Abstract
CXCR4 is a negative prognostic marker in acute myeloid leukemias (AMLs). Therefore, it is necessary to develop novel ways to inhibit CXCR4 expression in leukemia. AMD3100 is an inhibitor of CXCR4 currently used to mobilize cancer cells. CXCR4 is a target of microRNA (miR)-146a that may represent a new tool to inhibit CXCR4 expression. We then investigated CXCR4 regulation by miR-146a in primary AMLs and found an inverse correlation between miR-146a and CXCR4 protein expression levels in all AML subtypes. As the lowest miR-146a expression levels were observed in M5 AML, we analyzed the control of CXCR4 expression by miR-146a in normal and leukemic monocytic cells and showed that the regulatory miR-146a/CXCR4 pathway operates during monocytopoiesis, but is deregulated in AMLs. AMD3100 treatment and miR-146a overexpression were used to inhibit CXCR4 in leukemic cells. AMD3100 treatment induces the decrease of CXCR4 protein expression, associated with miR-146a increase, and increases sensitivity of leukemic blast cells to cytotoxic drugs, this effect being further enhanced by miR-146a overexpression. Altogether our data indicate that miR-146a and AMD3100, acting through different mechanism, downmodulate CXCR4 protein levels, impair leukemic cell proliferation and then may be used in combination with anti-leukemia drugs, for development of new therapeutic strategies.
Keywords: CXCR4, miR-146a, AMD3100, acute myeloid leukemia
Introduction
Several mechanisms regulate the interaction of normal and leukemic bone marrow (BM) stem/progenitor cells with the BM microenvironment, consisting of extracellular matrix and the stromal cells. This interaction has a key role in the control of survival, proliferation, self-renewal, chemoresistance and mobilization of stem/progenitor cells. Among the various mechanisms, the interaction between SDF-1α (also known as CXCL12) and its receptor is crucial for directing homing and engraftment of human hematopoietic stem cells into the BM after transplantation in immunodeficient mice.1, 2 Binding of SDF-1α induces CXCR4 phosphorylation and internalization, and subsequent incorporation into lipid rafts with generation of active cell signalling, ultimately leading to changes in gene transcription with consequent changes in survival, proliferation and migration of stem/progenitor cells.3 Several studies support the role of CXCR4 and its ligand, SDF-1α, in the regulation of the normal and leukemic stem cells motility in vivo,4, 5, 6 in leukemic cell survival and differentiation, and in the development of human acute myeloid leukemia (AML) stem cells.7, 8, 9 High level of CXCR4 expression on the surface of AML blasts is associated with poor relapse-free and overall survival9, 10, 11, 12 and may account for resistance to antileukemic drugs.6, 13 Blockage of the CXCR4 receptor by the small molecular inhibitor AMD3100 is currently being successfully employed to mobilize hematopoietic stem cell from the BM into the peripheral blood13 and to inhibit growth and metastasis of experimental tumors in animal models.12 Another strategy to downregulate CXCR4 expression in target cells is based on specific blocking of CXCR4 or SDF-1 expression at the mRNA level by employing small interfering RNAs or an oligonucleotide antisense strategy.14, 15, 16
MicroRNAs (miRs) are small non-coding RNAs that regulate gene expression through translational repression;16 recently, we have identified CXCR4 as a target of miR-146a that may represent a new tool to downmodulate CXCR4 expression at the protein level in target cells.17
Here we analyzed miR-146a and CXCR4 expression in primary blast cells obtained from M1-M5 AML cases. We provide evidence that in M1 to M5 AMLs, CXCR4 expression is regulated by miR-146a through a mechanism similar to that observed in leukemic cell lines and in normal megakaryocytes.17 The absent/low expression of miR-146a represents a major determinant of the high CXCR4 expression observed in some AMLs, mainly pertaining to the M3, M4 and, in particular, M5 subtypes.
Accordingly, we investigated and confirmed the role of miR-146a in regulating CXCR4 protein expression at protein level, in normal and leukemic monocytic cells.
We then analyzed the effect of AMD3100 treatment, in combination or not with miR-146a overexpression, on the miR-146a/CXCR4 regulatory pathway in leukemic cells. AMD3100 treatment of leukemic cells impair cell proliferation, whereas the CXCR4 protein level is downmodulated and miR-146a expression increases. Importantly, AMD3100 treatment increases leukemic blast cell sensitivity to anticancer drug treatment by downmodulating membrane CXCR4 protein levels, this phenomenon being potentiated by miR-146a overexpression.
Altogether our data indicate that the miR-146a/CXCR4 regulatory pathway operates during monocytopoiesis and in primary AMLs, but that it is deregulated in some AMLs. These observations shed light on the molecular mechanisms regulating CXCR4 expression by miR-146a in normal and leukemic cells.
Materials and methods
Primary AML blast cells
Primary AMLs were obtained after informed consent.17 RNA and protein samples were previously prepared as described.18
Cell cultures
Human myelomonocytic and monocytic leukemic cell lines, U937 and THP-1 respectively (American Type Culture Collection, Manassas, VA, USA), were grown in RPMI medium supplemented with 10% fetal calf serum (Gibco, Carlsbad, CA, USA). Human cord blood was obtained from healthy donors after informed consent. Cord blood CD34+ HPC purification, unilineage monocytic liquid cultures in BIT 9500 serum-free medium (Stemcell Technologies Inc., Vancouver, BC, Canada) supplemented with human low density lipoprotein (40 μg/ml), FLT3 ligand (100 ng/ml), interleukin 6 (1 μg/ml) and macrophage colony-stimulating factor (50 ng/ml) were performed as previously described.19
For morphologic analysis, cells were smeared on glass slides by cytospin centrifugation, stained with May–Grunwald Giemsa and analyzed at 400 magnification under a microscope (Eclipse 1000; Nikon, Tokyo, Japan) equipped with a digital camera.
Monocytic differentiation of leukemic cells
U937 cells were cultured with 1α,25-dihydroxyvitamin D3, VitD3 (250 n), whereas THP-1 cells were cultured with tetradecanoyl phobol acetate (TPA) (32 n) (Sigma-Aldrich, St Louis, MO, USA) for induction of monocytic differentiation/maturation. Cell growth analysis and Mo membrane marker (CD11b, CD14, CD18) expression by flow cytometry analysis were performed as described.20
AMD3100, SDF-1α and drug treatment of cells
U937 and THP-1 cell lines were cultured at 50 000 cells/ml and treated with AMD3100 (10 μ) (Sigma-Aldrich), added every 2 days of culture, then with or without cytosine arabinoside,21 Ara-C (Sigma-Aldrich), at 1 and 3 μ for 24–48 h, compared with cultures with Ara-C alone or without any treatment as control (C). Cell growth, apoptosis and CXCR4 protein expression were then analyzed by growth curve, annexin-test (Annexin V-FITC apoptosis detection kit, BD Pharmingen, San Diego, CA, USA) and flow cytometry analysis (FACS) respectively, as described below. Blast cells from primary AML samples were cultured for only 24 h with (i) SDF-1α (100 ng/ml) (Sigma-Aldrich), or (ii) AMD3100 (1 μ), or (iii) Ara-C (1 μ) or doxorubicinhydrochloride (Sigma-Aldrich), Doxo, at 1 μ, alone or in combination as compared with cultures without any treatment for control (C). The percentage of apoptotic cells (annexin-V-positive cells) and FACS analysis of CXCR4 protein level were analyzed. The murine stromal cell line MS5 was obtained from ATCC. These cells were currently grown in α-minimum essential medium. For chemotherapy sensitivity experiments these cells were grown in 12-wells cell culture plates (Costar, Corning, NY, USA) until the formation of a cell layer; then, culture supernatant was removed and replaced with leukemic cells (U937-E or U937-miR146a) resuspended in 1 ml of tissue culture medium.
CXCR4 mRNA analysis
Quantitative reverse transcription and real-time PCR analysis for GAPDH and CXCR4 were performed using commercial ready-to-use primers/probe mixes (Assays on Demand Products, Applied Biosystems, Foster City, CA, USA), as previously described.17
MiR-146a expression analysis
Quantitative real-time PCR analysis of miR-146a was performed by TaqMan technology, using the kit ABI PRISM 7700 DNA Sequence Detection System specific for miR-146a reverse transcription and PCR analysis (Applied Biosystems), according to the manufacturer's procedure and as previously described.17
FACS
To evaluate total CXCR4 protein expression, cells were fixed and permeabilized with Permi-Fix (BD Pharmingen) before incubation with either a phycoerythrin-conjugated anti-CXCR4 monoclonal antibody, clone 1245 (BD Pharmingen) or a control phycoerythrin-conjugated monoclonal antibody of the same isotype. Cells were analyzed using the FACSCAN flow cytometer and the Cell Quest software (BD Pharmingen) for acquisition and analysis.17 To evaluate CXCR4 membrane expression, 20 000 cell aliquots were washed in phosphate-buffered saline and then labelled on the membrane using the same anti-CXCR4-conjugated antibody described above and fluorescence emission was analyzed as described above. Intracytoplasmic CXCR4 protein expression was then evaluated by subtraction between total and membrane CXCR4 protein levels. All experiments were performed with the same batch of anti-CXCR4 conjugated antibody. The reported mean fluorescence intensity indicates the ratio between the geometric mean fluorescence values observed in CXCR4 labeled cells and cells labeled with a negative control, gated on FSC/ SSC dot plot to select viable cells.
Enforced expression of miR-146a in U937 cells
The human miR-146a precursor cloned into a pCDH expression lentivector, PMIRH146aPA-1 (System Biosciences (SBI), Mountain View, CA, USA), was transfected into 293TN producer cells with the pPACKH1 packaging Plasmid mix, LV500A-1, (SBI), using Lipofectamine and Plus Reagents (Invitrogen, Carlsbad, CA, USA) to produce pseudoviral particles. The pseudoviral titer was estimated to perform the transduction of the packaged lentivector expression constructs: (i) the empty pCDH expression lentivector (pCDH-empty, pCDH-E) used for internal control; (ii) the PMIRH146aPA-1 construct referred as (pMIRNA-146a), in the target U937 cells, according to the SBI manufacturer's instructions. Infected-U937 cells were selected for copGFP expression by cell sorting analysis. Stable transduced cell lines, U937-pMIRNA-146a (U937-miR146a) and U937-pCDH-E (U937-E), were then maintained in RPMI/fetal calf serum 10%. MiR-146a overexpression was controlled by specific miR-146a real-time PCR analysis (Applied Biosystems) in U937-miR146a cells as compared with control U937-E and U937 wild-type (wt) cells.
Results
MiR-146a and CXCR4 expression in AML samples
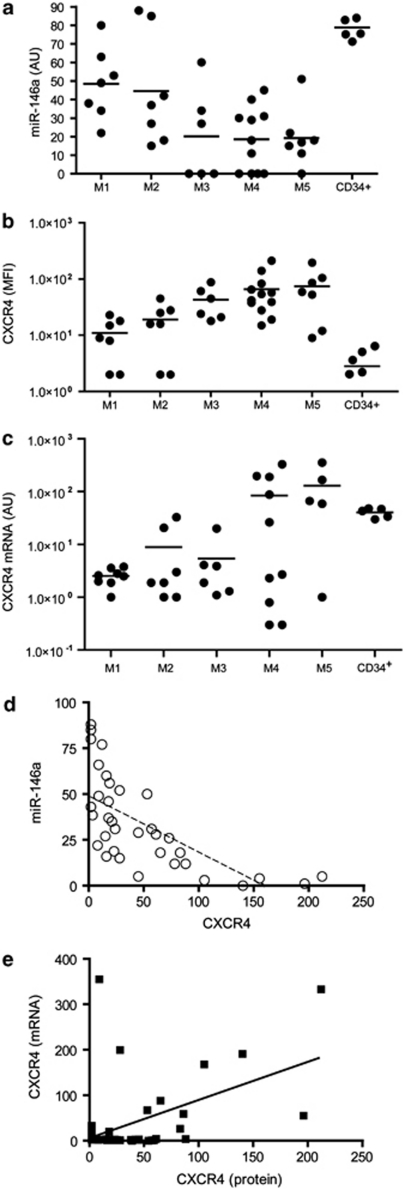
We investigated miR-146a and CXCR4 expression among 38 AMLs mainly pertaining to five subgroups from M1 to M5 FAB subsets (7 M1; 7 M2; 6 M3; 11 M4; 7 M5), as compared with normal CD34+ HPCs.
The levels of miR-146a expression analyzed by quantitative real-time PCR were quite heterogeneous for each group of FAB-classified AMLs; however, despite this heterogeneity lower miR-146a expression was detected in the M4/M5 subtypes, as compared with higher miR-146a expression detected in normal CD34+ HPCs (Figure 1a). In AMLs, a high CXCR4 membrane protein expression, as evaluated by FACS analysis, has been mostly associated with M4 and M5 AMLs;9, 10, 11 furthermore, we observed higher levels of CXCR4 protein expression in the group of M4/M5 AMLs as compared with M1/M2 AMLs (Figure 1b). Indeed, we found a highly significant inverse correlation between miR-146a expression and CXCR4 membrane protein level, indicative of a posttranscriptional control of gene expression; in fact, the plotting of miR-146a and CXCR4 protein levels showed that these two parameters are inversely related (r2=0.45; P=0.0001) (Figure 1d). These data strongly suggest that the regulatory network involving miR-146a regulation of CXCR4 protein expression could operate in primary AML samples. It is also of interest to note the following: (i) a total absence of miR-146a expression was observed in 8/38 AML cases, all pertaining to the M3, M4 and M5 subtypes; (ii) some M1 and M2 AMLs express membrane CXCR4 at high levels, associated with a low miR-146a expression.
Figure 1.
High level of CXCR4 membrane protein expression is correlated with a low level of miR-146a expression in AMLs. (a) Quantitative real-time (qRT)-PCR analysis of miR-146a expression levels in primary AML samples (from M1 to M5 of FAB classification) compared with normal CD34+ HPCs. (b) FACS of CXCR4 membrane protein expression levels in the same AML samples. (c) RT-PCR analysis of CXCR4 mRNA expression in AMLs samples. (d) Negative correlation between CXCR4 levels and miR-146a expression (P⩽0.0001). (e) Direct correlation between CXCR4 mRNA and protein levels (P⩽0.05).
We also analyzed CXCR4 mRNA expression level as compared with CXCR4 protein levels in AML samples (Figures 1c and b). Our results showed a correlation between CXCR4 mRNA and CXCR4 protein level (P⩽0.05) (Figure 1e), however, less significant than that observed between miR-146a and CXCR4 protein expression.
Altogether, our data indicate that in primary AML samples CXCR4 expression is under, both, transcriptional and posttranscriptional control. Indeed, M4 and M5 AMLs expressing the highest level of CXCR4 protein, expressed also miR-146a at lower level.
MiR-146a and CXCR4 expression in unilineage Mo culture
CXCR4 expression in myelomonocytic leukemias M4/ M5 prompted us to evaluate CXCR4 at the mRNA and protein level, and miR-146a expression in human cord blood CD34+ HPCs induced in serum-free suspension culture to unilineage differentiation and maturation through the Mo pathway.19, 20 Mo proliferation and differentiation were monitored by cell growth (Figure 2a) and increasing expression of CD11b antigen (Figure 2b).
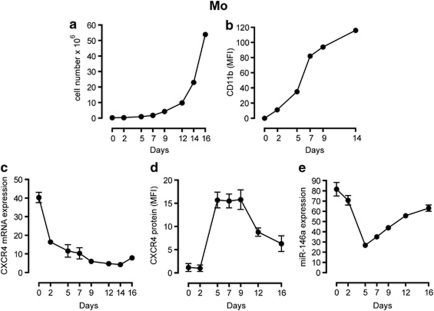
Figure 2.
MiR-146a and CXCR4 expression in purified CD34+ HPCs induced to monocytic (Mo) differentiation. (a, b) CD34+ HPCs (day 0) proliferation and differentiation were controlled, respectively, by growth curve analysis (a) and expression of CD11b (b). (c) Real time PCR analysis of CXCR4 mRNA expression in HPCs grown in liquid-phase unilineage Mo cultures. (d) FACS of CXCR4 membrane protein expression in HPCs during unilineage Mo cultures. The geometric mean fluorescence intensity is indicated. (e) Real time PCR analysis of miR-146a expression in HPCs during Mo differentiation. (a, b) One representative experiment out of three is shown. (c–e) The results of three independent experiments (mean±s.e.m. values) are shown.
CXCR4 mRNA expression is high in CD34+ HPCs (day 0) then is rapidly downmodulated during the first days of culture (days 0–2), being progressively lower from day 5 onwards, during Mo differentiation/maturation (Figure 2c). Conversely, CXCR4 membrane (Figure 2d) and total (not shown) protein expression is low at days 0–2 but is thereafter markedly upregulated, with a peak at day 5 and remaining constant through day 9, when it tends to decrease again from day 9 to 16 of culture (Figure 2d). In the same Mo cultures, miR-146a expression level is high in the first days of culture (days 0–2), then it is downmodulated (days 2–5), increasing thereafter during Mo differentiation (days 5–16) (Figure 2e). The rise of CXCR4 membrane and total proteins, in the presence of a decreased mRNA, suggests that the sharp decline of miR-146a could effectively unblock CXCR4 mRNA translation.
Overall, during Mo proliferation and differentiation, miR-146a expression is inversely related to the CXCR4 protein level (Figures 2d and e). The results hence suggest a post-transcriptional control of miR-146a on CXCR4 protein level, without excluding a coupled transcriptional control of CXCR4 protein level in this lineage.
MiR-146a overexpression reduces CXCR4 expression in U937 cells, whereas VitD3 or TPA-induced monocytic differentiation of U937 or THP-1 cells increases miR-146a expression and inhibits CXCR4 protein expression
To strengthen the link between miR-146a and CXCR4 expression in the Mo pathway, we used U937 cells as a model of Vitamin D3 (VitD3)-inducible Mo differentiation and evaluated whether overexpression of miR-146a and consequent changes in CXCR4 expression affect proliferation, differentiation and maturation of these cells.
Lentivectors, such as pMIRNA-146a to increase miR-146a expression level and pCDH-E, the empty vector used as a control, were prepared to infect U937 cells. U937-pMIRNA-146a cells (U937-miR146a) and U937-pCDH-E control cells (U937-E) were then cultured as compared with U937 wt cells.
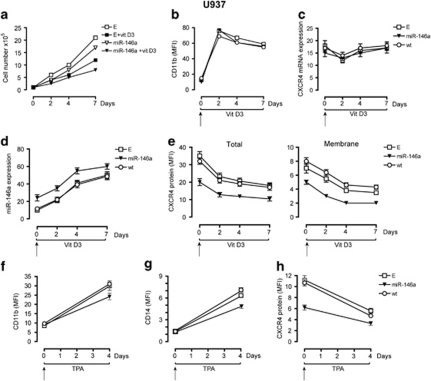
MiR-146a overexpression in U937 cells induces a decrease of CXCR4 protein, as evidenced through analysis of total and membrane CXCR4 expression levels (Figure 3e). In control cells U937-E or U937 wt (not shown), VitD3 treatment impairs cell proliferation (Figure 3a) and increases CD11b (Figure 3b) and CD14, CD18 (not shown) Mo marker expression. We also found that VitD3 upmodulates miR-146a expression (Figure 3d), whereas CXCR4 protein expression decreases (Figure 3e), without any significative modulation of CXCR4 mRNA expression (Figure 3c). These data mimic those observed in HPCs induced to Mo unilineage differentiation and maturation (Figures 2c–e), indicating that VitD3-induced U937 cells represent a useful model system to evaluate the role of the miR-146a/CXCR4 interaction during Mo differentiation.
Figure 3.
Overexpression of miR-146a in U937 cells downregulates CXCR4 protein expression and impairs cell growth induced by vitamin D3 or TPA. (a) Growth curve of U937-miR146a (miR-146a) and U937-Empty (E) cells grown either in the absence or in the presence of vitamin D3 (VitD3). (b) FACS of CD11b expression in U937-miR146a (miR-146a) cells, as compared with control cells U937-wt (wt) and U937-E cells (E), treated with VitD3 to induce Mo differentiation, compared with VitD3-untreated cells (day 0). (c, d) Quantitative real-time PCR analysis of CXCR4 mRNA and miR-146a expression were performed at the indicated days, in U937-miR146a cells as compared with control cells (wt and E), treated by VitD3 for Mo differentiation compared with untreated cells (day 0). (e) FACS of CXCR4 protein expression during VitD3 treatment of U937- miR146a and control cells (wt and E) compared with untreated cells (day 0). CXCR4 protein expression was evaluated at both, total (left panel) and membrane (right panel) level, on permeabilized and intact cells, respectively. (f, g) FACS of CD11b and CD14 expression in U937-miR146a (miR-146a) cells, as compared with control cells U937-wt (wt) and U937-E cells (E), treated with TPA to induce Mo differentiation, compared with TPA-untreated cells (day 0). (h) FACS of CXCR4 membrane protein expression during TPA treatment of U937-miR146a and control cells (wt and E) compared with untreated cells (day 0). (a, b) One representative experiment out of three is shown. (c–h) The results of three independent experiments (mean±s.e.m. values) are shown. (e–h) The geometric MFI is shown as mean±s.e.m. values from three independent experiments.
FACS evaluation revealed that in VitD3-treated (from day 0 to 7) or untreated U937 cells (day 0), CXCR4 was downmodulated by miR-146a overexpression (Figure 3e), whereas we did not observe any significant modulation of CD11b (Figure 3b) and CD14 or CD18 (not shown) Mo markers, as compared with U937-E and wt control cells (Figure 3b). However, miR-146a clearly impairs cell proliferation (miR-146a) and exerts a synergistic effect in combination with VitD3 (miR-146a + VitD3) to decrease the proliferation rate growth of U937-miR146a cells, as compared with control cells (Figure 3a). Thus, miR-146a diminishes CXCR4 protein expression and impairs Mo proliferation of U937 cells, without any apparent modulation of Mo differentiation.
However, we evaluated the effect of miR-146a overexpression in U937 cells induced to monocytic differentiation using a different monocytic inducer, TPA. We treated U937-miR146a cells, as compared with control cells (wt and U937-E) by TPA and observed (a) a marked inhibitory effect on cell growth, major in U937-miR146a cells, as compared with U937-wt (P<0.05) and U937-E (P<0.05) cells (data not shown); (b) a downmodulation of CXCR4 membrane protein expression (Figure 3h), concomitantly with the induction of Mo membrane differentiation markers: interestingly, CD11b and CD14 expression was lower in U937-miR146a than in control cells U937-wt (for both P<0.05) and U937-E (for both P<0.05) cells (Figures 3f and g).
Altogether, these data indicate that using another inducer of Mo differentiation, miR-146a overexpression inhibits cell growth and Mo differentiation of U937 cells treated by TPA.
We also used another cellular model of Mo differentiation, TPA-induced THP-1 cells to analyze cell growth, Mo differentiation, CXCR4 membrane protein expression and miR-146a expression, as compared with untreated THP-1 cells (Supplementary Figure S1). Altogether, our data were in line with those observed during VitD3-induced Mo differentiation of U937 cells (Supplementary Figure S1).
Furthermore, we could not detect any modulation of previously described miR-146a target genes, such as tumor necrosis factor receptor-6 (ref. 22) (data not shown), indicating CXCR4 as a specific target of miR-146a in VitD3- or TPA- treated U937 cells.
AMD3100 treatment impairs leukemic cell proliferation and differentiation, induces miR-146a expression and inhibits CXCR4 expression
AMD3100 is a non-peptide antagonist of the CXCR4 receptor currently used to inhibit CXCR4 function and to mobilize hematopoietic stem cells.12 Here we analyzed the response to AMD3100 treatment of U937-miR146a cells, as compared with control cells U937-E and U937-wt.
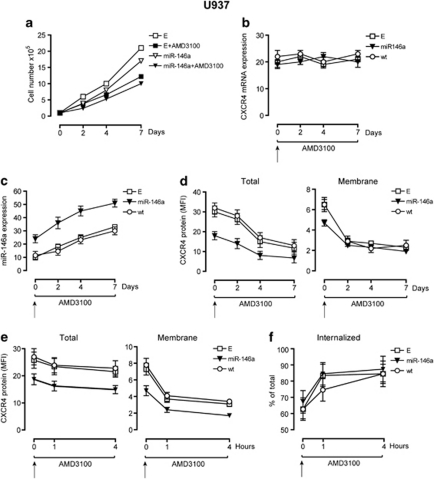
We first observed a decrease of the proliferation rate of U937-E cells upon AMD3100 treatment, as compared with untreated cells (Figure 4a), which is further enhanced by miR-146a (Figure 4a). The cell growth inhibition induced by AMD3100 is related to a reduced cell cycling (the proportion of S+G2+M cells was at day of culture: in U937-wt C 48.5±3.7 vs AMD3100 41.2±3.3, P<0.03; in U937-E 47±3.5 vs AMD3100 41.8±3.6, P=0.05; in U937-miR-146a C 44.9±3.5 vs 38±3.7, P<0.05) (mean values±s.d. observed in five separate experiments).
Figure 4.
AMD3100 treatment, downmodulates CXCR4, activates miR-146a and impairs leukemic cell proliferation. (a) Growth curve of U937-miR146a (miR-146a) and control U937-Empty (E) cells treated with AMD3100 added every two days in cultures, compared with untreated cells. (b) Quantitative real-time PCR analysis of CXCR4 mRNA and (c) miR-146a expression were, respectively, performed on the indicated days, in U937-miR146a cells as compared with control cells (wt and E), treated by AMD3100 compared with untreated cells (day 0). (d) FACS analysis of CXCR4 protein expression at sequential days during AMD3100 treatment of U937-miR146a and control cells (wt and E) compared with untreated cells (day 0). CXCR4 protein expression was evaluated at both, total (left panel) and membrane (right panel) level, on permeabilized or intact cells, respectively. The geometric MFI is shown as mean±s.e.m. values from three independent experiments. (e) FACS analysis of total and membrane CXCR4 protein expression, as compared with (f) internalized CXCR4 protein levels during acute (1 and 4 h) AMD3100 treatment of U937-miR146a and control wt and E cells. (a) One representative experiment out of three is shown. (b–f) Results of three independent experiments are shown (mean±s.e.m. values).
In line with previous studies,6, 7 short AMD3100 treatment (days 0–2) decreases CXCR4 protein level expressed on the leukemic cell membrane (Figure 4d, right panel). Surprisingly, upon longer AMD3100 treatment (days 2–7) of control cells (U937-E and wt), total CXCR4 protein expression is downmodulated, as detected by FACS analysis in permeabilized cells (Figure 4d, left panel), whereas miR-146a increases (Figure 4c), without any significative modulation of CXCR4 mRNA level (Figure 4b), as compared with untreated cells (day 0). We also investigated the modulation of CXCR4 protein expression in U937 cells exposed to acute treatment (1–4 h) with AMD3100. Exposure to AMD3100 elicited a rapid downmodulation of CXCR4 membrane protein expression, due in large part to receptor internalization, a clearly detectable event after 1 h of incubation (Figure 4e). The extent of CXCR4 internalization induced by AMD3100 was comparable in all, U937-wt, U937-E and U937-miR146a, cell lines (Figure 4f).
We also show that AMD3100, used in a chronic treatment of leukemic cells, induces miR-146a expression (Figure 4c) and enhances the effects previously observed by miR-146a overexpression in leukemic cells, such as the decrease of CXCR4 protein level (Figure 4d) and leukemic cell growth inhibition (Figure 4a).
Although we explored a possible effect of AMD3100 treatment on Mo differentiation of U937 cells in the absence of a Mo differentiation inducer, we found that AMD3100 alone was unable to induce any significant change of morphological features and CD11b, CD14 membrane antigen expression levels (Supplementary Figures S2a, b: C). We then evaluated the effect of AMD3100 treatment on Mo differentiation of U937 cells, induced by chemical inducers, such as, TPA (Supplementary Figures S2a, b, c: TPA) and VitD3 (Supplementary Figures S2d, e, f: VitD3).
Our data showed that during TPA-induced Mo differentiation of U937 cells, CXCR4 membrane protein expression is downmodulated, whereas miR-146a is upmodulated (Supplementary Figure S2c and data not shown), as also observed in VitD3-treated cells (Supplementary Figure S2f). However, AMD3100 treatment of VitD3-treated cells only slightly impaired Mo differentiation as assessed by morphological features (not shown) and CD11b, CD14 Mo markers expression analysis (Supplementary Figure S2d; Figure 2e), whereas AMD3100 treatment of TPA-treated cells induced a marked inhibition of Mo differentiation, as shown by morphology and CD11b, CD14 expression analysis (Supplementary Figure S2a; Figure 2b). Similar results were observed when we treated THP-1 cells with TPA to induce Mo differentiation, in addition with AMD3100 treatment (Supplementary Figure S1: TPA+AMD3100).
It is also of interest to note that in U937-miR146a cells, AMD3100-mediated inhibition of U937 TPA-induced Mo differentiation is major than that observed in control U937-E and U937-wt cells (Supplementary Figure S2a).
Altogether, these data indicate that AMD3100 treatment has an effect on proliferation and differentiation of monocytic leukemic cell lines.
MiR-146a and AMD3100 downmodulate CXCR4 protein expression and stimulate the drug sensitivity of leukemic cells
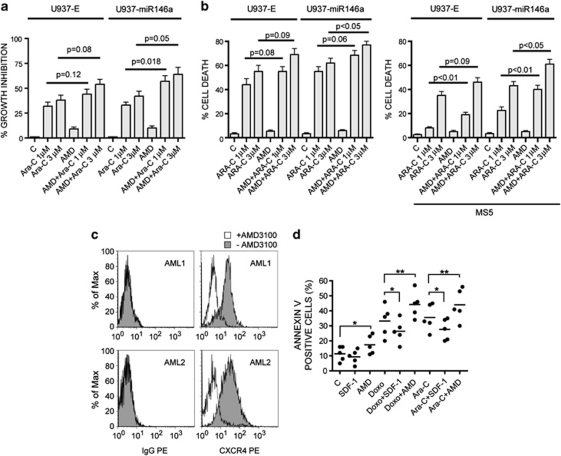
We investigated further the effect of AMD3100 for 24 h, in combination or not with an anti-leukemic drug, such as Ara-C21 (Figure 5a) or Doxorubicin21 (not shown) on leukemic cell proliferation. Our data indicate that, as expected, Ara-C induces a dose-independent growth inhibition of both U937-miR146a and U937-E control cells (Ara-C 1–3 μ), whereas AMD3100 alone has a mild inhibitory effect on the growth of these cells (Figure 5a, AMD). However, the effect of AMD3100 used in combination with Ara-C on the growth inhibition of leukemic cells is greater than that previously observed with Ara-C alone in both U937-miR146a and control U937-E cells (Figure 5a, AMD+Ara-C). Indeed miR-146a overexpression enhances, at a mild but significant level, the effect of AMD3100 used in combination with Ara-C, as a major growth inhibition of U937-miR146a cells is observed in combination treatment as compared with U937-E control cells (Figure 5a, U937-miR146a). The effect of AMD3100 in combination with Ara-C does not seem to be related to induction of cell differentiation, as indicated by analysis of cell morphology and of membrane differentiation antigens (data not shown).
Figure 5.
miR-146a and AMD3100 treatment downmodulates CXCR4, enhances the drug sensitivity of leukemic cells and reduces stromal-induced protection against chemotherapy. (a) Growth inhibition of U937-E and U937-miR146a cells after treatment (0–2 days) with AMD3100 (AMD) (10 μ) in combination, or not, with Ara-C (1 or 3 μ), as compared with untreated cells (C); (b) Induction of apoptosis in U937-E and U937-miR146a cells grown in liquid cultures as in a, either in the absence (left panel) or in the presence (right panel) of a layer of MS5 stromal cells; (c) FACS of CXCR4 surface protein expression in two representative primary AML samples treated 24 h with AMD3100 (1 μ) (white shadow) or not untreated (gray shadow). Fluorescence histograms immunoglobulin G-phycoerythrin (IgG-PE) and CXCR4-PE indicate, respectively, PE-conjugated control mouse IgG labelling and surface PE-conjugated CXCR4 labelling of both AML samples. (d) Percentage of apoptotic cells, indicated as annexin-V-positive cells, is detected by FACS performed in five primary AML samples cultured at 24 h: (i) without any treatment as a control (C); or with (ii) 100 ng/ml of the CXCR4 ligand, SDF-1; (iii) or 1 μ AMD3100 (AMD); (iv) or 1 μ Ara-C; (v) or 1 μ Doxorubicin (Doxo), (iv and v) alone or in combination with AMD. The bars reported in the scattergrams represent the mean values. *P<0.05; **P<0.01.
Altogether, we show that AMD3100 treatment increases the drug sensitivity of leukemic cells and miR-146a, moderately, but significantly enhances this effect.
Combined CXCR4 inhibition, mediated by miR-146a and AMD3100, reduces stromal-induced protection against chemotherapy
Previous studies have shown that BM stromal cells are able to modulate the chemosensitivity of leukemic blasts through mechanisms involving CXCR4/SDF-1α, VLA-4/fibronectin and other mediators/receptors.7 The growth in vitro of leukemic cells, on the stromal MS5 cells in part mimics this BM microenvironment, affecting the sensitivity of leukemic cells to antineoplastic agents.7 We then grew U937-E and U937-miR146a cells in the presence of a layer of MS5 cells (Figure 5b, right panel), as compared with liquid cultures (left panel), and evaluated their sensitivity to Ara-C, by analyzing the induction of apoptosis in U937-E and U937-miR146a cells (Figure 5b). We showed that (i) the sensitivity of both U937-E and U937-miR146a cells to Ara-C, particularly when added at 1 μ, is markedly reduced when cultured on MS5 cells; however, this decrease is more pronounced in U937-E than in U937-miR146a cells; (ii) AMD3100 in part rescued this decrease in drug sensitivity: this rescue being more pronounced in U937-miR146a than in U937-E cells (Figure 5b). These results indicate that the combined downmodulation of CXCR4 expression mediated by miR-146a overexpression and AMD3100 treatment is more efficacious than AMD3100 treatment alone to restore chemosensitivity of leukemic cells when these cells are grown in the presence of stromal cells.
Altogether, our data suggest that inhibition of CXCR4 expression by miR-146a overexpression and AMD3100 represent a strategy to enhance the drug sensitivity of leukemic cells.
AMD3100 enhances the sensitivity of CXCR4-positive AMLs to chemotherapy in vitro
We investigated further the functional role of CXCR4 blockade by AMD3100 on primary AMLs. We first selected five primary AMLs (two M4, two M5 and one M2) for both their high levels of CXCR4 protein expression and a low rate of spontaneous apoptosis, to analyze the ability of AMD3100 to compete with the anti-CXCR4 monoclonal antibody 12G5 for CXCR4 binding.22, 23 Our data show that, as previously observed in leukemic cells using high concentration (10 μ) of AMD3100 (Figure 5a), AMD3100 treatment, at lower concentration such as 1 μ, induces a marked decrease of surface staining of CXCR4 protein, of our selected primary AMLs (Figure 5c).
We then analyzed whether the inhibition of CXCR4 by acute AMD3100 treatment (24 h) modulates the sensitivity of AML blast cells to chemotherapeutic agents, by treating the CXCR4-positive AML samples with AMD3100 in combination, or not, with chemotherapy (Ara-C or Doxorubicin, Doxo) for 24 h (Figure 5d). Our data indicate that SDF-1α treatment did not significantly modify the rate of spontaneous apoptosis of AML blasts; in contrast, AMD3100 induces a slight but significant increase of the percentage of apoptotic cells as compared with untreated cells (C) (Figure 5d). However, AMD3100 treatment enhances significantly Ara-C or Doxorubicin-induced apoptosis of the leukemic blast cells, as compared with untreated control cultures, whereas SDF-1a reduces it (Figure 5d).
Altogether, our data show that acute AMD3100 treatment of primary AML blast cells downmodulates surface CXCR4 protein expression and enhances the sensitivity of AML blasts to chemotherapy-induced apoptosis.
Discussion
We have recently identified the mechanism of CXCR4 regulation by miR-146a targeting in megakaryocytic cells.17 Previous studies have shown that a high CXCR4 expression level is a negative prognostic marker in AMLs,2, 9, 10, 11 independently of some other negative prognostic factors.10, 11, 24 However, the molecular mechanisms responsible for the high CXCR4 expression observed in some AML cases are currently unknown.
Here, we have investigated miR-146a expression as compared with CXCR4 expression levels in AMLs and found that the miR-146a/CXCR4 regulatory pathway seems to operate in various AML subtypes independently of the differentiation capacities of these cells. Altogether, our study suggests that the absence of or low miR-146a expression could represent one of the mechanisms responsible for the elevated CXCR4 expression in leukemic cells. Indeed, high CXCR4 expression was associated with low miR-146a expression in all AML subtypes, including M1, M2, M3, M4 and M5. In line with previous reports,9, 24 CXCR4 expression was particularly high in myelomonocytic (M4) and monocytic (M5) AMLs. Therefore, it seemed of particular interest to explore the expression and regulation of the miR-146a/CXCR4 pathway in the normal monocytic lineage. The importance of this issue is also supported by a recent study showing that SDF-1α, by an autocrine/paracrine mechanism, regulates a peculiar set of the genetic program of macrophage differentiation.25 In CD34+ HPCs cultures undergoing monocytic differentiation, we show that CXCR4 mRNA is downmodulated during monocytopoiesis, whereas CXCR4 protein expression is markedly upmodulated. This finding strongly suggests that unblocking CXCR4 translational is one of the main molecular mechanisms controlling CXCR4 protein levels during monocytopoiesis. In line with this hypothesis, we observed that miR-146a levels are clearly downmodulated during the initial stages of monocytic differentiation. At later, terminal stages of monocytic differentiation, miR-146a markedly increases, while CXCR4 protein levels decrease.
An autocrine SDF-1α/CXCR4 loop was recently shown to be involved in the control of monocyte to macrophage differentiation.25 Furthermore, previous studies have shown a possible effect of AMD3100 on leukemic cell differentiation.6 Our results showed that a reduction of CXCR4 expression/function does not modify the spontaneous differentiation of leukemic cells, while it impairs the monocytic differentiation of these cells triggered by chemical inducers, particularly by TPA.
Altogether we show that in normal monocytopoiesis miR-146a regulates CXCR4 expression, as previously reported by our group in the megakaryocytic lineage.17 According to these findings, a low or absence of miR-146a expression, often observed among AMLs, may represent a key factor responsible for the high CXCR4 expression in these cells. Furthermore, our previous17 and present findings are in line with studies that have shown that miR-146a expression is deregulated in leukemia.26, 27, 28 Garzon et al.26 in a screening carried out on 122 untreated AMLs showed that miR-146a is one of the miRNAs most downmodulated in AML blasts, compared with normal CD34+ cells. M4/M5 AMLs, with MLL rearrangement and with the majority of leukemic cells being monoblasts, display miR-146a levels clearly lower than other AMLs.27 Finally, miR-146a was found to be markedly downmodulated in 5q- syndrome CD34+ cells, compared with normal CD34+ cells.28 Importantly, knockdown of miR-146a in mouse hematopoietic stem cells resulted in the development of some hematological abnormalities, such as neutropenia and megakaryocytic dysplasia, typically observed in myelodysplastic syndromes.28 Furthermore, knockdown of miR-145 and miR-146a results in long-latency myeloid leukemia in mice.29
Why high CXCR4 expression is a negative prognostic factor for AML patient survival after chemotherapy remains to be elucidated. This phenomenon could be related, to a certain extent, to the regulation of AML cell migration by SDF-1α/CXCR4.30 However, recent reports suggest an alternative mechanism related to the effect of CXCR4 signalling on drug sensitivity. In fact, evidence has been provided that CXCR4 activation by SDF-1α (either as soluble chemokine or as stromal cell-bound chemokine) reduces the sensitivity of AML blasts to various anti-leukemic drugs, whereas an opposite effect is elicited by CXCR4 inhibitors.6, 7, 31, 32
In the present study we have explored the effect of a CXCR4 inhibitor, AMD3100, recently registered as a hemopoietic stem cell mobilizing agent, on AML blasts. In line with a previous report,30 acute exposure of leukemic cells to this agent determines the rapid and massive downmodulation of surface CXCR4 due to receptor internalization, although the total receptor expression remains virtually unmodified.32 However, prolonged exposure of leukemic cells to AMD3100 induces a decrease of total CXCR4, a phenomenon possibly related to intracellular receptor degradation and also to reduced receptor translation. In fact, we observed that AMD3100 induces an increased expression of miR-146a. Furthermore, in line with previous studies,7 we observed that disruption of the CXCR4/SDF-1α interaction by the CXCR4 inhibitor AMD3100 clearly increases the sensitivity of leukemic cells to Ara-C. This effect was further potentiated in leukemic cells where CXCR4 expression is decreased by miR-146a overexpression. These findings suggest that the optimal therapeutic effect potentially deriving from inhibition of the CXCR4/SDF-1α axis requires a combined effect both at the level of CXCR4 content (that is, decreasing it by miR-146a overexpression) and of SDF-1α/CXCR4 interaction (that is, inhibiting this interaction using AMD3100 or similar agents). In this context, it is of interest to note that a recent study reported the synergistic antileukemic activity of a histone deacetylase inhibitor that markedly upmodulates miR-146a levels and AMD3100.33
In addition to CXCR4, miR-146a was shown to target in hematopoietic cells other mRNAs and particularly those encoding some factors involved in innate immune signalling, such as tumor necrosis factor receptor-6,22 toll-like receptor 234 and through these mechanisms it seems to have a critical role for endotoxin-induced tolerance by monocytes.35 Although in our cellular models we have failed to demonstrate an effect of miR-146a overexpression on tumor necrosis factor receptor-6 protein expression levels, we cannot exclude that other miR-146a targets may have a relevant role to explain its effect at the level of AMLs.
Altogether, our study indicates that a low miR-146a expression may contribute to the elevated CXCR4 expression observed in some AMLs. Furthermore, modulating the levels of miR-146a, as also recently shown for some solid tumors, including breast cancer36 and pancreatic cancer,37 could have a therapeutic potential, together with CXCR4 inhibitors, to improve drug sensitivity of leukemia cells.
Acknowledgments
We thank G Loreto for graphics. This work was supported from Associazione Italiana per la Ricerca sul Cancro (AIRC) to CL and RF, from ‘Fondazione Roma' Rome, Italy to UT and by institutional grants from the Italian Ministry of Health (Italy-USA) to UT and CL.
The authors declare no conflict of interests.
Footnotes
Supplementary Information accompanies the paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Broxmeyer HE. Chemokines in hematopoiesis. Curr Opin Hematol. 2008;15:49–58. doi: 10.1097/MOH.0b013e3282f29012. [DOI] [PubMed] [Google Scholar]
- Gazitt Y. Homing and mobilization of hematopoietic stem cells and hematopoietic cancer cells are mirror image processes, utilizing similar signalling pathway and occurring concurrently: circulation cancer cells constitute an ideal target for concurrent treatment with chemotherapy and antilineage-specific antibodies. Leukemia. 2004;18:1–10. doi: 10.1038/sj.leu.2403173. [DOI] [PubMed] [Google Scholar]
- Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768:952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavor S, Petit I, Porozov S, Avigdor A, Dar A, Leider-Trejo L, et al. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. 2004;64:2817–2824. doi: 10.1158/0008-5472.can-03-3693. [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–1924. doi: 10.1038/sj.leu.2404357. [DOI] [PubMed] [Google Scholar]
- Tavor S, Eisenbach M, Jacob-Hirsch J, Golan T, Petit I, Benzion K, et al. The CXCR4 antagonist AMD3100 impairs survival of human AML cells and induces their differentiation. Leukemia. 2008;22:2151–2158. doi: 10.1038/leu.2008.238. [DOI] [PubMed] [Google Scholar]
- Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, et al. Chemosensitization of AML following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–6214. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco G, Belmont JW, Konopleva M, Andreeff M, Tavor S, Petit I, et al. Correlation between CXCR4 and homing or engraftment of acute myelogenous leukemia. Cancer Res. 2004;64:6832–6833. doi: 10.1158/0008-5472.CAN-04-1936. [DOI] [PubMed] [Google Scholar]
- Spoo AC, Lubbert M, Wierda WG, Burger JA. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood. 2007;109:786–791. doi: 10.1182/blood-2006-05-024844. [DOI] [PubMed] [Google Scholar]
- Rombouts EJ, Pavic B, Lowenberg B, Ploemacher RE. Relation between CXCR4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood. 2004;104:550–557. doi: 10.1182/blood-2004-02-0566. [DOI] [PubMed] [Google Scholar]
- Konopiev S, Rassidakis GZ, Estey E, Kantarjian H, Liakou CI, Huang X, et al. Overexpression of CXCR4 predicts adverse overall and event-free survival in patients with unmutated FLT3 acute myeloid leukemia with normal karyotype. Cancer. 2007;109:1152–11510. doi: 10.1002/cncr.22510. [DOI] [PubMed] [Google Scholar]
- Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- Bonig H, Chudziak D, Priestley G, Papayannopoulou T. Insights into the biology of mobilized hematopoietic stem/progenitor cells through innovative treatment schedules of the CXCR4 antagonist AMD3100. Exp Hematol. 2009;37:402–415. doi: 10.1016/j.exphem.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4804. [PubMed] [Google Scholar]
- Lapteva N, Yang AG, Sanders DE, Strube RW, Chen SY. CXCR4 knockdown by small interfering RNA abrogates breast tumor growth in vivo. Cancer Gene Ther. 2005;12:84–89. doi: 10.1038/sj.cgt.7700770. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- Spinello I, Quaranta MT, Pasquini L, Pelosi E, Petrucci E, Pagliuca A, et al. PLZF-mediated control on c-kit expression in CD34(+) cells and early erythropoiesis. Oncogene. 2009;28:2276–2288. doi: 10.1038/onc.2009.87. [DOI] [PubMed] [Google Scholar]
- Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- Hughes PJ, Marcinkowska E, Gocek E, Studzinski GP, Brown G. Vitamin D3-driven signals for myeloid cell differentiation--implications for differentiation therapy. Leuk Res. 2010;34:553–565. doi: 10.1016/j.leukres.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robak T, Wierzbowska A. Current and emerging therapies for acute myeloid leukemia. Clin Ther. 2009;31:2349–2370. doi: 10.1016/j.clinthera.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRMA miR-146a, an inhibitor targeted to signalling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Kong P-Y, Shi Z-Z, Zeng D-F, Chen X-H, Chang C, et al. Effects of anti-CXCR4 monoclonal antibody 12G5 on proliferation and apoptosis of human acute myelocytic leukemia cell line HL-60. J Med Colleges PLA. 2007;22:17–22. [Google Scholar]
- Mohle R, Schittenhelm M, Failenschmid C, Bautz F, Kratz-Albers K, Serve H, et al. Functional response of leukaemic blasts to stromal cell-derived factor-1 correlates with preferential expression of the chemkine receptor CXCR4 in acute myelomonocytic and lymphoblastic leukaemia. Br J Haematol. 2000;110:563–572. doi: 10.1046/j.1365-2141.2000.02157.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin L, Estecha A, Samaniego R, Sanchez-Ramon S, Vegas MA, Sanchez-Mateos P. The chemokine CXCL12 regulates monocyte-macrophage differentiation and RUNX3 expression. Blood. 2011;117:88–97. doi: 10.1182/blood-2009-12-258186. [DOI] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukaemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starczynowski D, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- Starczynowski D, Morin R, McPherson A, Lam J, Chari R, Wegzzyn J, et al. Genome-wide identification of human micro-RNAs located in leukemia-associated genomic alterations. Blood. 2011;117:595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- Kittang AO, Hatfield K, Sand K, Reikvam H, Bruserud O. The chemokine network in acute myelogenous leukemia: molecular mechanisms involved in leukemogenesis and therapeutic implications. Curr Top Microbiol Immunol. 2010;870:149–172. doi: 10.1007/82_2010_25. [DOI] [PubMed] [Google Scholar]
- Fiegl M, Samudio I, Clise-Dwyer K, Burks JK, Mnjoyan Z, Andreeff M. CXCR4 expression and biologic activity in acute myeloid leukemia are dependent on oxygen partial pressure. Blood. 2009;113:1504–1512. doi: 10.1182/blood-2008-06-161539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Shi YX, Samudio I, Wang RY, Ling X, Frolova O, et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–6224. doi: 10.1182/blood-2008-05-158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandawat A, Fiskus W, Buckley KM, Robbins K, Rao R, Balusu R, et al. Pan-histone deacetylase inhibitor panobistat depletes CXCR4 levels and signaling and exerts synergistic antimyeloid activity in combination with CXCR4 antagonists. Blood. 2010;116:5306–5315. doi: 10.1182/blood-2010-05-284414. [DOI] [PubMed] [Google Scholar]
- Jurkin J, Schichl YM, Koeffel R, Bauer T, Richter S, Konradi S, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin induced tolerance. Implication in innate immunity. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, VandenBoom TG, II, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.