Abstract
Maintaining genome integrity in germ cells is important, given that the germ cells produce the next generation of offspring. Base excision repair is a DNA repair pathway that is responsible for the repair of most endogenous DNA damage. A key enzyme that functions in this repair pathway is DNA polymerase beta (Pol β). We previously used conditional gene targeting to engineer mice with sperm deleted of the Pol B gene, which encodes Pol β. We characterized mutagenesis in the sperm of these mice and compared it to wild-type and mice heterozygous for the Pol B gene. We found that sperm obtained that were heterozygously or homozygously deleted of the Pol B gene exhibited increased mutation frequencies compared to wild-type sperm. We identified an increase in transition mutations in both heterozygously and homozygously deleted sperm, and the types of mutations induced suggest that a polymerase other than Pol β functions in its absence. Interestingly, most of the transversions we observed were induced only in heterozygous, compared with wild-type sperm. Our results suggest that haploinsufficiency of Pol β leads to increased frequencies and varieties of mutation. Our study also shows that Pol β is critical for genome stability in the germline.
Keywords: DNA Polymerase Beta, Mutagenesis, Sperm Cells
1. Introduction
Germ line genomic stability is an important factor in reproductive health, with approximately 20% of genetic diseases attributed to new germ line mutations [1]. Maintenance of germ cell genetic integrity is fundamental to the development of healthy offspring. However, DNA is constantly exposed to endogenous sources of damage. If left unrepaired, DNA damage can result in errors during replication, and lead to the generation of de novo germ line mutations. Such mutations can result in genetic diseases. It has been suggested that most spontaneous germ line mutations occur in male gametes more often than female gametes [2].
Expression of Pol β is highest in the testes compared to other organs, and Pol β is expressed in zygotene and pachytene spermatocytes [3,4]. Pol β is a 39 kDa enzyme that has both polymerase and dRP lyase activities and is known to function in base excision repair [5]. The DNA base excision repair pathway is responsible for the repair of spontaneous base damage caused by oxidation/alkylation and protects cells against the effects of endogenous and exogenous agents [6]. BER is initiated by a damage-specific DNA glycosylase that removes a modified base; removal of the damaged base creates a baseless (AP) site. AP endonuclease (APE1) nicks the DNA backbone, usually leaving a 5′ deoxyribose-5-phosphate (dRP) residue and a single-strand break (SSB) [7]. Bifunctional glycosylases, which recognize oxidative lesions, generate an abasic site and then catalyze its removal via β-elimination to generate a 3′dRP and 5′phosphate. APE1 then catalyzes removal of the 3′dRP, leaving a 3′OH, to which Pol β can bind and fill in the resulting single nucleotide gap. In both cases, the XRCC1/DNA ligase IIIα complex catalyzes ligation of the resulting ends [8]. In the short-patch pathway, Pol β inserts a single nucleotide into the gap and removes the dRP group using its dRP lyase activity. The gap is sealed by DNA ligase I or a complex of X-ray Cross-Complementing 1 (XRCC1)/DNA ligase IIIα [9] [10] [11]. In the long-patch pathway, DNA polymerase δ/ε along with Proliferating Cell Nuclear Antigen (PCNA) adds 2–10 nt into the gap by displacing the dRP residue as part of an oligonucleotide flap; this flap is removed by flap endonuclease I, followed by sealing the gap with DNA ligase I [12]. Pol β is essential for long patch repair and is suggested to add the first base [13].
It has been suggested that BER proteins must be tightly regulated because imbalances in BER lead to genomic instability [14]. Pol β is essential for embryonic development, and thus Pol β null mice die immediately after birth [15,16]. The impact of altered Pol β levels on mutagenesis has been studied in cultured cells, Pol β heterozygous mice and Pol β null mouse embryonic fibroblast cells [17,18]. However, the relationship between a complete Pol β deficiency and in vivo mutagenesis during sperm development remains unexplored. In this study, we characterized mutagenesis in sperm cells obtained from wild type, Pol β deficient and heterozygous mice to determine the role of Pol β in sperm cell genomic stability. Because deletion of the PolB gene from mice results in death of all newborn mice shortly after birth [15], we used a Cre-loxP conditional gene targeting approach that limited deletion of PolB to germ cells. Briefly, mice carrying a floxed allele of the PolB gene [19] were crossed to mice in which the Cre+ gene was expressed from the tissue non-specific alkaline phosphatase promoter (TNAP), resulting in deletion of part of the promoter and exons 1 and 2 in primordial germ cells. These cells give rise to spermatocytes and oocytes. Using this mouse model, we recently showed that Pol β is critical for synapsis during meiosis in the mouse [20]. During Prophase I of meiosis, double-strand breaks (DSBs) are introduced by the Spo11 complex, and Spo11 remains associated with the 5′ end of the break [21]. Removal of Spo11 and subsequent 5′ resection are compromised in Pol β-deficient germ cells [20]. Although the use of the conditional gene targeting system did not result in complete deletion of the PolB gene during spermatogenesis, it did lead to complete deletion of PolB in sperm [20].
Our results indicate that the mutation frequency is significantly higher in Pol β deficient (Cre+PolB flox/Δ) and Pol β heterozygous (Cre−PolB flox/Δ) sperm cells compared to Pol β proficient sperm cells. Importantly, specific types of transversions occur in the Pol B heterozygous sperm at higher frequencies than what is observed in the WT or homozygous sperm, suggesting that suboptimal levels of Pol β in sperm contribute to genomic instability. Therefore, our work points to a critical role of Pol β in maintaining genomic stability.
2. Materials and methods
2.1 Chemicals
[γ-32P]ATP (>6,000 Ci/mmol, 150 mCi/mL) was purchased from Perkin-Elmer. Uracil DNA glycosylase (UDG; M0280S), human AP endonuclease I (APE1; M0282S) and T4 polynucleotide kinase (M0201S) were purchased from New England Biolabs.
2.2 Mouse breeding and genotyping
CD1 mice (AVF81) that contain the lambda cII transgene were crossed with C57Bl/6 PolB flox/flox mice to generate the F1 generation and genotyped for the cII PolB flox/+ markers. The cII PolB flox/+ mice were backcrossed with C57B6flox/flox to obtain mice with cII transgene and that have the PolBflox/flox genotype. These mice were then mated with TNAP-Cre mice, in which the Cre transgene is expressed from the tissue nonspecific alkaline phosphatase promoter (TNAP). Tail genotyping was performed on DNA isolated from small tail clippings using a DNA isolation kit (Viagen). For the cII gene, the following set of primers (5′-ACC ACA CCT ATG GTG TAT GCA-3′ and 5′-GTC ATA ATG ACT CCT GTT GA-3′) were used to obtain the genotype. To determine the Cre and PolB status, DNA was amplified using the polymerase chain reaction (PCR) with allele-specific primers as described previously [21] We denote wild-type sperm cells as WT, Pol β heterozygous sperm cells as Cre−PolBflox/Δ and Pol β deleted sperm cells as Cre+PolBflox/Δ. In our earlier publication [21] we showed that 94% of offspring of Cre+PolBflox/Δ X WT mice carried the PolB deleted allele, suggesting that almost all sperm derived from these mice carried the deleted allele.
2.3 Isolation of Sperm DNA
Male mice at the age of 8 weeks were euthanized and the sperm DNA was isolated [22]. Briefly, the sperm were gently squeezed out of both Cauda epididymides and pooled together to obtain enough sperm cells in 1ml dounce buffer (12mM Na2HPO4, 136mM NaCl, 2.6mM KCl, 1.5mM KH2PO4, 0.5M EDTA). Sperm DNA was isolated by incubating the released sperm with 100 mg/ml RNase at 37°C for 1 hour and subsequently adding 1 ml Dounce Buffer with 2% SDS, 100 mM EDTA, pH 7.5 and 2 mg/ml of proteinase K and incubating at 50°C for 2 hours. The DNA was extracted once using phenol/chloroform/isoamyl alcohol followed by a single chloroform extraction. The DNA was then precipitated using 3mM Sodium Acetate and 100% ethanol and washed with 70% ethanol and dried. Each DNA sample was resuspended in 30μl Tris-EDTA buffer (pH7.5) and stored at 4°C until packaged.
2.4 λcII Mutagenesis Assay
High molecular weight genomic DNA was purified as described previously [22]. Packaging extracts were prepared as described previously [23,24]. The Escherichia. coli K12 strain, NM759 with the genotype recA56 (mcrA) e14° λ (mrr-hsd-mcr)(λ imm434 cIts b2 red3 Dam15 Sam7/) was used for the preparation of the sonication extract [25] and BHB2688 [N205 recA−,(λimm434 clts, b2, red3, Eam4, Sam7)/λ] was used for the preparation of the freeze-thaw extracts. Phage packaging was carried out as described [24]. Lambda cII mutants were obtained from three independent packaging reactions. The number of plaques obtained from packaged DNA of Cre+PolBflox/Δ, Cre−PolBflox/Δ and wild-type mice (WT) were counted and mutation frequency was calculated by taking the ratio of number of plaques arising at 24°C (cII mutants) divided by the number of plaques at 37°C (total). To identify the types of mutations produced in the sperm, each cII mutant plaque was isolated, and the cII gene was amplified by PCR with the following primers: 5′-ACC ACA CCT ATG GTG TAT GCA-3 and 5′-GTC ATA ATG ACT CCT GTT GA-3′. After DNA purification, the DNA sequences of these mutants were determined by the Keck Center for Biotechnology at Yale School of Medicine.
2.5 Statistical analysis
All data were collected and recorded in an Excel spreadsheet and data were analyzed using the GraphPad Prism (GraphPad Software, Inc.) program to quantify the mutation frequency differences between different genotypes.
2.6 Preparation of cellular extracts from sperm
Eight week-old mice of each genotype (Wild type, Cre−PolBflox/Δ, Cre+PolB flox/Δ), were euthanized, the cauda epididymides was collected, and sperm were squeezed into 1xPBS. Sperm extracts were prepared based on a modified version of the protocol [26]. Briefly, the cell suspensions were filtered through 70μm and 40μm nylon filters and the cells were centrifuged for 15 min at 3000rpm. The cells were suspended in 20mM HEPES, pH 7.9, 10mM NaCl, 1.5mM MgCl2, 10% glycerol 1mM PMSF and a protease inhibitor cocktail and incubated for 1hr on a rocking platform at 4°C. The whole cell lysate was collected after centrifugation at 13,000rpm at 4°C for 20 minutes.
2.7 Preparation of DNA substrate for base excision repair
Oligonucleotides were synthesized by the WM Keck facility at Yale University. The substrates used are shown in Table 4. The primer oligonucleotide (45AG substrate) and LPSD template which has a single U at position 19 were labeled at the 5′-end with T4 polynucleotide kinase and [γ-32P]ATP. The downstream oligonucleotide of the 45AG substrate was 5′-end phosphorylated with the kinase and cold ATP. After purification on a Bio-Rad spin column to remove unincorporated deoxynucleotide triphosphates, annealing was performed by mixing phosphorylated template, radiolabeled primer, and phosphorylated downstream oligos in 50 mM Tris-HCl (pH 8.0) containing 0.25 mM NaCl. The mixture was incubated sequentially at 95°C (5 min), slowly cooled to 50°C (for 30 min) and 50°C (for 20 min), and immediately transferred to ice. To verify proper hybridization, the product was analyzed on an 18% native polyacrylamide gel followed by autoradiography to assess the quality of annealing.
Table 4.
DNA substrates employed in primer extension and base excision repair assays.
| Substrate | Sequence |
|---|---|
| 45AG | 5′ GCCTCGCAGCCGTCCAACCAAC CAACCTCGATCCAATGCCGTCC 3′ 3′ CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG 5′ |
| LPSD | 5′ CTGCAGCTGATGCGCUGTACGGATCCCCGGGTAC 3′ 3′ GACGTCGACTACGCGGCATGCCTAGGGGCCCATG 5 |
The template base is bold and underlined.
2.8 In vitro polymerase assay
A primer extension experiment was conducted in a solution containing 50 mM Tris-HCl buffer (pH 8.0), 5 mM MgCl2, 2 mM DTT, 20 mM NaCl, 10% glycerol, and 25 μM dTTP. Sperm extract was mixed with 1-bp–gapped DNA (30ug/5nM), was incubated with dTTP-MgCl2 and reactions were carried out at 37°C for 5 to 20 min, after which they were stopped by addition of an equal volume of 90% formamide dye and 0.3 M EDTA. Samples were resolved by electrophoresis on 20% polyacrylamide gels containing 8 mol/L urea and then visualized and quantified using a Storm 860 PhosphorImager (Molecular Dynamics, Inc.).
2.9 Base excision repair assay
5′-End–labeled LPSD substrate was used as a BER substrate. For reconstituted BER with purified proteins, we followed the method as described by Haracska and colleagues [27] with some minor modifications. 5 nM UDG–treated substrate was incubated for 20 min with commercially available APE1 as a control for BER reaction. Approximately 5 nM UDG–treated substrate was incubated with 10, 20 and 45ug of sperm extract in BER buffer [45 mM HEPES (pH 7.8), 70 mM KCl, 2 mM DTT, 7.5 mM MgCl2, 0.5 mM EDTA, 2 mM ATP, and 20 μM dCTP for 20 min at 37°C.
To determine if the polymerase activity observed in our BER assay is from Pol β, Lithocholic acid (LCA), a specific inhibitor of this polymerase was used during the assay. Typically, 300uM LCA was mixed with 20ug extract. We also supplemented the Cre+ extract with purified Pol β as a control for LCA inhibition specificity. EDTA containing formamide dye was added to stop the reaction. The repaired product was resolved on a 20% denaturing polyacrylamide gel followed by visualization on a Storm 860 PhosphorImager.
3. Results
3.1 Mutation frequency in DNA polymerase beta deficient sperm
DNA lesions and the absence of DNA repair functions can result in mutations and/or unique mutation spectra in DNA isolated from sperm cells. To evaluate these possibilities in wild type, heterozygous and Pol β null sperm cells, mutation frequencies using the cII gene as a target were quantified, and independent mutants were selected and characterized by DNA sequencing. The frequencies of mutations observed in these spontaneous mutant spectra are summarized in Table 1. The mutation frequency of Pol β deficient (mean± SEM; 3.9×10−5± 0.5) and Pol β heterozygous sperm (5.9×10−5 ±1.5) was ~3 and 4 fold higher than that of WT (1.4×10−5 ±0.3) respectively. The increase in mutation frequency between WT and Pol β deficient sperm was significant (P<0.005). In addition, we have observed a significant increase of the mutation frequency in sperm derived from heterozygous mice as compared with WT (P<0.01). In contrast, there is no statistically significant difference in mutation frequencies between sperm derived from the heterozygous as compared to Pol β deficient mice (P>0.74).
Table 1. Mutation frequency is increased in Pol β deficient sperm cells.
A statistically significant difference was observed between WT and Cre+PolBflox/Δ and WT versus Cre−PolBflox/Δ. In contrast no statistical significance difference was observed between Cre−PolB flox/Δ and Cre+PolBflox/Δ sperm cells, using the Mann-Whitney test.
| Genotypes | Mutation Frequency | Mean±SEM | 95%CI |
|---|---|---|---|
| WT (n=6) | 1.7x10−5 | 1.4x10−5± 0.29 | 0.8<CI<2.29 |
| 1.8x10−5 | |||
| 0.7x10−5 | |||
| 2.4x10−5 | |||
| 1.9x10−5 | |||
| 0.7x10−5 | |||
|
| |||
| Cre−PolB flox/Δ(n=11) | 2.0x10−5 | 5.9 x10−5± 1.45 | 2.68<CI<9.17 |
| 15.6x10−5 | |||
| 13.2x10−5 | |||
| 4.7x10−5 | |||
| 1.8x10−5 | |||
| 8.9x10−5 | |||
| 7.4x10−5 | |||
| 1.6x10−5 | |||
| 4.1x10−5 | |||
| 2.4x10−5 | |||
| 3.4x10−5 | |||
| P < 0.01 vs. WT | |||
|
| |||
| Cre+PolB flox/Δ(n=11) | 4.3X10−5 | 3.9 x10−5± 0.52 | 2.73<CI<5.02 |
| 6.5x10−5 | |||
| 4.9x10−5 | |||
| 6.3x10−5 | |||
| 5.4x10−5 | |||
| 2.7x10−5 | |||
| 2.6x10−5 | |||
| 3.8x10−5 | |||
| 1.6x10−5 | |||
| 2.5x10−5 | |||
| 2.1x10−5 | |||
| P < 0.005 vs. WT; P > 0.74 vs. Cre−PolBflox/Δ | |||
3.2 Types and Frequencies of Base Substitutions
The mutation pattern reflects the balance of DNA damage and repair for nine types of base substitutions plus small and large deletions and insertions. The mutation spectra for WT, heterozygous, and Pol β deleted sperm cells are shown in Figure 1. The cII mutation frequencies for the types of base substitutions analyzed from sperm DNA of Pol β proficient, deficient and heterozygous mice are presented in Table 2. We detected a significant increase of transition mutations, including both GC to AT and AT to GC, in Pol β deficient and heterozygous sperm cells in comparison to WT sperm cells (Table 2). The frequencies of GC to CG and AT to CG transversions were increased in sperm from both the Cre− and Cre+ PolBflox/Δ mice in comparison to WT. However, the frequencies of GC to TA and AT to TA transversions were increased in the Cre− sperm cells in comparison to WT (4- and 7- fold, respectively) and Cre+ sperm cells (7- and 6- fold, respectively).. The frequency of single base frameshifts was similar in all groups of mice (Table 3).
Figure 1. Mutation Spectra of sperm generated from WT, Cre−PolBflox/Δ and Cre+PolBflox/Δ mice.
The DNA sequence is that of the lambda cII gene. The mutations induced in the WT, Cre−PolBflox/Δ, and Cre+PolBflox/Δ mice are shown in red, blue, and green, respectively. Mutations induced by WT are shown below the cII DNA sequence. Numbers next to specific mutations, for example, 3x, denote the numbers of that specific mutation at that site in the cII gene. Base insertions are denoted by (◆) and deletions by (Δ).
Table 2. Mutation frequency and types of base substitutions in Pol β deficient and proficient sperm cells.
Types of mutations are shown in the first column. Total numbers of mutants (Total no mut) and mutation frequencies (MF) are shown for each type of mutation for each genotype. The columns at the right compare each of the genotypes with WT and with each other on a mutation frequency basis. The frequency of GC to AT transitions is significantly increased in Cre−PolBflox/Δ and Cre+PolBflox/Δ as compared to WT. However the frequencies of GC to TA and AT to TA transversions were increased only in the Cre-sperm in comparison to WT.
| Mutation type | Type | Wild type | Cre−PolBflox/Δ | Cre+PolBflox/Δ | M.F ratio b/n genotypes | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total No. mut=90(%) | M.F (1x10−7) | Total No. mut =79(%) | M.F (1x10−7) | Total No. mut=50(%) | M.F (1x10−7) |
Cre−PolBflox/Δ |
Cre+PolBflox/Δ |
Cre−PolB flox/Δ |
||
| WT | WT | Cre+PolB flox/Δ | ||||||||
| G:C to A:T | transition | 24(27) | 37 | 34(43) | 260* | 26(52) | 200** | 7 | 5.4 | 1.3 |
| A:T to G:C | transition | 2(2) | 3.1 | 3(3.8) | 23 | 3(6) | 23 | 7.6 | 7.6 | 1 |
| G:C to T:A | transversion | 8(8.9) | 12 | 7(8.8) | 53 | 1(2) | 7.8 | 4.4 | 0.7 | 6.8 |
| G:C to C:G | transversion | 5(5.6) | 7.8 | 2(2.5) | 15 | 2(4) | 16 | 1.9 | 2 | 0.9 |
| A:T to T:A | transversion | 4(4.4) | 6.2 | 6(7.6) | 45 | 1(2) | 7.8 | 7.3 | 1.3 | 5.8 |
| A:T to C:G | transversion | 3(3.3) | 4.7 | 10(12.7) | 75* | 3(6) | 23 | 16 | 4.9 | 3.3 |
Compared with wild type for each muation type
P<0.05;
P<0.01
Table 3. Mutation Frequency and types of frameshift mutations.
Types of mutations are shown in the first column. Total numbers of mutants (Total no mut) and mutation frequencies (MF) are shown for each type of mutation for each genotype. The columns at the right compare each of the genotypes with WT and with each other on a mutation frequency basis. The frequency of total transition mutations is significantly increased in Cre+PolBflox/Δ and Cre−PolBflox/Δ as compared to wild type sperm cells. There were no significant differences in the frameshift frequencies among the groups.
| Types of Muations | Total M.F (1x10−7) | Total M.F ratio b/n genotypes | ||||
|---|---|---|---|---|---|---|
| Wild type | Cre−PolBflox/Δ | Cre+PolBflox/Δ | Cre−PolBflox/Δ |
Cre+PolBflox/Δ |
Cre−PolBflox/Δ |
|
| WT | WT | Cre+PolBflox/Δ | ||||
| Transitions | 40 | 280* | 230** | 7 | 5.8 | 1.2 |
| Transversions | 31 | 190* | 55 | 6.1 | 1.8 | 3.4 |
| Single base | 72 | 470 | 280 | 6.5 | 3.9 | 1.7 |
| One base insertions | 32 | 68 | 86 | 2.1 | 2.7 | 0.8 |
| one base deletions | 34 | 60 | 23*** | 1.8 | 0.7 | 2.6 |
| Frameshits | 67 | 130 | 110 | 1.9 | 1.6 | 1.2 |
| Large deletions | 0 | 0 | 0 | 0 | 0 | 0 |
Total type of muations compared with wild type
P<0.05;
P<0.001; Total type of mutations compared with Cre−PolBflox/Δ
P<0.0001
3.3 In vitro polymerase beta activity in sperm cell extracts
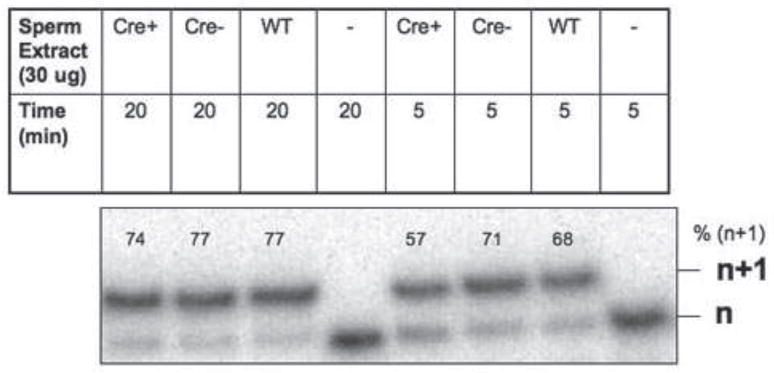
To determine if DNA polymerase activity was present in the protein extracts isolated from sperm, we performed an in vitro polymerase assay with mouse sperm extract and a single base pair gapped DNA substrate as described in Materials and Methods. As shown in Figure 2, Cre+PolB flox/Δ mice retain overall polymerase activity and the n+1 product formed with almost equal efficiency in extracts prepared from mice with all three genotypes. This is not surprising because the sperm extracts likely express polymerases other than Pol β.
Figure 2. Sperm extracts exhibit polymerase activity.
5 nM of 45AG DNA substrate was incubated with sperm extract as described in Materials and Methods. Formation of the n+1 product was analyzed via gel electrophoresis.
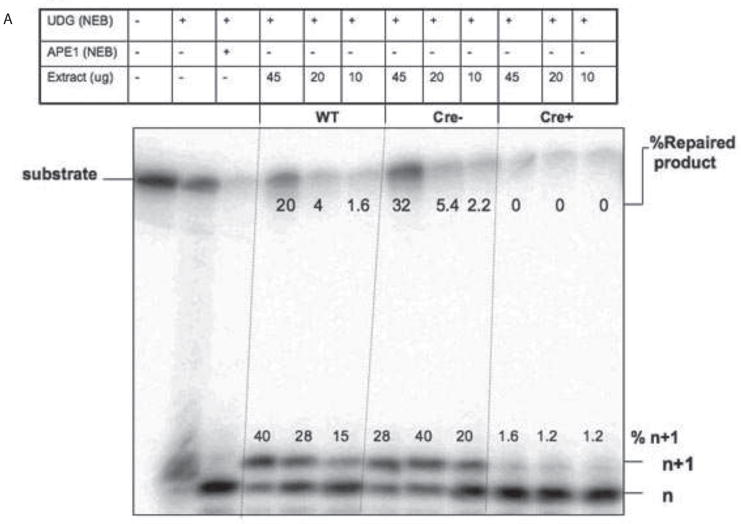
To determine if active Pol β was expressed in sperm cells from mice, we employed an in vitro BER assay, using nuclear extracts prepared from the sperm of animals of the various genotypes we have used in this study (Figure 3). We used an oligonucleotide with a uracil. This is based upon studies showing that UDG activity is higher than other glycosylase activity in testes [28] and also based on Illumina Gene Atlas database (http://biogps.gnf.org), which shows that UDG is more highly expressed than other DNA glycosylases in testis. Upon incubation with nuclear extracts, we could not detect a band that would correspond to excision of this base and incision by APE1 (data not shown). We also did observe a product of appropriate size after addition of UDG to the extracts. These results suggest that UDG and backup glycosylases such as SMUG1 are not present at concentrations high or active enough in our extracts to catalyze excision of the uracil. This is not surprising since a previous report suggested that there is low abundance of UDG in male germ cell nuclear extracts [35], even though it is more abundant than other DNA glycosylases. After treatment of the uracil-containing DNA substrate with UDG, nuclear extract from sperm was added. Endonuclease activity and single nucleotide gap-filling, as represented by the n+1 band, and completion of BER were supported by extracts prepared from WT and Pol B haploinsufficient sperm, but not from sperm isolated from the Cre+ Pol Bflox/Δ mice. In addition, WT and Pol B haploinsufficient sperm extracts exhibit completion of BER, whereas extracts from Cre+ Pol Bflox/Δ mice do not support a fully repaired product likely due to the absence of Pol β. If we supplement with purified Pol β, polymerase activity and repair activity was restored to extracts prepared from Cre+ Pol Bflox/Δ mice (Figure 3B). These results suggest that active Pol β is not present in sperm isolated from Cre+ Pol Bflox/Δ mice.
Figure 3. Sperm extracts prepared from Cre+PolBflox/Δ mice do not exhibit Pol β activity.
A. 100 nM of LPSD DNA substrate was incubated with UDG (2 Units/pMol) for 10 min (lane 2) or with UDG (2 units/pMol) for 10 min and APE1 (2 units/pMol) (lane 3) for 20 min at 37°C. 5 nM of UDG-treated substrate was treated with increasing concentrations of extract prepared from either WT, Cre− PolBflox/Δ, and Cre+PolBflox/Δ mice. The enzymes added and their amounts are shown above the lanes of the gel. Formation of the n+1 product and repaired BER product was analyzed via gel electrophoresis. The n+1 and fully repaired product were quantified by phosphorimager analysis and are reported in the Figure. Note that there was no statistically significant difference between WT and Cre−PolBflox/Δ repaired BER product band intensity with three different concentrations of sperm extract (P>0.05).
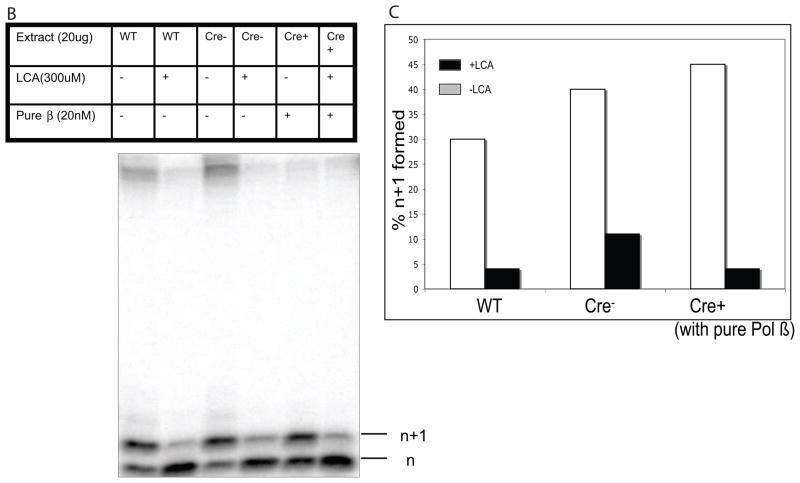
B. 5 nM of UDG treated LPSD substrate was incubated with 20 ug of extract of WT, Cre− and Cre+ mice in presence of 300 uM of lithocholic acid. In experiments with the or Cre + extract, 20 nM pure rat polymerase beta was added and then incubated in presence of 300 uM Lithocolic acid. The percentage of the (n+1) band is shown with LCA (black bar) and without LCA (grey bar).
C. Quantification of the product formed in B in the presence and absence of lithocholic acid (LCA).
To be certain that BER in extracts from WT and Cre− mice was supported by Pol β, we added LCA, a specific inhibitor of Pol β [29,30]. Addition of LCA to extracts from WT and Cre− mice resulted in inhibition of 90–95% of the activity. Thus, BER in these extracts is supported by Pol β. To be certain of the specificity of LCA in our assay, we also added LCA to Cre+ extracts supplemented with purified Pol β. Again, we observed inhibition of the BER activity, as shown in Figure 3B and quantified in Figure 3C.
4. Discussion
Germ line DNA directs the development of the next generation and, as such, is profoundly different from somatic cell DNA. We found that both partial and complete deletion of Pol β from sperm results in an increased mutation frequency. Transitions are increased in sperm that are either haploinsufficient or completely deleted of Pol β. Therefore, we conclude that Pol β protects sperm from transition mutations. Interestingly, most types of transversions were increased in Pol β haploinsufficient in comparison to sperm completely deleted of Pol β. This suggests that the presence of insufficient quantities of Pol β leads to increases in specific types of mutations that are less frequently observed in WT sperm.
4.1 Pol β maintains genome stability in germ cells
Wilson and colleagues used the lambda cII system to measure the mutation frequencies of immortalized WT mouse embryo fibroblasts (MEFs) and those derived from embryos deleted of the PolB gene [18]. These authors reported a lower than two-fold increase in mutation frequency between the Pol β deleted versus WT MEFs but showed that the frequency of transversions was increased in the Pol β nullizygous MEFs. Another study of mutagenesis showed no increase in mutation frequency of brain cells from Pol β knock-out mouse embryo in comparison to WT mouse embryo [16]. Two previous studies measured mutation frequencies in Pol β haploinsufficient mice. Cabelof and colleagues did not detect an increased mutation frequency in somatic tissues of Pol β haploinsufficient versus WT mice using the LacZ transgenic mouse system [31]. However, these authors demonstrated Pol β heterozygosity resulted in a 2.5-fold increase in hypoploid cells. Walters and colleagues have shown that heterozygosity of Pol β leads to a two-fold increase in mutation frequency at the lacI locus in a population of mixed male germ cells [32] but they did not generate mutation spectra to determine the types of mutations that were induced. The mutation frequency we observe in sperm that are heterozygous for Pol β is slightly increased over that obtained from a mixed population of male germ cells in the Walters study. The slight increase in mutation frequency we observe might be due to our use of sperm in comparison to a mixed population of germ cells. In combination, the mutagenesis studies of Pol β haploinsufficient and completely deleted cells suggest that Pol β is critically important in the maintenance of genomic stability in sperm.
4.2 Pol β protects against transitions
In sperm isolated from both the Cre+ and Cre− mice, we observed an increase in the frequency of transitions. The increase in the frequency of GC to AT and AT to GC transition mutations in comparison to WT is similar in sperm from the Cre+ and Cre− mice. The majority of transitions are GC to AT mutations that arise at CpG sequences, suggesting that they arose as a result of deamination of 5-methylcytosine to thymine. There are at least two ways that this event could give rise to the GC to AT transition. The first would be during replication, where dATP would be inserted opposite the T that arose via deamination of 5-methylcytosine. The fact that we observe an increase in GC to AT transitions in cells with either less Pol β than WT cells or no Pol β suggests that Pol β itself plays an active role in the prevention of this type of mutation. Thus, it is more likely that once the deamination event occurs, the T opposite template G is removed by either the thymidine or MBD4 DNA glycosylase, followed by incision by APE1, removal of the dRP group, and misincorporation of dTTP opposite template G by a polymerase other than Pol β. This polymerase could be DNA polymerase iota (Pol ι) given that it possesses dRP lyase activity [33] and is highly expressed in testes [34]. It is unlikely to be DNA polymerase lambda (Pol λ) because this polymerase induces predominantly I base frameshift mutations [33] and these types of mutations are not increased in Pol β haploinsufficient or deficient germ cells. It is theoretically possible that the haploinsufficiency or complete lack of Pol β could somehow result in favoring mutation fixation during DNA replication through an as yet unknown mechanism that might be active in germ cells.
There are few AT to GC transitions observed compared to the GC to AT mutations. The AT to GC transitions could have arisen during BER of an A lesion, perhaps ethenoadenine, by removal of the lesion and misincorporation of dGTP opposite template T by a polymerase other than Pol β. Pol iota is the best candidate not only because it possesses dRP lyase activity, but mainly because the most common error committed by this enzyme is misincorporation of dGTP opposite template T. In fact, Pol ι misinserts dG opposite template T at much higher frequencies than other DNA polymerases, including Pol β.
4.3 Transversions are increased in Pol β haploinsufficient cells
AT to CG transversions are increased in both Pol β haploinsufficient and deficient germ cells when compared to WT. However, GC to TA and AT to TA transversions are increased only in haploinsufficient cells in comparison to WT. Both of these types of transversions are observed at 6-fold higher frequencies in Pol β haploinsufficient versus deficient cells. This suggests that the presence of lower amounts of Pol β than what is normally present in WT cells leads to the induction of GC to TA and AT to TA transversions.
G to T transversions are known to arise from the inaccurate bypass of the 8-oxo-G adduct. Interestingly, even in the presence of RPA and PCNA, Pol β is able to insert dC or dA opposite 8-oxo-G at nearly equal frequencies [35], suggesting that the presence of Pol β itself in the Pol β haploinsufficient cells induces this mutation. This leads us to speculate that germ cells and perhaps other types of cells might be more prone to the induction of G to T transversions in the presence of abnormally low levels of Pol β by an unknown mechanism. AT to TA transversions are also increased in Pol β heterozygous cells 6-fold over what is observed in PolB deleted cells, suggesting that the presence of lower than normal levels of Pol β contributes to the induction of this mutation.
4.4 Mutagenesis is induced predominantly in cells that are not haploid with respect to Pol B
Mice that are completely deleted of the PolB gene die just after birth, likely due to massive apoptosis of postmitotic neurons [15]. In our case, we used a conditional gene targeting system to delete the PolB gene specifically in germ cells [20]. In our system, the Cre gene is expressed from the Tissue Non-specific Alkaline Phosphatase (TNAP) promoter in primordial germ cells, resulting in deletion of the PolB gene in these cells and all cells descended from them including spermatocytes [21]. This strategy resulted in a deletion index of ~80% in spermatocytes. The PolB-deleted spermatocytes do not complete meiosis due to defects in synapsis resulting from incomplete removal of the Spo11 complex from the DNA [20]. Pol β deleted spermatogonia also appear to die, perhaps by apoptosis, and the spermatogonia become enriched for Cre+PolBflox/Δ cells as the mice age. These cells were able to progress through meiosis due to the presence of one allele of the PolB gene, resulting in fertility of the Cre+PolBflox/Δ mice. Importantly, we showed that in Cre+ mice, a second Cre-mediated deletion of the remaining PolB floxed allele occurs some time after meiosis is completed because we were unable to detect the floxed allele of the PolB gene in spermatids [20]. In agreement with our results, Griswold and colleagues have used transcription profiling to show that the TNAP gene is not only expressed in primordial germ cells, but also in secondary spermatocytes and round spermatids [36]. Thus, the Cre protein would also be expressed in these cells and result in deletion of the remaining PolB floxed allele in these cells.
Base excision repair activity was observed at premeiotic, meiotic, and postmeiotic spermatogenic cell types and modestly elevated in pachytene spermatocytes and round spermatids [37]. Mutations observed in sperm can be induced at any point during the process of germ cell development, in spermatogonia, spermatocytes, spermatids, or the sperm themselves. In the case of the conditional gene targeting approach we have used, none of the cells that are completely deleted of the PolB gene progress through meiosis but undergo apoptosis instead [20], so the mutations we observe in the Pol BΔ/Δ sperm could arise from germ cells in the Cre+ mice that are PolBflox/Δ or in sperm that are completely deleted of the PolB gene. If the mutations arise in the PolBflox/Δ germ cells, we would expect to observe similar types of mutations in sperm obtained from the Cre+ versus Cre− mice. Although the mutation frequencies are similar to each other, the sperm isolated from the Cre−PolBflox/Δ mice, which retain heterozygosity at the PolB locus, carry specific types of transversions at higher frequencies than what was observed in the sperm isolated from the Cre+PolBflox/Δ mice, which became completely nullizygous at the PolB locus. Therefore, the simplest explanation of these results is that the presence of abnormally low abundance of Pol β in germ cells results in the observed transversions. It follows that these specific mutations are unlikely to have arisen in haploid germ cells, i.e spermatids, but must have arisen in either tetraploid (primary spermatocytes) or diploid (secondary spermatocytes) cells. Haploid germ cells carrying the floxed allele of the PolB gene are likely equivalent to haploid Pol β WT germ cells with regard to their phenotypic properties and it is thus unlikely that the transversion mutations arose in these cells. This is in agreement with extensive results that show that DNA repair is very low in haploid germ cells [38]. A caveat to this explanation is that the turnover of the Pol β protein may be slow in germ cells such that when cell division takes place, some active Pol β protein may be present in the haploid cells that received the deleted allele of the PolB gene, essentially rendering the cell “haploinsufficient” for Pol β protein. The results of our assay showing that sperm extracts prepared from Cre+ mice cannot support BER argue against this idea. Alternatively, it is possible that the floxed allele of the PolB gene confers properties to Pol β that the WT enzyme does not possess and that these properties lead to the induction of specific types of transversion mutations. We do not favor this explanation given that previous studies [18] have not yielded results consistent with this explanation.
Most important, our studies clearly demonstrate that haploinsufficiency of Pol beta results in spontaneous mutagenesis. These results also suggest that in its complete absence, other polymerases may functionally substitute for Pol β, resulting in a mutation pattern different from that found in wild type control mice.
5. Conclusions
In conclusion, we have shown that Pol β is important for maintaining a low spontaneous mutation frequency in the male germ line. Our results suggest that haploinsufficiency of Pol β in other types of cells may lead to an increase in mutational variety due to the inability of functional substitution by other DNA polymerases. Although the Pol B heterozygotes are fertile, it is possible that there is an increase of mutations in their offspring that could have wide ranging consequences.
Acknowledgments
This work was supported by CA116753 to JBS. JBS is a Donaghue Investigator.
Footnotes
Conflict of interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crow JF. The high spontaneous mutation rate: is it a health risk? Proc Natl Acad Sci U S A. 1997;94:8380–8386. doi: 10.1073/pnas.94.16.8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 3.Hirose F, Hotta Y, Yamaguchi M, Matsukage A. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocyte. Exp Cell Res. 1989;181:169–180. doi: 10.1016/0014-4827(89)90191-2. [DOI] [PubMed] [Google Scholar]
- 4.Alcivar AA, Hake LE, Hecht NB. DNA polymerase-beta and poly(ADP)ribose polymerase mRNAs are differentially expressed during the development of male germinal cells. Biol Reprod. 1992;46:201–207. doi: 10.1095/biolreprod46.2.201. [DOI] [PubMed] [Google Scholar]
- 5.Sobol RW, Wilson SH. Mammalian DNA beta-polymerase in base excision repair of alkylation damage. Prog Nucleic Acid Res Mol Biol. 2001;68:57–74. doi: 10.1016/s0079-6603(01)68090-5. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T, Karran P, Wood RD. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 7.Piersen CE, McCullough AK, Lloyd RS. AP lyases and dRPases: commonality of mechanism. Mutat Res. 2000;459:43–53. doi: 10.1016/s0921-8777(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 8.Tomkinson AE, Chen L, Dong Z, Leppard JB, Levin DS, Mackey ZB, Motycka TA. Completion of base excision repair by mammalian DNA ligases. Prog Nucleic Acid Res Mol Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- 9.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DPA. Abbondandolo and E. Dogliotti Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 10.Tanabe K, Bohn EW, Wilson SH. Steady-state kinetics of mouse DNA polymerase beta. Biochemistry. 1979;18:3401–3406. doi: 10.1021/bi00582a029. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Widen SG, Williams KR, Kedar P, Karpel RL, Wilson SH. Studies of the domain structure of mammalian DNA polymerase beta. Identification of a discrete template binding domain. J Biol Chem. 1990;265:2124–2131. [PubMed] [Google Scholar]
- 12.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 13.Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J Biol Chem. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- 14.Chan KK, Zhang QM, Dianov GL. Base excision repair fidelity in normal and cancer cells. Mutagenesis. 2006;21:173–178. doi: 10.1093/mutage/gel020. [DOI] [PubMed] [Google Scholar]
- 15.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. Embo J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niimi N, Sugo N, Aratani Y, Gondo Y, Katsuki M, Koyama H. Decreased mutant frequency in embryonic brain of DNA polymerase beta null mice. Mutagenesis. 2006;21:55–59. doi: 10.1093/mutage/gei074. [DOI] [PubMed] [Google Scholar]
- 17.Canitrot Y, Cazaux C, Frechet M, Bouayadi K, Lesca C, Salles B, Hoffmann JS. Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc Natl Acad Sci U S A. 1998;95:12586–12590. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobol RW, Watson DE, Nakamura J, Yakes FM, Hou E, Horton JK, Ladapo J, Van Houten B, Swenberg JA, Tindall KR, Samson LD, Wilson SH. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proc Natl Acad Sci U S A. 2002;99:6860–6865. doi: 10.1073/pnas.092662499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 20.Kidane D, Jonason AS, Gorton TS, Mihaylov I, Pan J, Keeney S, de Rooij DG, Ashley T, Keh A, Liu Y, Banerjee U, Zelterman D, Sweasy JB. DNA polymerase beta is critical for mouse meiotic synapsis. EMBO J. 2009 doi: 10.1038/emboj.2009.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tinwell H, Lefevre P, Williams CV, Ashby J. The activity of ENU, iPMS and MMS in male mouse germ cells using the Muta Mouse positive selection transgenic mutation assay. Mutat Res. 1997;388:179–185. doi: 10.1016/s1383-5718(96)00115-5. [DOI] [PubMed] [Google Scholar]
- 23.Xu XS, Narayanan L, Dunklee B, Liskay RM, Glazer PM. Hypermutability to ionizing radiation in mismatch repair-deficient, Pms2 knockout mice. Cancer Res. 2001;61:3775–3780. [PubMed] [Google Scholar]
- 24.Glazer PM, Sarkar SN, Summers WC. Detection and analysis of UV-induced mutations in mammalian cell DNA using a lambda phage shuttle vector. Proc Natl Acad Sci U S A. 1986;83:1041–1044. doi: 10.1073/pnas.83.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunther EJ, Murray NE, Glazer PM. High efficiency, restriction-deficient in vitro packaging extracts for bacteriophage lambda DNA using a new E.coli lysogen. Nucleic Acids Res. 1993;21:3903–3904. doi: 10.1093/nar/21.16.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Har-Vardi I, Mali R, Breietman M, Sonin Y, Albotiano S, Levitas E, Potashnik G, Priel E. DNA topoisomerases I and II in human mature sperm cells: characterization and unique properties. Hum Reprod. 2007;22:2183–2189. doi: 10.1093/humrep/dem170. [DOI] [PubMed] [Google Scholar]
- 27.Haracska LPL, Prakash S. A mechanism for the exclusion of low-fidelity human Y-family DNA polymerases from base excision repair. Genes Dev. 2003;17:2777–2785. doi: 10.1101/gad.1146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 29.Stachelek GC, Dalal S, Donigan KA, Campisi Hegan D, Sweasy JB, Glazer PM. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase beta is accentuated by BRCA2 mutation. Cancer Res. 2010;70:409–417. doi: 10.1158/0008-5472.CAN-09-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa A, Murate T, Suzuki M, Nimura Y, Yoshida S. Lithocholic acid, a putative tumor promoter, inhibits mammalian DNA polymerase beta. Jpn J Cancer Res. 1998;89:1154–1159. doi: 10.1111/j.1349-7006.1998.tb00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabelof DC, Guo Z, Raffoul JJ, Sobol RW, Wilson SH, Richardson A, Heydari AR. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res. 2003;63:5799–5807. [PubMed] [Google Scholar]
- 32.Allen D, Herbert DC, McMahan CA, Rotrekl V, Sobol RW, Wilson SH, Walter CA. Mutagenesis is elevated in male germ cells obtained from DNA polymerase-beta heterozygous mice. Biol Reprod. 2008;79:824–831. doi: 10.1095/biolreprod.108.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 34.McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase eta. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- 35.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 36.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 37.Intano GW, McMahan CA, McCarrey JR, Walter RB, McKenna AE, Matsumoto Y, MacInnes MA, Chen DJ, Walter CA. Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Mol Cell Biol. 2002;22:2410–2418. doi: 10.1128/MCB.22.7.2410-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchetti F, Essers J, Kanaar R, Wyrobek AJ. Disruption of maternal DNA repair increases sperm-derived chromosomal aberrations. Proc Natl Acad Sci U S A. 2007;104:17725–17729. doi: 10.1073/pnas.0705257104. [DOI] [PMC free article] [PubMed] [Google Scholar]