Abstract
MicroRNAs (miRNAs) are involved in the regulation of many cellular processes including hematopoiesis, with the aberrant expression of differentiation-stage specific miRNA associated with lymphomagenesis. miRNA profiling has been essential for understanding the underlying biology of many hematological malignancies; however the miRNA signature of the diverse tumor clone associated with Waldenstrom's macroglobulinemia (WM), consisting of B lymphocytes, plasmacytes and lymphoplasmacytic cells, has not been characterized. We have investigated the expression of over 13 000 known and candidate miRNAs in both CD19+ and CD138+ WM tumor cells, as well as in their malignant and non-malignant counterparts. Although neither CD19+ nor CD138+ WM cells were defined by a distinct miRNA profile, the combination of all WM cells revealed a unique miRNA transcriptome characterized by the dysregulation of many miRNAs previously identified as crucial for normal B-cell lineage differentiation. Specifically, miRNA-9*/152/182 were underexpressed in WM, whereas the expression of miRNA-21/125b/181a/193b/223/363 were notably increased (analysis of variance; P<0.0001). Future studies focusing on the effects of these dysregulated miRNAs will provide further insight into the mechanisms responsible for the pathogenesis of WM.
Keywords: microRNA, Waldenstrom's macroglobulinemia, B lymphocyte, plasma cell
Introduction
Waldenstrom's macroglobulinemia (WM) is a rare, non-Hodgkin's lymphoma, marked by a monoclonal immunoglobulin (Ig)M gammopathy and tumor infiltration into the bone marrow and lymphatic tissue.1 Classified as a lymphoplasmacytic lymphoma, the WM tumor clone comprised of B lineage cells across a morphological continuum that includes B lymphocytes, plasmacytoid cells and plasma cells, prompting many to postulate that WM tumors arise during the differentiation of B-lymphocytes into IgM-secreting plasma cells.2 As a consequence of this heterogeneity, it is not surprising that WM tumors share biological characteristics and clinical manifestations with other malignancies of B lineage cells such as chronic lymphocytic leukemia (CLL) and multiple myeloma (MM). As with CLL, WM tumors are indolent and proliferate slowly, whereas the hypersecretion of Ig, also common to MM, is responsible for much of the morbidity associated with WM. Yet, unlike CLL and MM, the pathogenesis of WM has not been commonly associated with the presence of cytogenetic or epigenetic abnormalities, suggesting that other processes may be regulating the behavior of WM tumor cells.3, 4, 5
Comparing and contrasting the clonal compartments of WM tumors with their lymphocytic and plasmacytic counterparts in malignancies with more homogeneous tumor clones has begun to enhance our understanding of the potential mechanisms driving the development and maintenance of this disease. Recently, gene expression profiles of B cells and plasma cells from WM have been compared with B cells from CLL and plasma cells from MM.6, 7 These analyses identified an mRNA signature for WM tumor cells that displayed a higher degree of similarity to CLL and non-malignant B cells than MM or non-malignant plasma cells. Yet the level of mRNA expression in the different WM samples included in the analyses was highly dependent on the extent of tumor involvement of the bone marrow, with the transcription profile of cases demonstrating low bone marrow infiltration clustering with normal plasmacytes. Because of this variability, the expression of only a small set of genes uniquely characterized WM. Similarly, a recent proteomics analysis identified the differential expression (>two-fold difference in expression) of only 10 proteins in WM cells as compared to non-malignant controls out of a total of 512 proteins assessed.8 Taken together, these results suggest that identifying unique factors responsible for the biological behavior of WM tumors may be more challenging than in other hematological malignancies.
MicroRNA (miRNA) are endogenous, short (19–22 nucleotides), non-coding RNA molecules that act as negative regulators of eukaryotic gene expression.9 This regulation is accomplished at the post-transcriptional level by the binding of miRNA to 3′-untranslated regions of target transcripts.10 Substantial evidence exists, supporting the crucial role of miRNA in controlling important biological processes, such as proliferation, apoptosis, cellular signaling and metabolism.11, 12, 13 Additionally, the dynamic and temporal expression of lineage-specific miRNAs during hematopoiesis is critical for the differentiation of hematopoietic cells.14, 15 Ectopic expression of these miRNAs, including miR-181a and -223, substantially altered lineage commitment and differentiation and was associated with lymphomagenesis in animal models, underscoring the important regulatory role of miRNA in controlling normal hematopoiesis.14
The aberrant expression of miRNAs has been frequently implicated in the development of malignancy, including hematological disorders, leading to the classification of many miRNAs as oncogenes or oncomiRs.16 The activation of oncomiRs is often a direct result of genomic instability, with many miRNA genes localized to fragile chromosomal sites, minimal regions of amplification or on common breakpoints associated with human cancers.17 Additionally, many miRNAs become dysregulated during the process of cellular transformation. Yet, the miRNA profiles of neoplastic cells often retain inherited expression patterns reflective of their non-malignant progenitors, including the expression of cell type-dependent and differentiation stage-specific miRNAs.18 Thus, the unique miRNA signature of a tumor may provide clues as to both the origin of the tumor itself, as well as the mechanisms controlling tumorigenesis.
Recently, the miRNA expression profile of WM B cells was characterized. A distinct molecular signature, defined by the dysregulation of seven miRNAs, distinguished B cells of patients diagnosed with WM from those of healthy subjects.19 However, the morphology of WM tumors is heterogenous by nature, consisting of plasmacytes and lymphoplasmacytic cells, in addition to B lymphocytes. Thus, it is possible that the differential expression of other miRNAs not previously identified, specifically those involved in the regulation of Ig production, may be detected in a more diverse WM tumor cell population. Even more, comparisons between the miRNA profiles of WM tumors and those of CLL and MM may reveal differentially regulated miRNAs involved in abnormal differentiation of B lineage cells. Identifying these miRNAs and their potential gene targets will deepen our understanding of the biological processes involved in WM pathogenesis.
Materials and methods
Patient specimens
Malignant B lineage cells were isolated from the bone marrow and/or peripheral blood of patients diagnosed with WM, CLL or MM. As a control, B lineage cells were also obtained from 13 healthy donors (non-malignant, NM). The collection of all specimens was approved by the Mayo Foundation Institutional Review Board, and informed patient consent was obtained in accordance with the Declaration of Helsinki. The collected samples were isolated using CD19- and/or CD138-positive selection beads and a RoboSep Cell Separator per the manufacturer's instructions (Stem Cell Technologies; Vancouver, BC, Canada). The groups were as follows: WM 19+138+ (n=8), WM 19+ (n=6), WM 138+ (n=3), NM 19+138+ (n=4), NM 19+ (n=3), NM 138+ (n=6), CLL 19+ (n=5) and MM 138+ (n=5).
RNA isolation and microarray profiling
RNA was isolated from all specimens using the mirVANA RNA Isolation kit (Ambion Inc., Austin, TX, USA) according to the manufacturer's protocol for total RNA isolation. Extracted RNA was quantified using a Nanodrop ND-1000 spectrophotometer (Nanodrop; Wilmington, DE, USA) and stored at −80 °C until needed. Total RNA of 1 μg from each sample was shipped to Asuragen Services for miRNA profiling via their DiscovArray miRNA Expression Service (Asuragen Services; Austin, TX, USA). miRNA expression analysis was performed through the hybridization of RNA to a custom-manufactured Affymetrix Gene Chip (miRChip V1, Ambion Inc.) that targets 14 215 orthologous candidate miRNAs including 452 confirmed human miRNAs derived from the Sanger miRBase version 9.2 (http://www.mirbase.org/). The 14 215 miRNAs were measured by two probes each (probes A and B for each miRNA). The signal processing method utilized for the miRChip V1 array involved multiple steps including probe specific signal detection calls, corrections for background estimates and global normalization across all signals detected. More specifically, a set of G-C-matched anti-genomic oligonucleotides was employed as a control for each probe. The signal derived from each probe on an array was compared with the signal obtained from the probe-specific anti-genomic control with a Wilcoxon rank-sum test. According to Asuragen, a probe with detection P⩽0.06 is considered ‘detected above background.' Probes with P-values >0.06 have insufficient signal to discriminate from the background and were thus considered not detected Additionally, the median signal arising from the oligonucleotide controls served as an estimated background value and was subtracted from the probe-specific signal to yield a normalized value used in further analysis. Global normalization of all arrays within a specific analysis experiment was achieved through the use of the variance stabilization normalization method described by Huber et al.20 The post-normalized data is reported as generalized log2 (glog2).
Identification of target genes
Putative miRNA binding sites were identified in the 3′-untranslated regions of human target genes and their orthologs, using the target prediction algorithms specific to TargetScan v5.1 (http://www.targetscan.org/, Whitehead Institute for Biomedical Research, Cambridge, MA, USA), TarBase v5.c (http://diana.cslab.ece.ntua.gr/tarbase/, Alexander Fleming Biomedical Sciences Research Center, Varkiza, Greece) and miRanda (http://www.microrna.org/micror na/home.do, Memorial Sloan-Kettering Cancer Center, New York, NY, USA) web sites.21, 22, 23 Specifically, genes with known association to either Ig production or hematopoietic differentiation and also possessing a high predicted efficacy of binding score were included for further discussion.
Statistical analysis
For statistical hypothesis testing, a two-sample t-test, with assumption of equal variance was applied, and one-way ANOVA was used for multiple group comparisons. If a probe set was not detectable in any of the samples, the set was filtered out. Specific miRNA were considered to be significantly differentially expressed based on a cutoff P-value <0.001 and an absolute fold-change in expression of greater than two (absolute log2 difference >1).
Results
A total of 16 specimens were isolated from patients with a known diagnosis of WM. At the time of diagnosis, all patients presented with symptomatic disease. The median patient age was 74.5 years (range 54–85) as compared with a median age of 72 years for patients diagnosed with either CLL (n=5, range 45–89 years) or MM (n=5, range 63–83 years). The median level of serum IgM in WM patients was 1.65 g/dl (range 0–6.4 g/dl), with a median bone marrow involvement of 60% (range 0–80%). Cytogenetic findings were unremarkable for all patient samples, except for one specimen with a reported 13q deletion.
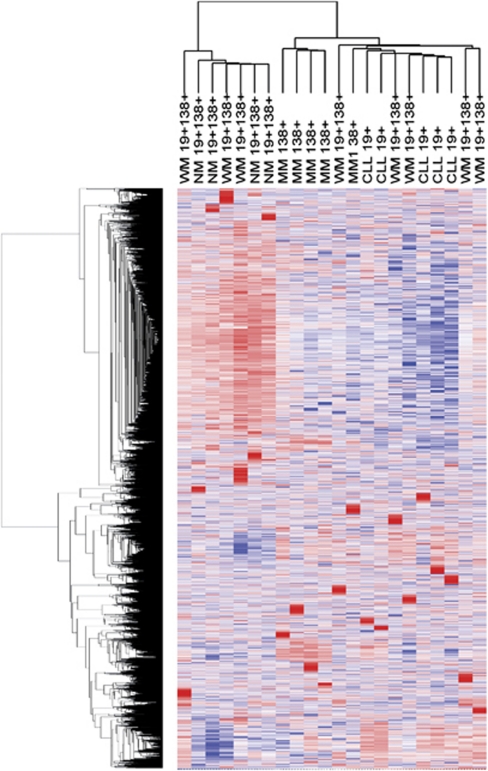
Expression of 14 125 putative and confirmed miRNAs (28 430 total probes) was initially assessed in malignant lymphoplasmacytic cells (CD19+CD138+) derived from patients diagnosed with WM. There were 4272 probes including all species with at least one P-value ⩽0.06, which were considered detectable above background and included in the analyses. Comparisons were then made with the detected probes of malignant B cells from patients with CLL (CD19+), malignant plasma cells from patients with MM (CD138+) and B-lineage cells from healthy donors. Initial unsupervised hierarchical clustering, with average linkage method and Pearson's correlation as the distance metric, was performed all on ‘detectable' probes. The heat map generated from this analysis did not indicate a distinct cluster corresponding to the double-sorted (CD19+CD138+) WM cells despite near perfect segregation of CLL and MM cells (Figure 1). Instead, subsets of the CD19+CD138+ WM cells had patterns of expression similar to CLL, whereas the signatures of other CD19+CD138+ WM samples were more consistent with MM or non-malignant cells.
Figure 1.
Profiling of miRNA expression in WM (CD19+CD138+), CLL (CD19+), MM (CD138+) and non-malignant B lineage cells (CD19+CD138+). Unsupervised hierarchical clustering was performed using all miRs detected in at least one sample. Both candidate miRNAs and known human miRNAs derived from the Sanger miRBase have been included for analysis. Generalized log ratios (glog2) for each miRNA are represented. Gradations of blue and red mean lower and higher expression levels, respectively. WM indicates Waldenstrom's macroglobulinemia; CLL, chronic lymphocytic leukemia; MM, multiple myeloma; NM, non-malignant.
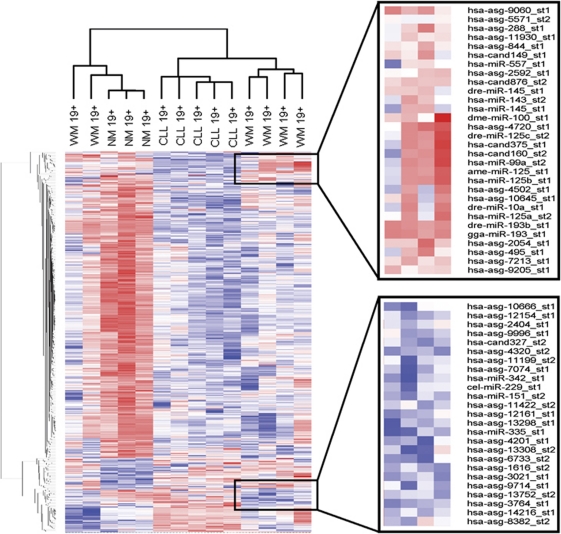
The lack of a clear miRNA signature for lymphoplasmacytic (CD19+138+) WM cells was not unexpected and is most likely due to the tumor cell's dual nature, possessing features of both B lymphocytes and plasma cells. Therefore, separate analyses for B cells (CD19+) and plasma cells (CD138+) derived from WM tumors were performed and again compared with their malignant and non-malignant counterparts. In the analysis for B cells, the 531 probes that were different between any two groups of CD19+ WM, NM or CLL cells (group comparison P-value ⩽0.01) were used in the clustering analysis. As shown in Figure 2, the majority of WM B lymphocyte samples clustered distinctly on a separate dendrogram branch from both non-malignant and CLL B-cell populations, but more closely resembled the miRNA expression profile of CLL cells. However, two WM samples, both derived from patients with very low percentages of bone marrow involvement, clustered with the non-malignant B cells. With the removal of these two samples, the miRNA signature of WM B cells was clearly defined by a number of consistently downregulated miRNAs as compared with CLL and non-malignant B cells, including miR-151, miR-335 and miR-342, whereas miRNA-373 was notably increased in WM B cells (P<0.001, Figure 2). As with previous reports, overall expression levels of miRNAs were higher in non-malignant B cells as compared with malignant B cells, but a small subset of miRNAs were routinely downregulated in the non-malignant population, including miR-32, miR-142b, miR-156, miR-215 and miR-338 (P<0.001).24 Interestingly, the most abundantly expressed miRNAs in CD19+ B cells remained largely constant regardless of pathology, with miR-15, miR-26a and miR-150 being the three most highly expressed miRNA in WM B cells, CLL B cells and normal B cells, respectively. Conversely, miR-155 was expressed highly in CD19+ CLL cells as has been reported previously, but was not as abundant in either non-malignant or WM B cells (Table 1).25
Figure 2.
miRNA expression profiling in CD19+ B cells and derived from WM, CLL and NM donors. Heatmaps were generated using unsupervised hierarchical clustering of all miRs detected in at least one sample. Only known human miRNAs from the Sanger miRBase have been included for analysis. Generalized log ratios (glog2) for each miRNA are represented. Gradations of blue and red mean lower and higher expression levels, respectively.
Table 1. Ten most abundantly expressed miRNAs in WM (CD19+, CD138+ and CD19+CD138+), CLL (CD19+), MM (CD138+) and NM B lymphocytes and plasma cells.
| 19+ NM | 138+ NM | 19+138+ NM | 19+ WM | 138+ WM | 19+138+ WM | 19+ CLL | 138+ MM |
|---|---|---|---|---|---|---|---|
| 150 | 638 | 638 | 15 | 15 | 150 | 150 | 148a |
| 15 | 148a | 150 | 150 | 638 | 15 | 26a | 29a |
| 26a | 223 | 15 | 26a | 223 | 26a | 15 | 15 |
| 638 | 26a | 223 | 638 | 150 | 29a | 29a | 26a |
| let-7f2 | 29a | 26a | 21 | 26 | let-7f2 | let-7g | 29b |
| let-7g | 15 | 148 | 223 | 29a | 223 | 21 | let-7f2 |
| let-7a3 | 150 | 29a | let-7g | 23a | let-7g | let-7f2 | 638 |
| 29a | let-7g | let-7f2 | let-7f2 | 21 | 21 | 29b | let-7g |
| 17 | 29b | let7g | let-7a | let-7g | 29b | 155 | 29c |
| 181a | let-7f2 | let-7a3 | 29a | let7f2 | let-7a3 | let-7a3 | 150 |
Abbreviations: CLL, chronic lymphocytic leukemia; MM, multiple myeloma; NM, non-malignant; WM, Waldenstrom's macroglobulinemia.
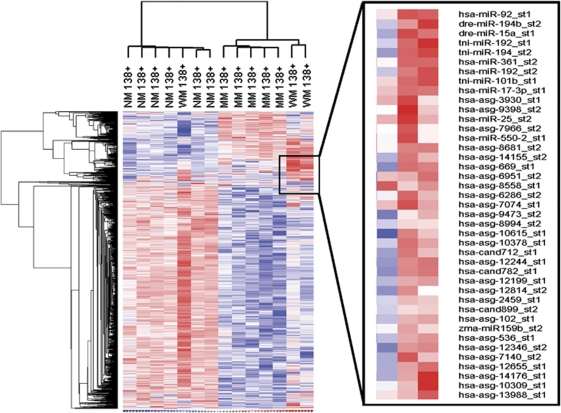
Similarly, microarray analysis yielded individual signatures for MM plasma cells and non-malignant plasma cells as well (Figure 3). There were 937 probes that were different (P-value ⩽0.01) in at least one of the pair-wise group comparisons between CD138+ WM, NM and MM cells, and these 937 probes were used in the clustering analysis. Again, the majority of CD138+ WM cells clustered with MM plasma cells, yet retained a distinct miRNA profile of their own, characterized by the increased expression of more than 40 candidate miRNAs (Figure 2b). One CD138+ WM sample was defined by an miRNA signature more similar to that of the non-malignant plasma cells, which in general, was characterized by higher levels of miRNA expression as compared with the malignant plasma cells. This particular WM sample was found to have a lower percentage of cellular involvement of the bone marrow as compared with the other two cases in the WM 138+ set. No differentially expressed miRNA were detected when CD138+ WM cells were compared with CD19+ WM cells. However, a unique set of 48 miRNA discriminated between non-malignant plasma cells and B cells, of which 43 were downregulated in plasma cells. The five miRNA identified as being more highy expressed in non-malignant plasma cells included miR-152 (7.2-fold, P<0.0001), miR-148a (12.8-fold, P<0.0001), miR-182 (13.45-fold, P=0.0006), miR-193b (18.1-fold, P<0.0001) and miR-96 (24.3-fold, P<0.0001).
Figure 3.
miRNA expression profiling in CD138+ plasma cells derived from WM, MM and NM donors. Heatmaps were generated using unsupervised hierarchical clustering of all miRs detected in at least one sample. Only known human miRNAs from the Sanger miRBase have been included for analysis. Generalized log ratios (glog2) for each miRNA are represented. Gradations of blue and red mean lower and higher expression levels, respectively.
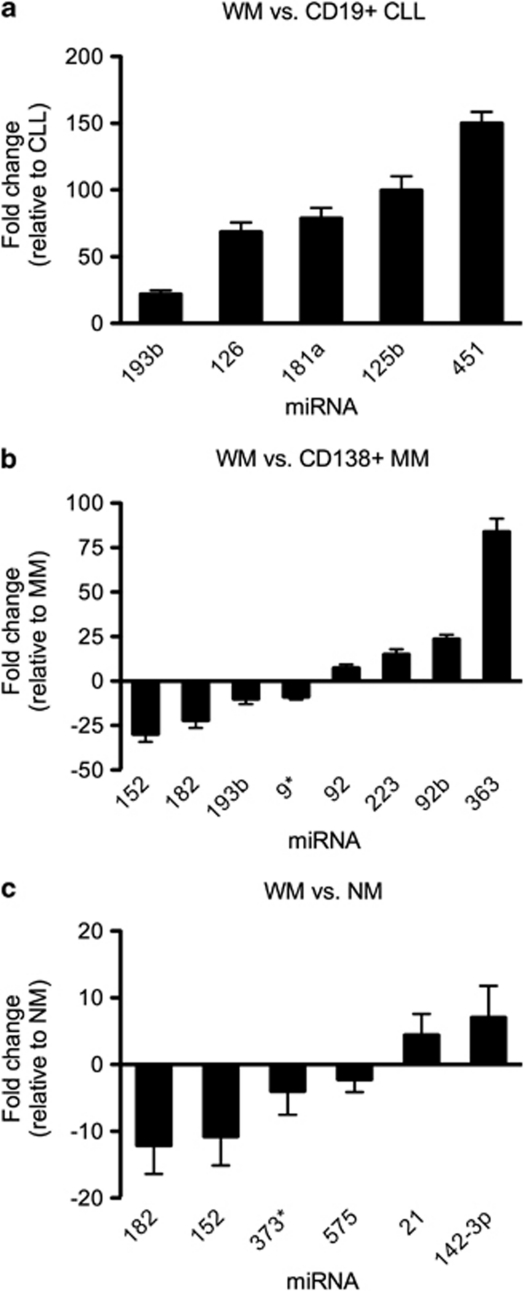
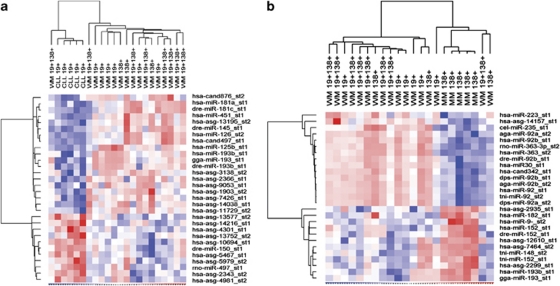
Lastly, to account for the heterogeneity in the WM tumor clone, all WM samples (B lymphocytes, plasma cells and lymphoplasmacytic cells, n=17) were analyzed collectively, and the miRNA profile obtained was compared with that of all non-malignant B lineage cells (n=8), as well as CLL B cells and MM plasma cells. This analysis identified six miRNAs with significantly altered expression between WM cells and the non-malignant group. Between these two cell populations, miR-152 (10.7-fold, P<0.0001), miR-182 (12.1-fold, P<0.0001), miR-373* (3.9-fold, P=0.0007) and miR-575 (2.2-fold, P=0.0003) were observed to be expressed at a higher level in non-malignant cells relative to WM, with the opposite pattern for miR-21 (4.4-fold, P=0.0002) and miR-142-3p (7.1-fold, P=0.0007) (Figure 4). When compared with CLL B lymphocytes, miR-125b (99.7-fold, P=0.0003), miR-126 (68.5-fold, P=0.0001), miR-181 (78.8-fold, P=0.0003), miR-193b (21.8-fold, P=0.0003) and miR-451 (150.1-fold, P<0.0001), demonstrated significantly higher expression in WM cells (Figures 4 and 5a). A set of eight miRNAs were differentially expressed between the combined WM cells and MM cells, with multiple probes identifying the upregulation of miR-92 and miR-92b in WM cells (Figures 4 and 5b). After differentially expressed miRNAs had been identified in WM, predicted gene targets for each deregulated miRNA were determined. Putative mRNA targets were selected based on a high sequence complementarity and binding efficacy between the target gene and the deregulated miRNA, as well as a known relevance to either Ig production and/or B lineage differentiation. Selected targets are presented in Table 2.
Figure 4.
Differentially expressed miRNAs in WM. All samples derived from patients with WM (CD19+, CD138+, CD19+CD138+) were combined into one group (n=17) for analysis and compared with miRNA expression in (a) CD19+ CLL, (b) CD138+ MM and (c) NM (CD19+, CD138+, CD19+CD138) cells. Values are fold-change in expression relative to CLL, MM or NM cells, respectively. Specific miRNA were considered to be significantly differentially expressed based on a cutoff P-value <0.001 and an absolute fold-change in expression of greater than two. Only known human miRNAs derived from the Sanger miRBase were considered for analysis.
Figure 5.
miRNA expression profiling in B lymphocytes and plasmacytes from WM tumors (CD19+, CD138+, CD19+CD138+) and (a) CD19+ B lymphocytes from CLL and (b) CD138+ plasmacytes from MM. Heatmaps were generated using unsupervised hierarchical clustering of all differentially expressed miRNAs (P<0.001). Both human and orthologous miRNAs are included. Generalized log ratios (glog2) for each miRNA are represented. Gradations of blue and red mean lower and higher expression levels, respectively.
Table 2. Dysregulated miRNAs in WM with putative mRNA targets involved in immunoglobulin secretion and/or B-cell differentiation.
| miRNA | Chromosomal location | Expression level | Putative mRNA targetsa | Role in lymphoma |
|---|---|---|---|---|
| 9* | 1q22 | Low vs MM | XBP1, BCL6, PRDM1, SOCS4, SOCS5, JAK2 | WM,19, 30 HL,42 BL,50 B-cell differentiation51 |
| 21 | 17q23.1 | High vs NM | IL12A, TIMP3, PTEN, IL12A | NHL,52 BCL,28 NK,53 WM30 |
| 125b | 21q21.1 | High vs CLL | IRF4, PRDM1, IL6R, CCR5 | B-cell differentiation18, 27, 29 |
| 152 | 17q21.32 | Low vs MM, NM | PTEN, SOCS3, STAT1, JAK1, IL15 | SzS,36 ALL35 |
| 181a | 1q32.1 | High vs CLL | TIMP3, DUSP5, BCL2, PRDM1, BCL6, IL2, STAT3, PAX5, IRF4 | NHL,38 B-cell differentiation,14 MDS,54 T-cell maturation55 |
| 182 | 7q32.2 | Low vs MM, NM | IL2, PRDM1 | HL,34 MCL,56 expansion of T-helper cells57 |
| 193b | 16p13.12 | High vs CLL Low vs MM | SOCS3, IL17R, SOCS6 | MM,58 HSC29 |
| 223 | Xq12 | High vs MM | LMO2, STAT1, PRDM1, IL17R, MYBL1, IRF4 | B-cell differentiation,14, 15, 18, 51 CLL,59 WM,30 DLBCL60 |
| 363 | Xq26.2 | High vs MM | TACI, LMO2, BCL6, DUSP5, CIITA | WM19 |
Abbreviations: WM, Waldenstrom's macroglobulinemia; HL, Hodgkin's lymphoma; BL, Burkitts lymphoma; NHL, non-Hodgkin's lymphoma; BCL, B-cell lymphoma; NK, natural killer cell lymphoma; ALL, acute lymphocytic leukemia; MDS, myelodysplastic syndromes; MCL, mantle cell lymphoma; MM, multiple myeloma; CLL, chronic lymphocytic leukemia; HSC, hematopoietic stem cells; DLBCL, diffuse large B-cell lymphoma.
Predicted target genes identified from the TargetScan, TarBase and MirRanda databases.
Experimentally validated target genes are in bold.
Discussion
The tumor clone of WM is unique in its morphologic diversity, consisting of B lymphocytes and plasmacytes, as well as lymphoplasmacytoid cells. Phenotypically, WM cells maintain characteristics of both B lymphocytes and plasmacytes, consistent with the hypothesis that the WM tumor clone arises from a mature, memory-like B-cell that has, through some degree of differentiation, acquired the ability to secrete IgM.26 Thus, clues to understanding the mechanisms driving WM biology may lie within this lymphoplasmacytic differentiation process itself.
Normal hematopoietic differentiation is tightly regulated by both extrinsic signals derived from the cellular microenvironment as well as intrinsic modulators, including miRNA. Recently, distinctive miRNA expression signatures have been defined for specific stages of B-cell development. Forced expression of differentiation stage-specific miRNAs in healthy B cells has been associated with a multitude of outcomes including the inhibition of B-cell maturation, premature plasma differentiation and lymphomagenesis.14, 27, 28, 29 Initial miRNA profiling in WM B cells identified a small set of dysregulated miRNAs, many of which have been implicated in the normal progression of germinal center B cells to mature plasma cells.19, 30 To elaborate upon these findings, we profiled miRNA expression in WM B cells and plasma cells, prototypic malignant B cells and plasma cells derived from patients with CLL and MM, respectively, and in their non-malignant counterparts.
Unlike previous analyses, our study revealed a unique signature for neither CD19+ WM cells nor CD138+ MM. Some samples clustered more closely with non-malignant B cells or plasma cells, whereas others more closely resembled CLL or MM cells, each of which, as previously reported, clustered as a homogenous group.31, 32 Although we acknowledge that this variability may be largely due to the relatively small sample sizes of our respective groups, our findings are also consistent with earlier mRNA profiling studies, whereby the variability in expression pattern was directly related to the extent of WM disease in the patients from which the samples were derived. Namely, the expression profiles of samples obtained from patients with higher bone marrow involvement were more similar to their malignant counterparts, whereas patients with less involvement had samples that tended to cluster with the respective non-malignant cells.6, 7 In our CD19+ B-cell analysis, the two WM samples with miRNA profiles bearing a stronger resemblance to normal as opposed to malignant B cells were derived from previously treated patients with subsequently lower levels of bone marrow involvement, suggesting that a patient's miRNA signature may change in response to treatment. Similar post-treatment alterations in miRNA expression have been demonstrated in vitro in BCWM.1 cells after exposure to perifosine, bortezomib and rituximab.19 Although the current study was not designed to examine the effect of treatment on miRNA expression, these initial cases suggest that patients who respond to therapy with a reduction in bone marrow infiltration may display an miRNA profile more similar to non-malignant cells, as compared with either non-responders or untreated patients. However, this is merely speculative and must be confirmed in a larger study.
Despite the interpatient variability and lack of a distinct miRNA transcriptome, dysregulation of miRNAs was still observed in WM, even after combining CD19+, CD138+ and CD19+CD138+ cells together as one group in an attempt to mimic the heterogeneity observed in WM tumors. Six miRNAs were differentially expressed in WM as compared with non-malignant B lineage cells, many of which have been mentioned previously for their involvement in hematopoiesis and lymphomagenesis, including miR-182, miR-21 and miR-142-3p.28, 33, 34 The downregulation of miR-152, which demonstrated 10.7-fold lower expression in WM, has been reported in other lymphocytic malignancies as well, often as a result of epigenetic silencing.35, 36 One potential gene target of miR-152 based on sequence complementarity is the Janus kinase family member, Jak1, whose corresponding protein product is highly involved in cytokine-mediated signaling.37 Intriguingly, Jak1 was one of the few proteins identified in a recent proteomics analysis as being overexpressed in WM.8 As Jak family proteins are frequently implicated in the development of hematological malignancies, it may be of interest in future studies to examine the relationship between the downregulation of miR-152 and the upregulation of Jak1 in WM.
In addition to non-malignant B lineage cells, the miRNA expression profile of the WM tumor clone was also compared with the profiles of CLL and MM cells. Both miR-125b and miR-181a were expressed at significantly higher levels in WM than in CLL (Table 2).38 During normal hematopoiesis, the expression of both of these miRNAs is much higher in germinal center lymphocytes than in memory B cells or plasma cells, and both miRNAs appear to be involved in preventing premature plasmacytic differentiation through the repression of specific transcription factors.18, 27, 39 Namely, functional studies have validated the binding of both miR-125b and miR-181a to the 3′-untranslated regions of PDRM1 and IRF4, with other predicted gene targets including IL6R, PAX5 and DUSP5 (Table 2).18, 27 Coexpression of PDRM1, which encodes for the protein B lymphocyte-induced maturation protein-1 (BLIMP-1) and IRF4 is essential for the post-GC differentiation of B cells into Ig-secreting plasma cells.40 Increases in BLIMP-1 and IRF4 produce reciprocal decreases in the expression of Bcl-6, which, in serving as a crucial regulator of the GC reaction, must be repressed for differentiation towards memory B cells and plasma cells to occur.41 Forced expression of miR-181a in activated primary B cells prevented the induction of PDRM1 and IRF4 and ultimately increased Bcl-6, which, in compromising plasmacytic differentiation, produced a substantial increase in CD19+ B cells and a decrease in the survival of cultured myeloma cells.14, 18, 27 Similarly, when introduced into a cultured B-cell line, miR-125b expression was associated with a decrease in IgM secretion of more than 70%, whereas having no effect on B-cell viability.27
Yet, consistent with previous reports, our analysis also detected the downregulation of miRNA-9* in WM cells as compared with CD138+ MM cells.19, 30 Curiously, miRNA-9* is also a validated repressor of PDRM1 expression,42 and downregulation of miRNA-9* would appear to counteract the effects of increased miR-125b and miR-181a. However, evidence suggests that disease states, such as lymphoma, are unlikely to be driven entirely by one dysregulated miRNA and that instead, it is subtle changes in the expression of multiple miRNAs with overlapping targets that control pathogenesis.43 Under this assumption, one could hypothesize that if the increased expression of miR-181a and miR-125b in WM was not sufficient to counterbalance the effects of downregulated miRNA-9*, a tumor cell might develop with the ability to hypersecrete IgM while maintaining a B-cell phenotype. With potentially hundreds of biologically active miRNAs interacting in this manner within neoplastic cells, it becomes difficult to elucidate the effects of a single, isolated, dysregulated miRNA on a disease state without fully appreciating the role of the miRNA within the entire transcriptome.
Another challenge in studying miRNA activity in WM is that unlike with other malignancies, we were not able to associate any of the dysregulated miRNAs in WM cells with previously described cytogenetic abnormalities identified in WM tumors.44, 45 Furthermore, all but one WM patient sample included in this analysis maintained a normal cytogenetic profile and the miRNA signature of the one patient with a reported 13q deletion was not significantly different than the other samples. This suggests that the dysregulation of miRNAs in WM may have a stronger extrinsic component, whereby soluble factors within the microenvironment regulate the expression of intratumoral miRNAs. Recent studies have defined a mechanism for cytokine-induced upregulation of mioRNAs occurring through activation of the Jak/Stat signaling pathway.46, 47, 48 Expression of these miRNAs, which include miR-let7a and miR-21, is linked to increased cell survival and is further potentiated through their own ability to heighten Stat phosphorylation by inducing downstream transcription factors. In myeloma cells, two Stat3 binding sites were detected on the upstream enhancer of miR-21. Addition of exogenous IL-6 led to activation of Stat3, an increase in miR-21 expression and a subsequent reduction in apoptosis.46 In WM, IL-6 and its soluble receptor have been identified as two of the most highly expressed genes, and serum levels of IL-6 are significantly higher in patients with WM than in healthy controls.6, 49 Interestingly, our analysis has confirmed the findings of Hunter et al.30 by also detecting significantly higher levels of miR-21 in WM cells as compared to non-malignant controls (Figure 3). This evidence may indicate the presence of an autocrine or paracrine mechanism by which high concentrations of IL-6 in the WM tumor microenvironment are responsible for the dysregulation of miR-21. However, the origins and ultimate effects of elevated miR-21 expression on WM tumor biology remain to be examined.
In conclusion, our genome-wide analysis has identified a unique set of differentially expressed miRNA in a heterogeneous sample of WM cells. Consistent with the results of earlier miRNA-profiling studies in WM B cells, many of the dysregulated miRNA in WM are also critical for normal hematopoiesis, suggesting that subtle changes in the expression of these regulatory miRNA may be sufficient to drive lymphomagenesis. Additionally, as the target genes for many of these miRNAs overlap, it seems unlikely that single miRNAs predict the behavior of a WM tumor, but instead, it is the intricate balance that exists between these miRNAs that controls the fate of a B lymphocyte. Future investigations into the effects of these dysregulated miRNAs, both individually and in combination, will provide further insight into the mechanisms responsible for the pathogenesis of WM.
Acknowledgments
This work is supported in part by grants from the Jensen Foundation, the International Waldenstrom's Macroglobulinemia Foundation (IWMF) and the Predolin Foundation.
The authors declare no conflict of interest.
References
- Owen RG, Treon SP, Al-Katib A, Fonseca R, Greipp PR, McMaster ML, et al. Clinicopathological definition of Waldenstrom's macroglobulinemia: consensus panel recommendations from the Second International Workshop on Waldenstrom's macroglobulinemia. Semin Oncol. 2003;30:110–115. doi: 10.1053/sonc.2003.50082. [DOI] [PubMed] [Google Scholar]
- Remstein ED, Hanson CA, Kyle RA, Hodnefield JM, Kurtin PJ. Despite apparent morphologic and immunophenotypic heterogeneity, Waldenstrom's macroglobulinemia is consistently composed of cells along a morphologic continuum of small lymphocytes, plasmacytoid lymphocytes, and plasma cells. Semin Oncol. 2003;30:182–186. doi: 10.1053/sonc.2003.50073. [DOI] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Bener A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. NEJM. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Liebisch P, Dohner H. Cyogenetics and molecular cytogenetics in multiple myeloma. Eur J Cancer. 2006;42:1520–1529. doi: 10.1016/j.ejca.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Schop RFJ, Jalal SM, Van Wier SA, Ahmann GJ, Bailey RJ, Kyle RA, et al. Deletions of 17p13.1 and 13q14 are uncommon in Waldenstrom macroglobulinemia clonal cells and mostly seen at the time of disease progression. Cancer Genet Cytogenet. 2002;132:55–60. doi: 10.1016/s0165-4608(01)00526-x. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Schop RF, Price-Troska T, Ghobrial I, Kay N, Jelinek DF, et al. Gene-expression profiling of Waldenstrom macroglobulinemia reveals a phenotype more similar to chronic lymphocytic leukemia than multiple myeloma. Blood. 2006;108:2755–2763. doi: 10.1182/blood-2006-02-005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez NC, Ocio EM, de las Rivas J, Maiso P, Delgado M, Ferminan E, et al. Gene expression profiling of B lymphocytes and plasma cells from Waldenstrom's macroglobulinemia: comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma, and normal individuals. Leukemia. 2007;21:541–549. doi: 10.1038/sj.leu.2404520. [DOI] [PubMed] [Google Scholar]
- Hatjiharissi E, Ngo H, Leontovich AA, Leleu X, Timm M, Melhem M, et al. Proteomic analysis of Waldenstrom macroglobulinemia. Cancer Res. 2007;67:3777–3784. doi: 10.1158/0008-5472.CAN-06-3089. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucl Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Bio. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Saunder NJ, Soneji S, Palazzo S, Dunlop HM, Cooper CDO, et al. MicroRNA expression in lymphocyte development and malignancy. Leukemia. 2008;22:1440–1446. doi: 10.1038/sj.leu.2405083. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. OncomiRs: microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. PNAS. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccaro AM, Sacco A, Chen C, Runnels J, Leleu X, Azab F, et al. MicroRNA expression in the biology, prognosis, and therapy of Waldenstrom macroglobulinemia. Blood. 2009;30:4391–4402. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarry data. Stat Appl Genet Mol Biol. 2003;2:3. doi: 10.2202/1544-6115.1008. [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2008;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The mircoRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschi T, Sander C, DS M. Human microRNA targets. PLoS Biol. 2005;3:e264. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Wang M, Tan LP, Dijkstra MK, van Lom K, Robertus JL, Harms G, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-159 and high BIC/mIR-155 expression. J Pathol. 2008;215:13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- Stone MJ, Pascual V. Pathophysiology of Waldenstrom's macroglobulinemia. Haematologica. 2010;95:359–364. doi: 10.3324/haematol.2009.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–91. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Gibson WSJ, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematoipoietic output. PNAS. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter Z, Cao Y, Lewicki M, Sun J, Tseng H, Hanzis C, et al. Aberrant expression of regulatory miRNAs and transcripts for IRS-PI3K growth and survival signaling in Waldenstrom's macroglobulinemia American Society of HematologyOrlando, FL, 2010
- Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. PNAS. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- Stumpel DJPM, Schotte D, Lange-Turenhout EAM, Schneider P, Seslija L, de Menezes RX, et al. Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: major matters at a micro scale. Leukemia. 2010;25:429–439. doi: 10.1038/leu.2010.282. [DOI] [PubMed] [Google Scholar]
- Ballabio E, Mitchell T, van Kester MS, Taylor S, HM D, Chi J, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood. 2010;116:1105–1113. doi: 10.1182/blood-2009-12-256719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in respnose to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Lwin T, Lin J, Choi YS, Zhang X, Moscinski LC, Wright KL, et al. Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood. 2010;116:5228–5236. doi: 10.1182/blood-2010-03-275925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with blimp-1 expression in vivo. J Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. JEM. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LHC, et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131–140. [PMC free article] [PubMed] [Google Scholar]
- Kern W, Haferlach T, Schnittiger S, Haferlach C.Cytogenetic aberrations in lymphoplasmacytic lymphoma (LPL): a study of 166 cases American Society for HematologyOrlando, FL2010
- Braggio E, Keats JJ, Leleu X, Van Wier S, Jimenez-Zepeda VH, Valdez R, et al. Identification of copy number abnormalities and inactivating mutations in two negative regulators of nuclear factor-kB signaling pathways in Waldenstrom's macroglobulinemia. Cancer Res. 2009;69:3579–3588. doi: 10.1158/0008-5472.CAN-08-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6-dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The microRNA let-7a modulates interleukin-6-dependent Stat3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256–8264. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, et al. MicroRNA-21 expression in CD4+ T cells is regulated by Stat3 and is pathologically involved in Sezary Syndrome. J Invest Dermatol. 2011;131:762–768. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- Hatzmichael EC, Christou L, Bai M, Kolios G, Kefala L, Bourantas KL. Serum levels of IL-6 and its soluble receptor (sIL-6R) in Waldenstrom's macroglobulinemia. Eur J Haematol. 2001;66:1–6. doi: 10.1034/j.1600-0609.2001.00152.x. [DOI] [PubMed] [Google Scholar]
- Onnis A, De Falco G, Antonicelli G, Onorati M, Bellan C, Sherman O, et al. Alteration of microRNAs regulated by c-Myc in Burkitt lymphoma. PLoS One. 2010;5:12960. doi: 10.1371/journal.pone.0012960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Tan LP, Harms G, Schakel RN, de Jong D, Blokzijl T, et al. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–176. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- Pons A, Nomdedeu B, Navarro A, Gaya A, Gel B, Diaz T, et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50:1854–1859. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, MIn H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Di Lisio L, Gomez-Lopez G, BSanchez-Beato M, Gomez-Abad C, Rodriguez ME, Villuendas R, et al. Mantle cell lymphoma: transcriptional regulation by microRNAs. Leukemia. 2010;24:1335–1342. doi: 10.1038/leu.2010.91. [DOI] [PubMed] [Google Scholar]
- Stittrich AB, Haftmann C, Hegazy A, Floessdorf M, Dong J, Fuhrmann F, et al. MicroRNA-182 promotes clonal expansion of activated T helper cells. Ann Rheum Dis. 2010;69:A65. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- Unno K, Zhou Y, Zimmerman T, Platanias LC, Wickrema A. Identification of a novel microRNA cluster miR-193b-365 in multiple myeloma. Leuk Lymphoma. 2009;50:1865–1871. doi: 10.3109/10428190903221010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, et al. microRNA-29c and imcroRNA0223 downregulation has in vivo significance in chronic lymphocytic leukemia and improve disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- Li C, Kim SW, Rai D, Bolla AR, Adhvaryu S, Kinney MC, et al. Copy number abnormalities, MYC activity, and the genetic fingerprint of normal B cells mechanistically define the microRNA profile of diffuse large B-cell lymphoma. Blood. 2009;113:6681–6690. doi: 10.1182/blood-2009-01-202028. [DOI] [PMC free article] [PubMed] [Google Scholar]