Abstract
In the past 5 years, a convergence of studies has resulted in a broad appreciation in the cancer research community that reprogramming of cellular metabolism may be more central to cancer than appreciated in the past 30 years. The re-emergence of cancer metabolism stems in part from discoveries that a number of common oncogenes and tumor suppressor genes more directly control cell metabolism than previously thought. In addition, a number of what would previously have been called “card-carrying” metabolic enzymes have been identified as human tumor suppressors or oncogenes, causally mutated in a variety of human cancers. This growing appreciation of the role of altered cell metabolism has led to further investigation into the rate-limiting proteins involved in different aspects of the unique metabolism of tumor cells. Targeting cancer metabolism with drugs requires a therapeutic window in which tumor cells, compared to normal tissues, have a greater dependence on specific metabolic enzymes. Themes that have emerged in the past decade of developing oncogene-targeted cancer therapeutics suggest that tumors with distinct oncogenic lesions are likely to require drugs that target distinct metabolic pathways. Ultimately, the hope is that detailed knowledge of oncogene and tumor suppressor gene functions and their effects on metabolism will lead to drug combinations that will be far more effective in treating cancers.
Introduction and context
During the process of tumorigenesis, genetic and epigenetic events allow the metabolism of the cancer cells to evolve in a manner that optimizes conditions for growth and survival in the tumor microenvironment. The observation by Otto Warburg in 1924 of increased reliance of tumor cells on aerobic glycolysis in the cytosol, and conversion of pyruvate to lactate rather than its entry into the mitochondrial pathway for oxidative phosphorylation, was one of the first identified biochemical distinctions of cancer cells [1]. Discoveries in the late 1970s and early 1980s revealed that cancers could result from somatic mutations that caused either gain of function (oncogenes) or loss of function (tumor suppressors) of normal genes involved in cellular regulation. Since none of the early oncogenes or tumor suppressor genes were metabolic enzymes, and since studies in the 1960s and 1970s revealed exceptions to the Warburg observation, enthusiasm for the study of cancer cell metabolism waned. In the past ten years, a dramatic resurgence in interest in cancer metabolism has occurred as a number of tumor suppressors and oncogenes have been identified that directly regulate cellular metabolism (see figure 1), indicating a more prominent role for metabolism in tumorigenesis than previously appreciated [2]. In particular, a number of connections between cellular metabolism and the phosphoinositide-3kinase/Akt pathway have been made, involving activation of the mammalian target of rapamycin (mTOR) signaling pathways [3]. mTOR has emerged as a coordinator of cell growth and metabolism in response to a wide variety of environmental cues including growth factors and nutrients such as oxygen, glucose, and amino acids [4]. The last 5 years have seen tens of new components of this critical pathway decoded, linking growth and metabolism [3]. Another unexpected genetic link between tumorigenesis and metabolism was the identification of the LKB1/STK11 tumor suppressor (which switches cells from ATP consumption to ATP production) as the critical kinase needed to activate the AMP-activated protein kinase (AMPK) under conditions of low cellular energy [5-7]. This direct linkage of a tumor suppressor with a central regulator of cellular energy, lipid and glucose metabolism, has led to a number of new findings showing the convergent mechanisms by which AMPK reprograms cell metabolism in opposition to mTOR signaling [8]. As described in detail below, a number of additional connections have now led to a large number of labs decoding the circuitry by which mitogenic signaling pathways control metabolism. Reflecting the accumulated evidence suggesting that altered cellular metabolism is a common feature of most cancers, in an update on their seminal “Hallmarks of Cancer” review, Hanahan and Weinberg added “deregulating cellular energetics” as an emerging seventh hallmark of cancer cells [9].
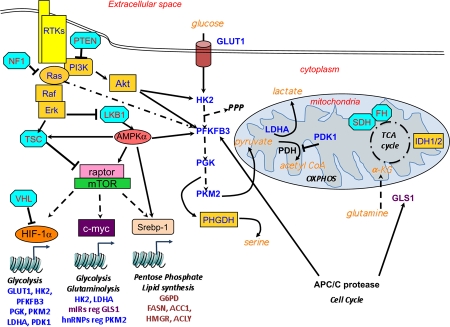
Figure 1. Oncogenic and tumor suppressor control of metabolism.
Oncogene (yellow) and tumor suppressors (light blue) control metabolism directly (isocitrate dehydrogenase, succinate dehydrogenase/fumarate hydratase, 3-phosphoglycerate dehydrogenase) or indirectly. Mechanisms including phosphorylation, and transcription, and proteolysis are illustrated. Key: broken lines indicate an indirect connection (i.e. non-direct biochemical events); unbroken lines indicate the two proteins interacting.
Abbreviations: FH, fumarate hydratase; GLS1, glutaminase 1; HK2, hexokinase 2; IDH, isocitrate dehydrogenase; LDHA, lactate dehydrogenase A; OXPHOS, oxidative phosphorylation; PDH: pyruvate dehydrogenase; PDK1, pyruvate dehydrogenase kinase, isozyme 1; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, isoform 3; PGK, phosphoglycerate kinase; PHGDH, 3-phosphoglycerate dehydrogenase; PKM2, pyruvate kinase M2 isoform; PPP, pentose phosphate pathway; SDH, succinate dehydrogenase.
The emergence of 18-flourodeoxyglucose positron emission tomography (FDG-PET) as a common imaging modality for visualizing a wide variety of human cancers pointedly illustrated Warburg’s observations, by showing the increased glucose utilization of tumors compared to most surrounding normal tissues. Initially it seemed counter-intuitive that tumor cells would switch towards the less-efficient ATP generation from aerobic glycolysis rather than from oxidative phosphorylation in mitochondria. However, increased glycolysis allows the diversion of glycolytic intermediates into intersecting biosynthetic pathways, including those generating amino acids and nucleosides, which fuel the biosynthesis of the macromolecules required for proliferating cells [2]. It should be pointed out that the increased glycolytic flux observed in some tumors does not remove the need for mitochondrial function, as mitochondria play key roles in biosynthetic pathways other than their role in ATP synthesis. Another broad finding has been the dependence of many cancer cells on elevated glutamine metabolism in addition to, or in some cases instead of, increased glucose metabolism. In fact, not all tumors exhibit high rates of glycolysis, as evidenced by their failure to be visualized by FDG-PET, and it is likely that many of these FDG-PET negative tumors are fueled by glutamine. The unique sensitivity of tumor cells to glutamine levels has been appreciated for a number of years, which is thought to due to the fact that like glucose, glutamine can be used to generate ATP as well as a number of key metabolic intermediates [10].
The past 5 years have witnessed an explosion of research in this area as more of the critical regulatory steps deregulated in tumors have been decoded. These advances have been accompanied by the hope that tumors may be targeted therapeutically by taking advantage of their unique metabolic needs and reprogramming, exposing a theoretical Achilles heel. As our knowledge of the key regulatory steps in metabolic reprogramming in different cancers advances, it also has begun to emerge that distinct oncogenes use complimentary molecular mechanisms to achieve the same ends of increased glucose and/or glutamine metabolism. Here we review a number of the recent advances in our understanding of the circuitry of cancer cell metabolism, and discuss how these pathways might be exploited therapeutically.
Transcriptional control allows for coordinated reprogramming of metabolic enzymes
Studies from several different lines of investigation have revealed that a handful of transcription factors directly bind to the promoters of a number of metabolic enzymes. This allows for coordinate up- and down-regulation of entire metabolic processes based on the activity of these transcription factors. Best-appreciated currently are the transcription factors HIF-1 (hypoxia-inducible factor 1), c-Myc, and Srebp-1. The HIF-1a was initially discovered to directly bind to the promoter of one isoform of nearly every protein responsible for a step of glycolysis [11] from the glucose transporter (GLUT1) all the way through to pyruvate kinase, inducing their expression under hypoxic conditions. In addition, HIF-1a induces lactate production via increases in lactate dehydrogenase A mRNA and suppresses flux of pyruvate into oxidative phosphorylation by inducing the PDK1 (pyruvate dehydrogenase kinase 1) mRNA, and inhibits entry of pyruvate into the oxidative phosphorylation pathway [12,13]. The discovery that the von-Hippel Lindau tumor suppressor protein encodes a ubiquitin ligase responsible for HIF-1a protein turnover under normoxic conditions provided a direct link between glucose metabolism and some forms of cancer [14]. This increased reliance on glycolysis and movement away from oxidative phosphorylation makes sense, as a physiological response to oxygen deprivation in which oxidative phosphorylation would not be functional. Furthermore, HIF-1a translation and transcription are increased under normoxic conditions by the mammalian target of rapamycin complex 1 (mTORC1), providing another mechanism for HIf-1a upregulation across a broad spectrum of human cancers. As a side note, HIF-1a and its closely related transcription factor HIF-2a are thought to be redundant in many, though not all, functions and are co-regulated by many of the same mechanisms. Importantly, in different tissue contexts, distinctions in their function do exist and studies suggest that HIF-2a is more oncogenic than HIF-1a, and that in some contexts HIF-1a may actually suppress the oncogenic effects of HIF-2a [15].
The oncogene c-Myc was also found to bind directly to many of the same promoters as HIF-1a (lactate dehydrogenase A, HK2), synergizing with HIF-1a and the closely related transcription factor, HIF-2a, depending on the context [16]. Notably, unlike HIF-1a, c-Myc was also shown to control glutamine metabolism [17]. Myc control of glutamine metabolism probably operates via multiple mechanisms, but one interesting development was the discovery that Myc repression of the mIR23a/b microRNAs results in increased expression of mitochondrial glutaminase (GLS1), one of the rate-limiting regulators of glutamine metabolism [18]. Myc is thought to be altered in up to 40% of all human cancers, and thus its dual control of glucose and glutamine metabolism makes this a common mechanism by which these processes are increased in cancer.
An additional commonality of c-myc and HIF-1a is that they both have been shown to be translationally upregulated by mTORC1-signaling. Strikingly, a third transcription factor that controls cell metabolism is also a target of mTORC1-dependent translational control: Srebp-1 [19]. Srebp-1 was originally discovered as the sterol-sensing transcription factor that induces the expression of fatty acid synthesis genes in the liver in response to insulin and sterols in the blood. Studies over the past 5 years have revealed that Srebp-1 has general roles in lipid synthesis across most cell types that may be essential for increased lipid production during cell proliferation [20]. More recently, transcriptional profiling revealed that in fibroblasts, Srebp-1 is one of the main targets of mTORC1 in regulating transcription, and here Srebp-1 induces not only lipogenic genes but also several enzymes in the oxidative branch of the pentose phosphate pathway [19]. Thus, three main transcription factors controlled by mTORC1: HIF, Myc, and Srebp-1, emerge as central regulators of cellular metabolism that may be deregulated in a majority of tumor cells. HIFs shift glucose metabolism towards anaerobic glycolysis, together with c-Myc, which also increases glutamine metabolism, while Srebp-1 increases lipid production.
Mechanisms controlling key metabolic regulators
As common nodes for metabolic control have been revealed, a plethora of mechanisms have emerged to explain how the expression and activities of these enzymes are controlled, and these mechanims are far more complex than the product inhibition models previously taught in introductory biochemistry classes. A few examples follow.
One of the newly appreciated steps by which mitogenic signals from oncogenic receptor tyrosine kinases connect to the glycolytic machinery is via inhibition of the M2 isoform of pyruvate kinase (PKM2) a key regulator of glycolysis [21,22]. This inhibition is accomplished by a novel mechanism in which binding of PKM2 to other tyrosine-phosphorylated proteins triggers release of the allosteric activator, fructose-1, 6-bisphosphate (FBP) which inhibits PKM2. Alternative splicing controls the swapping in and out of a single short exon that encodes residues in PKM2 that allow binding to phosphotyrosine and the linkage between phosphotyrosine binding and release of FBP. In contrast the PKM1 alternatively spliced form contains residues that maintain the enzyme in a highly active state, independent of FBP. Interestingly, the PKM1 isoform is expressed in most terminally differentiated, non-dividing cells, whereas all dividing cells that have been investigated, both normal and cancerous, express the M2 isoform. This splicing event is controlled, in part, by the expression of hnRNP A1 and A2 proteins whose promoters are in turn regulated by c-Myc [23,24]. In addition to the splicing to PKM2, the promoter is upregulated by HIF-1a in an mTORC1-dependent way, thus giving tumors with elevated mTOR activity higher levels of PKM2 [25]. Finally, the activity of PKM2 is also decreased by acetylation under high glucose conditions [26] consistent with suppression by pro-growth signals, such as tyrosine phosphorylation. Another unique function of the M2, but not M1, isoform of PKM was reported to be translocation to the nucleus and direct transactivation of HIF-1a [27]. In this capacity, PKM2 would be reinforcing its own expression and that of other HIF-1a targets reprogramming glucose metabolism.
Another example of multifaceted regulation is that of GLS1, which encodes a form of glutaminase that resides in the mitochondria and converts glutamine to glutaminate, for use in TCA cycle anaplerosis. As aforementioned, glutaminase mRNA is upregulated by c-Myc through an indirect mechanism involving repression of mIR23a/b, which targets GLS1. GLS1 mRNA has also been shown to be upregulated by NFkB, downstream of other oncogenic signals [28]. In addition, GLS1 is alternatively spliced to form a shorter protein, GAC, whose expression is elevated in certain cancers. Further links between call proliferation and metabolism come from the observation that glutaminase protein levels are targeted by the APC/C protease during the cell cycle, ensuring a careful temporal control over glutaminase activity during the cell cycle [29].
Another metabolic enzyme whose turnover is controlled by APC/C is the “inducible” form of phosphofructokinase 2 (iPFK2/PFKFB3) [29]. PFKFBP3 is a HIF-1a target gene, upregulated in Ras-dependent tumor cells, and also has the unique distinction of being directly phosphorylated by Akt and AMPK on different sites [30]. The convergence of tyrosine and serine/threonine kinases, acetylases, ubiquitin ligases, miRNA, splicing, and transcriptional controls onto these 3 enzymes (PKM2, GLS1, and iPFK2/PFKFBP3) gives a hint of the level of complexity that is likely to emerge for many of the proteins at the nodes of metabolic control.
Bona fide metabolic enzymes as tumor suppressors and oncogenes in human cancer
Positional cloning efforts in familial cancer syndromes led to the identification of many if not most of the known human tumor suppressors, ranging from retinoblastoma protein (Rb) to PTEN to the tuberous sclerosis complex proteins. Studies of inherited forms of human paraganglioma and pheochromocytosis led to identification of inactivating mutations in genes that encode the succinate dehydrogenase (SDH) complex subunits B, C, and D in these tumors. A second TCA cycle enzyme, fumarate hydratase, was also found to be responsible for rare inherited forms of leiomyoma and renal carcinomas [31]. These identifications fulfill Warburg’s prediction that defects in mitochondrial respiration may underlie human cancer, albeit only in these rare cases. Interestingly, a recent study identified mutations in the gene for a novel conserved protein required for flavination of SDH subunit A, termed SDH5, which was subsequently determined to account for some of the paraglioma cases not bearing mutations in other SDH genes [32].
More examples come from unbiased genomic sequencing in human glioblastomas, which first revealed missense mutations in isocitrate dehydrogenase genes, IDH1 and IDH2 [33]. Unexpectedly, the mutations found in IDH1 and IDH2 conferred a gain-of-function resulting in mutant forms of the enzyme capable of converting a-ketoglutarate into 2-hydroxyglutarate [28]. 2-hydroxyglutarate has been proposed to act as an “oncometabolite” [34], interfering with the function of the many enzymes that use a-ketoglutarate as a co-factor, including PHD proteins that degrade HIFs and many histone methyl transferases such as the TET1 and TET2 oncoproteins [35]. Collectively, these data suggest a mechanism by which a gain-of-function in a single metabolic enzyme can stimulate reprogramming of cell fate, more than just metabolic state.
Two recent studies discovered another example of a metabolic enzyme acting directly as an oncogene due to amplification in a subset of human breast cancers [36,37]. Phosphoglycerate dehydrogenase is an enzyme that diverts the glycolytic intermediate 3-phosphoglycerate into the serine synthesis pathway. Phosphoglycerate dehydrogenase was found to be amplified in a subset of primary tumors and breast cancer cell lines, and those bearing phosphoglycerate dehydrogenase amplification also exhibited elevated flux through the serine synthesis pathway. These same lines, but not those lacking phosphoglycerate dehydrogenase amplification, were uniquely sensitive to growth suppression by PHDGH shRNA, suggesting that agents targeting phosphoglycerate dehydrogenase may be useful in treating the subset of breast cancers that exhibit phosphoglycerate dehydrogenase gene amplification.
Therapeutically targeting metabolism
One of the exciting prospects for exploiting the unique metabolism of tumor cells is that there appear to be only a limited set of metabolic pathways that most cancer cells up-regulate to accomplish growth and survival. Altered metabolism in the tumor cells, compared with the tissue of origin creates a theoretical therapeutic window in which normal cells will not be affected by therapies targeting metabolism, though clearly those simply targeting the metabolism found in proliferating cells will not be any different than conventional chemotherapies targeting nucleotide synthesis enzymes needed by all cells going through S phase (e.g. 5FU, methotrexate). Similarly, metabolic drugs that inhibit lipogenic or other anabolic processes needed in all dividing cells or needed to maintain adult quiescent stem cells will not likely improve on existing therapies. However, as all advanced tumor cells reprogram their metabolism to some extent or another, decoding the specific enzymes and isoforms upregulated in each tumor type based on the genetic mutations present will allow a set of genotype-specific targeted therapeutics based on inhibition of metabolic enzymes. This is a largely unexplored space in current cancer research efforts. Based on our current understanding of pathways deregulated in cancer, investigation of a handful of promising therapeutic agents have begun, including agents targeting lactate dehydrogenase A and pyruvate dehydrogenase kinases and other glycolytic enzymes that have been validated by shRNA studies in tumor cells lines. It remains to be seen whether these early compounds have sufficient specificity and pharmaceutical properties to achieve target inhibition without off-target toxicity. Another challenge will be to identify the biomarkers that predict patients who are likely to respond to such therapies, as well as biomarkers that verify that the drug has hit its intended target.
Future Prospects
As the field of cancer cell metabolism continues to grow over the next decade, a better picture of what steps are universal across many tumor types and what metabolic reprogramming is unique to one particular tumor genotype or tissue type will emerge. Based on these findings, new tools for imaging tumors and, hopefully, new therapeutics will emerge. To get to the stage of new FDA-approved anti-cancer agents, much work is needed. It is likely that, through ongoing mutational analyses of human tumors and extensive functional RNAi screens, additional metabolic enzymes will emerge as candidates for therapeutic intervention. Once candidate genes are found, full genetic dissection of the requirement for these components in tumors from different tissues with different initiating mutations is needed. Even in cases where a particular metabolic enzyme is critical in a specific tumor type, detailed studies are also needed to define likely resistance mechanisms and effective combinations of targeted therapeutics and metabolic therapeutics in specific tumor types. In short, cancers are far more complex than was once imagined and it is clear that many weapons will be needed to combat this devastating disease. Targets in the metabolic arena offer a new opportunity to expand the arsenal.
Acknowledgments
Ruben J. Shaw was funded by grants from the American Cancer Society and NIH R01 DK080425 and P01CA120964. Lewis C. Cantley was funded by grants from the NIH and also by NCI P01CA120964. We apologize to authors of the many publications in this area we had to omit due to length restrictions.
Abbreviations
- AMPK
AMP-activated protein kinase
- FBP
fructose-1,6-bisphosphate
- FDG-PET
18-flourodeoxyglucose positron emission tomography
- HIF-1
hypoxia-inducible factor 1
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- PKM2
pyruvate kinase M2
- SDH
succinate dehydrogenase.
Competing interests
Lewis C. Cantley is a founder of Agios Pharmaceuticals, a company that focuses on drugs that target metabolic pathways.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/4/2
Contributor Information
Reuben J. Shaw, Email: shaw@salk.edu.
Lewis C. Cantley, Email: lewis_cantley@hms.harvard.edu.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 12Evaluated by Reuben Shaw 19 Dec 2011, John Kyriakis 19 Jun 2008
- 5.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRADalpha/beta and MO25alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Bruce Kemp 16 Oct 2003
- 6.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–8. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Reuben Shaw 19 Dec 2011
- 7.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–35. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Bruce Kemp 08 Mar 2004
- 8.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Philippe Soriano 27 Apr 2011
- 10.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 11.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–63. [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Reuben Shaw 19 Dec 2011
- 12.Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J Bioenerg Biomembr. 2007;39:231–4. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 13.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Robert Abraham 27 Mar 2006
- 14.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/S1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Ueli Schibler 12 Aug 2002, Peter Ratcliffe 16 Sep 2002
- 15.Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, Kaelin WG., Jr. Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene. Cancer Discovery. 2011;1:222–35. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 16.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 17.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 18.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Reuben Shaw 19 Dec 2011
- 19.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Reuben Shaw 19 Dec 2011
- 20.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Reuben Shaw 19 Dec 2011
- 21.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 22.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 23.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–9. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, Chang L, Zhang Y, Goto J, Onda H, Chen T, Wang MR, Lu Y, You H, Kwiatkowski D, Zhang H. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–34. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Vladimir Marshansky 27 Jun 2011
- 26.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, Wang G, Huang Y, Xiong Y, Guan KL, Lei QY. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–30. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 27.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Andrew D Sharrocks 04 Jul 2011
- 28.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–19. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Reuben Shaw 19 Dec 2011
- 29.Colombo SL, Palacios-Callender M, Frakich N, De Leon J, Schmitt CA, Boorn L, Davis N, Moncada S. Anaphase-promoting complex/cyclosome-Cdh1 coordinates glycolysis and glutaminolysis with transition to S phase in human T lymphocytes. Proc Natl Acad Sci U S A. 2010;107:18868–73. doi: 10.1073/pnas.1012362107. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Reuben Shaw 19 Dec 2011
- 30.Telang S, Yalcin A, Clem AL, Bucala R, Lane AN, Eaton JW, Chesney J. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006;25:7225–34. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Reuben Shaw 19 Dec 2011
- 31.Bayley JP, Devilee P. Warburg tumours and the mechanisms of mitochondrial tumour suppressor genes. Barking up the right tree? Curr Opin Genet Dev. 2010;20:324–9. doi: 10.1016/j.gde.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, Gygi SP, Winge DR, Kremer H, Rutter J. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–42. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Gilbert Cote 17 Aug 2009, Barry Nelkin 23 Sep 2009
- 33.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by John Lazo 15 Oct 2008
- 34.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–44. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 35.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Löwenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Lewis Cantley 19 Dec 2011
- 36.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Aurelio Teleman 26 Sep 2011