0.1%) detected (by flow cytometric analysis) in 12-week mouse bone marrow cell samples.
Flow cytometric analysis of 9-week mouse bone marrow cell samples.
Table 3 summarizes xenotransplantation experiments using the cells of different HSC/PC sub-populations purified from CML patient samples. No engraftment was observed when NOG mice were transplanted with either the Lin−CD34+CD38−CD90+CD45RA− cell fraction (385–3000 cells injected) or the more differentiated Lin−CD34+CD38−CD90−CD45RA− cell fraction (150–1800 cells injected). We also did not see any engraftment when the CD90+CD45− HSC population was further sorted into IL-3+/− or HLA-DR+/− subsets for transplantation. PCR analysis of colonies generated in vitro from the cells of the various subsets in Table 3 were 100% positive for Bcr-Abl (data not shown). Again, although limiting dilution experiments will have to be done using higher numbers of cells from these various purified HSC subsets to quantitate marrow-repopulating cell activity in NOG mice, our preliminary results do indicate that, similar to MPB, the frequency of LT-SRCs within the Lin−CD34+CD38−CD90+CD38− sub-population purified from hematopoietic tissue from CML patients is much lower than that seen in CB. The reduced engrafting ability of CML HSC/PC sub-populations is in keeping with the observations noted earlier that the great majority of CML total progenitors and precursors as well as highly enriched CML Lin− Blasts and CD34+ G0 cells have greatly reduced proliferative and cloning potential than comparable normal cells.10, 12, 23, 38, 70, 71, 72 Eaves et al.77 previously found that highly enriched primitive CML cells display increased cycling (relative to similarly enriched primitive cells from normal bone marrow) and that only the quiescent HSC subset within their primitive fraction repopulated NOD/SCID mice. Consequently, xenotransplantation studies with CML HSC/PC fractions could underestimate the frequency of CML stem cells. Unfortunately, we did not look at the expression of the Ki67 antigen in the HSC/PC cell fractions given in Table 3 to see if they contain an increased number of cycling cells (relative to the negligible number found in comparable HSC/PC sub-populations isolated from CB); however, this must be determined in future studies. Of note, we have maintained the CD90+CD45RA− and CD90−CD45RA− subsets, derived from at least three separate CML patient samples, in liquid culture for 3 weeks in the presence of the early acting growth factors KL, TPO and FLT-3 and have consistently observed that the CD90+ subset proliferates to a much greater degree than the CD90− subset, indicating that the latter cells are less primitive (data not shown). Our observation is consistent with the studies of Majeti et al.17 that demonstrated that the more primitive CD90+CD45RA− cells, not surprisingly, also proliferate to a much higher degree in vitro than do CD90−CD45RA− cells purified from CB.
Table 3. Mouse engraftment data of long-term bone marrow transplantation of CD90+CD45RA− (and HLA-DR and IL-3 subsets) and CD90− CD45RA− sub-populations purified from CML patients.
| Subset | Sample | No. of cells injected | % Chimerism |
|---|---|---|---|
| CD90+CD45RA− | 3 | 1500 | Na |
| 3 | 3000 | N | |
| 4 | 385 | N | |
| CD90−CD45RA− | 1 | 150 | N |
| 3 | 1500 | N | |
| 3 | 1500 | N | |
| 4 | 900 | N | |
| 4 | 1800 | N | |
| HLA-DR (−)b | 1 | 113 | N |
| HLA-DR (+)b | 1 | 150 | N |
| IL-3R (−)b | 2 | 1000 | N |
| 2 | 4000 | N | |
| IL-3R (+)b | 2 | 400 | N |
| 2 | 800 | N |
Abbreviations: CML, chronic myelogenous leukemia; IL-3R, interleukin-3 cytokine receptor.
N=no engraftment, and therefore no human CD45-positive cells (or <0.1%) detected by flow cytometric analysis in 12-week mouse bone marrow cell samples.
HLA-DR (−), HLA-DR (+), IL-3R (−) and IL-3R (+) subsets were isolated from the Lin−CD34+CD38−CD90+CD38− sub-population.
In summary, the studies presented here have extended the phenotypic characterization of the rare, primitive Lin−CD34+CD38−CD90+CD45RA− sub-population from CB17 to include HLA-DR, CD33, c-kitR, Tie2, IL-3R and the activation antigen CD69. In addition, we have also examined the expression of these antigens on the CD90+CD45RA− cells isolated from MPB and CML patient samples. Expression profiles were achieved by using Alexa Fluor 700-conjugated MoAbs directed against each of these antigens in conjunction with a slight modification of a previously published17 multicolor scheme designed to identify Lin−CD34+CD38−CD90+CD45RA− cells by flow cytometry. Furthermore, we have demonstrated LT-marrow-repopulating cell activity within the HLA-DR+/−, c-kitR+/− and Tie2+/− subsets of CB CD90+CD45RA− cells as well as within the IL-3R+/− subsets of MPB CD90+CD45RA− cells. Based on these preliminary findings, it appears that HLA-DR, c-kitR, Tie2 and possibly the IL-3R may not be suitable markers for further purification of normal hematopoietic HSCs. However, as HSCs are known to be functionally heterogeneous with respect to their self-renewal, proliferative and repopulating potential,75, 76 more extensive functional studies will be needed to determine if these various phenotypically defined HSC/PC subsets are also functionally distinguishable by the level of primitiveness of the HSCs residing within each subset. Our studies also indicate that the frequency of LT-SRCs within CD90+CD45RA− subsets isolated from MPB and CML patient samples is significantly lower than that seen within CD90+CD45RA− cell subsets isolated from CB. This finding is consistent with previous observations by others demonstrating that MPB is more enriched for committed hematopoietic PCs56 and that patients with CML exhibit elevated numbers of Ph+ committed PCs.78 Thus, the present findings reinforce those of Majeti et al.17 demonstrating that the Lin−CD34+CD38−CD90+CD45RA− phenotype is not exclusively assigned to HSCs but rather includes a substantial population of committed primitive PCs. That said, the fluorescence-activated cell sorting flow cytometric multicolor stain presently described for phenotypically analyzing and sorting of subsets of the primitive Lin−CD34+CD38−CD90+CD45RA− sub-population provides a useful strategy for investigating other candidate hematopoietic cell surface antigens for the further purification of human HSCs. Such candidate antigens include CD133, CXCR4 and Robo4, all of which have been reported to be expressed by human HSCs.79, 80, 81
Acknowledgments
This work was supported by National Cancer Institute Grant CA64593, The Albert C Bostwick Foundation, The Enid A Haupt Charitable Trust, The Andrew Sage Trust, The Einard and Sue Sundin Fund, The Westvaco Corp. and MeadWestvaco Corp. We are very indebted to Dr Darren Veach at MSKCC for valuable discussions of our data and expert assistance in preparing the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Smith C. Hematopoietic stem cells and hematopoiesis. Cancer Control. 2003;10:9–16. doi: 10.1177/107327480301000103. [DOI] [PubMed] [Google Scholar]
- Wang JC, Doedens M, Dick JE. Primitive human hematopoietic cells are enriched in cord blood compared with adult bone marrow or mobilized peripheral blood as measured by the quantitative in vivo SCID-repopulating cell assay. Blood. 1997;89:3919–3924. [PubMed] [Google Scholar]
- Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothot A, Pyatt R, McMahel J, Rice S, Srour EF. Functional heterogeneity of human CD34(+) cells isolated in subcompartments of the G0/G1 phase of the cell cycle. Blood. 1997;90:4384–4393. [PubMed] [Google Scholar]
- Schriber JR, Dejbakhsh-Jones S, Kusnierz-Glaz CR, Ginzton N, Still B, Negrin RS, et al. Enrichment of bone marrow and blood progenitor (CD34+) cells by density gradients with sufficient yields for transplantation. Exp Hematol. 1995;23:1024–1029. [PubMed] [Google Scholar]
- Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci USA. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Louis O, Clay D, Brunet de la Grange P, Blazsek I, Desterke C, Guerton B, et al. Dual SP/ALDH functionalities refine the human hematopoietic Lin-CD34+CD38− stem/progenitor cell compartment. Stem Cells. 2009;27:2552–2562. doi: 10.1002/stem.186. [DOI] [PubMed] [Google Scholar]
- Christ O, Lucke K, Imren S, Leung K, Hamilton M, Eaves A, et al. Improved purification of hematopoietic stem cells based on their elevated aldehyde dehydrogenase activity. Haematologica. 2007;92:1165–1172. doi: 10.3324/haematol.11366. [DOI] [PubMed] [Google Scholar]
- Wisniewski D, Platsoucas C, Strife A, Lambek C, Clarkson B. Enrichment of hematopoietic progenitor cells (CFUC and BFUE) from human peripheral blood. Exp Hematol. 1982;10:817–829. [PubMed] [Google Scholar]
- Strife A, Perez A, Lambek C, Wisniewski D, Bruno S, Darzynkiewicz Z, et al. Characterization of lineage-negative blast subpopulations derived from normal and chronic myelogenous leukemia bone marrows and determination of their responsiveness to human c-kit ligand. Cancer Res. 1993;53:401–409. [PubMed] [Google Scholar]
- Wisniewski D, Strife A, Arlin Z, Knowles R, Lambek C, Gulati S, et al. Analysis of the individual and combined reactivities of monoclonal antibodies H25, H366, and MY9 with normal progenitor cells and blast cells from patients with acute myeloblastic leukemia. Leukemia. 1989;3:446–452. [PubMed] [Google Scholar]
- Strife A, Lambek C, Wisniewski D, Arlin Z, Thaler H, Clarkson B. Proliferative potential of subpopulations of granulocyte-macrophage progenitor cells in normal subjects and chronic myelogenous leukemia patients. Blood. 1983;62:389–397. [PubMed] [Google Scholar]
- Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent DG, Copley MR, Benz C, Wohrer S, Dykstra BJ, Ma E, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113:6342–6350. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- Domen J, Wagers A, Weissman IL.‘Bone marrow (hematopoietic) stem cells' in Regenerative MedicineNational Institutes of Health, U.S. Department of Health and Human Services: Bethesda, MD; 200613–34. . http://stemcells.nih.gov/info/2006report/2006chapter2 . [Google Scholar]
- Broxmeyer HE, Srour E, Orschell C, Ingram DA, Cooper S, Plett PA, et al. Cord blood stem and progenitor cells. Methods Enzymol. 2006;419:439–473. doi: 10.1016/S0076-6879(06)19018-7. [DOI] [PubMed] [Google Scholar]
- Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civin CI, Strauss LC, Brovall C, Fackler MJ, Schwartz JF, Shaper JH. Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. J Immunol. 1984;133:157–165. [PubMed] [Google Scholar]
- Terstappen LW, Huang S, Safford M, Lansdorp PM, Loken MR. Sequential generations of hematopoietic colonies derived from single nonlineage-committed CD34+CD38− progenitor cells. Blood. 1991;77:1218–1227. [PubMed] [Google Scholar]
- Lansdorp PM, Sutherland HJ, Eaves CJ. Selective expression of CD45 isoforms on functional subpopulations of CD34+ hemopoietic cells from human bone marrow. J Exp Med. 1990;172:363–366. doi: 10.1084/jem.172.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson B, Strife A, Perez A, Lambek C, Wisniewski D. Integration of molecular and biological abnormalities in quest for selective treatment of chronic myelogenous leukemia (CML) Leuk Lymphoma. 1993;11 (Suppl 2:81–100. doi: 10.3109/10428199309064267. [DOI] [PubMed] [Google Scholar]
- Clarkson BD, Strife A, Wisniewski D, Lambek C, Carpino N. New understanding of the pathogenesis of CML: a prototype of early neoplasia. Leukemia. 1997;11:1404–1428. doi: 10.1038/sj.leu.2400751. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Fialkow PJ, Jacobson RJ, Papayannopoulou T. Chronic myelocytic leukemia: clonal origin in a stem cell common to the granulocyte, erythrocyte, platelet and monocyte/macrophage. Am J Med. 1977;63:125–130. doi: 10.1016/0002-9343(77)90124-3. [DOI] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Mauro MJ, Druker BJ. STI571: targeting BCR-ABL as therapy for CML. Oncologist. 2001;6:233–238. doi: 10.1634/theoncologist.6-3-233. [DOI] [PubMed] [Google Scholar]
- de Lavallade H, Apperley JF, Khorashad JS, Milojkovic D, Reid AG, Bua M, et al. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008;26:3358–3363. doi: 10.1200/JCO.2007.15.8154. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A, Kantarjian H, Jones D, Shan J, Borthakur G, Thomas D, et al. Delayed achievement of cytogenetic and molecular response is associated with increased risk of progression among patients with chronic myeloid leukemia in early chronic phase receiving high-dose or standard-dose imatinib therapy. Blood. 2009;113:6315–6321. doi: 10.1182/blood-2008-07-166694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye T, Riehm B, Berger U, Paschka P, Muller MC, Kreil S, et al. Response and resistance in 300 patients with BCR-ABL-positive leukemias treated with imatinib in a single center: a 4.5-year follow-up. Cancer. 2005;103:1659–1669. doi: 10.1002/cncr.20922. [DOI] [PubMed] [Google Scholar]
- Branford S, Rudzki Z, Walsh S, Grigg A, Arthur C, Taylor K, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99:3472–3475. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–2249. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- Clarkson B, Rubinow SI. Growth Kinetics in Human Leukemia. Williams & Wilkins: Baltimore; 1977. pp. 591–628. [Google Scholar]
- Redner RL. Why doesn't imatinib cure chronic myeloid leukemia. Oncologist. 2010;15:182–186. doi: 10.1634/theoncologist.2009-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DJ, Melo JV. Primitive, quiescent and difficult to kill: the role of non-proliferating stem cells in chronic myeloid leukemia. Cell Cycle. 2006;5:2862–2866. doi: 10.4161/cc.5.24.3573. [DOI] [PubMed] [Google Scholar]
- Clarkson B, Strife A, Wisniewski D, Lambek CL, Liu C. Chronic myelogenous leukemia as a paradigm of early cancer and possible curative strategies. Leukemia. 2003;17:1211–1262. doi: 10.1038/sj.leu.2402912. [DOI] [PubMed] [Google Scholar]
- Bhatia R, Holtz M, Niu N, Gray R, Snyder DS, Sawyers CL, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- Shackelford DA, Kaufman JF, Korman AJ, Strominger JL. HLA-DR antigens: structure, separation of subpopulations, gene cloning and function. Immunol Rev. 1982;66:133–187. doi: 10.1111/j.1600-065x.1982.tb00437.x. [DOI] [PubMed] [Google Scholar]
- Schlossman SF, Chess L, Humphreys RE, Strominger JL. Distribution of Ia-like molecules on the surface of normal and leukemic human cells. Proc Natl Acad Sci USA. 1976;73:1288–1292. doi: 10.1073/pnas.73.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MA, Broxmeyer HE, Sheridan AP, Meyers PA, Jacobsen N, Winchester RJ. Continuous human bone marrow culture: Ia antigen characterization of probable pluripotential stem cells. Blood. 1980;55:682–690. [PubMed] [Google Scholar]
- Lu L, Broxmeyer HE, Meyers PA, Moore MA, Thaler HT. Association of cell cycle expression of Ia-like antigenic determinations on normal human multipotential (CFU-GEMM) and erythroid (BFU-E) progenitor cells with regulation in vitro by acidic isoferritins. Blood. 1983;61:250–256. [PubMed] [Google Scholar]
- Sutherland HJ, Eaves CJ, Eaves AC, Dragowska W, Lansdorp PM. Characterization and partial purification of human marrow cells capable of initiating long-term hematopoiesis in vitro. Blood. 1989;74:1563–1570. [PubMed] [Google Scholar]
- Sharma S, Gurudutta GU, Satija NK, Pati S, Afrin F, Gupta P, et al. Stem cell c-KIT and HOXB4 genes: critical roles and mechanisms in self-renewal, proliferation, and differentiation. Stem Cells Dev. 2006;15:755–778. doi: 10.1089/scd.2006.15.755. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr Signaling by Kit protein-tyrosine kinase--the stem cell factor receptor. Biochem Biophys Res Commun. 2005;337:1–13. doi: 10.1016/j.bbrc.2005.08.055. [DOI] [PubMed] [Google Scholar]
- Kawashima I, Zanjani ED, Almaida-Porada G, Flake AW, Zeng H, Ogawa M. CD34+ human marrow cells that express low levels of Kit protein are enriched for long-term marrow-engrafting cells. Blood. 1996;87:4136–4142. [PubMed] [Google Scholar]
- Gunji Y, Nakamura M, Osawa H, Nagayoshi K, Nakauchi H, Miura Y, et al. Human primitive hematopoietic progenitor cells are more enriched in KITlow cells than in KIThigh cells. Blood. 1993;82:3283–3289. [PubMed] [Google Scholar]
- Sogo S, Inaba M, Ogata H, Hisha H, Adachi Y, Mori S, et al. Induction of c-kit molecules on human CD34+/c-kit < low cells: evidence for CD34+/c-kit < low cells as primitive hematopoietic stem cells. Stem Cells. 1997;15:420–429. doi: 10.1002/stem.150420. [DOI] [PubMed] [Google Scholar]
- Batard P, Sansilvestri P, Scheinecker C, Knapp W, Debili N, Vainchenker W, et al. The Tie receptor tyrosine kinase is expressed by human hematopoietic progenitor cells and by a subset of megakaryocytic cells. Blood. 1996;87:2212–2220. [PubMed] [Google Scholar]
- Hashiyama M, Iwama A, Ohshiro K, Kurozumi K, Yasunaga K, Shimizu Y, et al. Predominant expression of a receptor tyrosine kinase, TIE, in hematopoietic stem cells and B cells. Blood. 1996;87:93–101. [PubMed] [Google Scholar]
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Lindemann A, Mertelsmann R. Interleukin-3 and its receptor. Cancer Treat Res. 1995;80:107–142. doi: 10.1007/978-1-4613-1241-3_5. [DOI] [PubMed] [Google Scholar]
- Reddy EP, Korapati A, Chaturvedi P, Rane S. IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled. Oncogene. 2000;19:2532–2547. doi: 10.1038/sj.onc.1203594. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Upchurch D, Szilvassy SJ, Guzman ML, Howard DS, Pettigrew AL, et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia. 2000;14:1777–1784. doi: 10.1038/sj.leu.2401903. [DOI] [PubMed] [Google Scholar]
- Tjonnfjord GE, Steen R, Evensen SA, Thorsby E, Egeland T. Characterization of CD34+ peripheral blood cells from healthy adults mobilized by recombinant human granulocyte colony-stimulating factor. Blood. 1994;84:2795–2801. [PubMed] [Google Scholar]
- Andrews RG, Singer JW, Bernstein ID. Precursors of colony-forming cells in humans can be distinguished from colony-forming cells by expression of the CD33 and CD34 antigens and light scatter properties. J Exp Med. 1989;169:1721–1731. doi: 10.1084/jem.169.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srour EF, Zanjani ED, Cornetta K, Traycoff CM, Flake AW, Hedrick M, et al. Persistence of human multilineage, self-renewing lymphohematopoietic stem cells in chimeric sheep. Blood. 1993;82:3333–3342. [PubMed] [Google Scholar]
- Blair A, Hogge DE, Sutherland HJ. Most acute myeloid leukemia progenitor cells with long-term proliferative ability in vitro and in vivo have the phenotype CD34(+)/CD71(−)/HLA-DR. Blood. 1998;92:4325–4335. [PubMed] [Google Scholar]
- Yuasa H, Takakura N, Shimomura T, Suenobu S, Yamada T, Nagayama H, et al. Analysis of human TIE2 function on hematopoietic stem cells in umbilical cord blood. Biochem Biophys Res Commun. 2002;298:731–737. doi: 10.1016/s0006-291x(02)02524-x. [DOI] [PubMed] [Google Scholar]
- Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, Kelly G, et al. Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood. 2005;106:4086–4092. doi: 10.1182/blood-2005-03-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce DJ, Taussig DC, Bonnet D. Implications of the expression of myeloid markers on normal and leukemic stem cells. Cell Cycle. 2006;5:271–273. doi: 10.4161/cc.5.3.2393. [DOI] [PubMed] [Google Scholar]
- Testi R, D'Ambrosio D, De Maria R, Santoni A. The CD69 receptor: a multipurpose cell-surface trigger for hematopoietic cells. Immunol Today. 1994;15:479–483. doi: 10.1016/0167-5699(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Hantschel O, Gstoettenbauer A, Colinge J, Kaupe I, Bilban M, Burkard TR, et al. The chemokine interleukin-8 and the surface activation protein CD69 are markers for Bcr-Abl activity in chronic myeloid leukemia. Mol Oncol. 2008;2:272–281. doi: 10.1016/j.molonc.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strife A, Lambek C, Perez A, Darzynkiewicz Z, Skierski J, Gulati S, et al. The effects of transforming growth factor beta 3 on the growth of highly enriched hematopoietic progenitor cells derived from normal human bone marrow and peripheral blood. Cancer Res. 1991;51:4828–4836. [PubMed] [Google Scholar]
- Olofsson T, Gartner I, Olsson I. Separation of human bone marrow cells in density gradients of polyvinylpyrrolidone coated silica gel (Percoll) Scand J Haematol. 1980;24:254–262. doi: 10.1111/j.1600-0609.1980.tb01334.x. [DOI] [PubMed] [Google Scholar]
- Wisniewski D, Strife A, Clarkson B. Co-detection of chimeric bcr/abl (target) and beta-actin (control) messenger RNA in individual CFU-GM colonies derived from CML patients using the polymerase chain reaction. Leuk Res. 1991;15:867–874. doi: 10.1016/0145-2126(91)90471-5. [DOI] [PubMed] [Google Scholar]
- Savoldo B, Sammarelli G, Dotti G, Garau D, Regazzi E, Cilloni D, et al. Reverse transcription polymerase chain reaction is a reliable assay for detecting leukemic colonies generated by chronic myelogenous leukemia cells. Leukemia. 1998;12:434–440. doi: 10.1038/sj.leu.2400942. [DOI] [PubMed] [Google Scholar]
- Strife A, Wisniewski D, Liu C, Lambek CL, Darzynkiewicz Z, Silver RT, et al. Direct evidence that Bcr-Abl tyrosine kinase activity disrupts normal synergistic interactions between Kit ligand and cytokines in primary primitive progenitor cells. Mol Cancer Res. 2003;1:176–185. [PubMed] [Google Scholar]
- Strife A, Perez A, Lambek C, Wisniewski D, Bruno S, Darzynkiewicz Z, et al. Differences in the composition and in the efficiency of red cell production of normal and CML erythroid progenitor populations are highlighted by response to human c-kit ligand. Leuk Res. 1993;17:799–807. doi: 10.1016/0145-2126(93)90115-2. [DOI] [PubMed] [Google Scholar]
- Clarkson B, Strife A. Linkage of proliferative and maturational abnormalities in chronic myelogenous leukemia and relevance to treatment. Leukemia. 1993;7:1683–1721. [PubMed] [Google Scholar]
- Affer M, Dao S, Liu C, Olshen AB, Mo Q, Viale A, et al. Gene expression differences between enriched normal and chronic myelogenous leukemia quiescent stem/progenitor cells and correlations with biological abnormalities. J Oncol. 2011;2011:1–25. doi: 10.1155/2011/798592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter C, Duchrow M, Wohlenberg C, Becker MH, Key G, Flad HD, et al. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kobayashi K, Nakahata T. NOD/Shi-scid IL2rgamma(null) (NOG) mice more appropriate for humanized mouse models. Curr Top Microbiol Immunol. 2008;324:53–76. doi: 10.1007/978-3-540-75647-7_3. [DOI] [PubMed] [Google Scholar]
- McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- Muller-Sieburg CE, Sieburg HB. Clonal diversity of the stem cell compartment. Curr Opin Hematol. 2006;13:243–248. doi: 10.1097/01.moh.0000231421.00407.65. [DOI] [PubMed] [Google Scholar]
- Eaves AC, Barnett MJ, Ponchio L, Cashman JD, Petzer AL, Eaves CJ.Differences between normal and CML stem cells: potential targets for clinical exploitation Stem Cells 199816(Suppl 177–83.discussion 89. [DOI] [PubMed] [Google Scholar]
- Petzer A, Eaves CJ, Lansdorp PM, Ponchio L, Barnett MJ, Eaves AC. Characterization of primitive subpopulations of normal and leukemic cells present in the blood of patients with newly diagnosed as well as established chronic myeloid leukemia. Blood. 1996;88:2162–2171. [PubMed] [Google Scholar]
- Rosu-Myles M, Gallacher L, Murdoch B, Hess DA, Keeney M, Kelvin D, et al. The human hematopoietic stem cell compartment is heterogeneous for CXCR4 expression. Proc Natl Acad Sci USA. 2000;97:14626–14631. doi: 10.1073/pnas.97.26.14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Flint A, Dvorin EL, Bischoff J. AC133-2, a novel isoform of human AC133 stem cell antigen. J Biol Chem. 2002;277:20711–20716. doi: 10.1074/jbc.M202349200. [DOI] [PubMed] [Google Scholar]
- Shibata FG-KY, Morikawa Y, Komori T, Ito M, Fukuchi Y, Houchins JP, et al. Roundabout 4 is expressed on hematopoietic stem cells and potentially involved in the niche-mediated regulation of the side population phenotype. Stem Cells. 2009;27:183–190. doi: 10.1634/stemcells.2008-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.