Abstract
Summary
Background and objectives
In a single-center renal clinic, we have established routine mutation testing to diagnose UMOD-associated kidney disease (UAKD), an autosomal dominant disorder typically characterized by gout, hyperuricemia, and renal failure in the third to sixth decades.
Design, setting, participants, & measurements
Four probands and their multigeneration kindreds were assessed by clinical, historical, and biochemical means. Diagnostic UMOD sequencing was performed, and mutant uromodulin was characterized in vitro.
Results
All available affected members of the four kindreds harbored the same complex indel change in UMOD, which was associated with almost complete absence of gout and a later onset of CKD; the youngest age at ESRD or death was 38 years (range, 38 to 68 years) compared with 3 to 70 years in other reports. Three mutation carriers (all ≤35 years) are currently asymptomatic. The indel sequence (c.278_289del TCTGCCCCGAAGinsCCGCCTCCT; p.V93_G97del/ins AASC) results in the replacement of five amino acids, including one cysteine, by four novel residues, also including a cysteine. Uromodulin staining of the only available patient biopsy suggested disorganized intracellular trafficking with cellular accumulation. Functional characterization of the mutant isoform revealed retarded intracellular trafficking associated with endoplasmic reticulum (ER) retention and reduced secretion into cell culture media, but to a lesser extent than we observed with the previously reported C150S mutation.
Conclusions
The indel mutation is associated with a relatively mild clinical UAKD phenotype, consistent with our in vitro analysis. UAKD should be routinely considered as a causative gene for ESRD of unknown cause, especially where there is an associated family history or where biopsy reveals interstitial fibrosis.
Introduction
Familial juvenile hyperuricemic nephropathy (FJHN; MIM 162000) and medullary cystic kidney disease type 2 (MIM 603860) are autosomal dominant disorders characterized by progressive renal tubulointerstitial fibrosis, hyperuricemia, and gout. About 45% of cases of FJHN and medullary cystic kidney disease type 2 are caused by mutations in three genes, UMOD (1,2), REN (3), and HNF1β (4), with mutations in UMOD accounting for the majority of these. A further locus for FJHN at 2p22.1-p21 has also recently been described (5), but some FJHN families show no linkage to any of the known loci, suggesting further genetic heterogeneity.
UMOD encodes uromodulin (or Tamm-Horsfall protein). Uromodulin is a heavily glycosylated 640-amino acid protein containing an N-terminal signal peptide, four potential EGF-like domains, a central region of unknown function containing eight conserved cysteine residues, a zona pellucida domain, and a C-terminal glycosylphosphatidylinositol anchor site (Figure 1). It is secreted into the nascent urine in the loop of Henle, the only segment in which it is expressed, and is the most abundant urinary protein. However, there is no consensus as to its physiologic function. Postulated roles for uromodulin have included providing a water-tight boundary to the thick ascending limb of the loop of Henle (6); prevention of urinary tract infections (7) or renal calculi formation (8); and cilial signaling (9). One certainty is that UMOD does not encode a urate transporter (10), so the etiology of the hyperuricemia in FJHN remains unexplained.
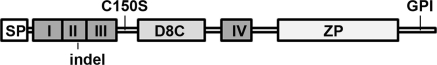
Figure 1.
Schematic representation of uromodulin. Uromodulin is a 640-amino acid protein comprising an N-terminal signal peptide (SP), four potential EGF-like domains (I to IV), a central region of unknown function containing eight conserved cysteine residues (D8C), a zona pellucida domain (ZP), and a C-terminal glycosylphosphatidylinositol (GPI) anchor site. Positions of uromodulin mutants analyzed in this study are shown.
Disease-causing UMOD mutations studied to date have mostly been shown to result in impaired trafficking and secretion of uromodulin, with intracellular accumulation in the ER (11–13). Because many are mis-sense changes and disrupt cysteine residues that are predicted to be important for correct domain folding, this is probably because of misfolding. Mutant uromodulin is likely acting in a dominant-negative manner, resulting in toxic effects independent of normal function. In contrast, other UMOD variants are reportedly associated with increased risk of developing CKD in the general population (14), suggesting that subtle alterations in expression of wild-type protein might also lead to impaired renal function as part of a complex trait.
Here, we report the identification of a recurrent UMOD mutation in a series of families ascertained from a single center renal out-patient service. This mutation (c.278_289delTCTGCCCCGAAGinsCCGCCTCCT; p.V93_G97del/ins AASC) is associated with a relatively mild clinical phenotype, and our in vitro analysis has revealed defective but milder abnormalities of intracellular trafficking and cellular localization compared with a previously reported severe mutation, C150S (15).
Materials and Methods
UMOD Patients and Kindreds
All probands (Figure 2 and Table 1) were referred to a regional renal genetics clinic (http://www.addenbrookes.org.uk/serv/clin/renal/services/renal_genetics/renal_genetics.html) because of a strong family history of renal disease for which no specific diagnosis had been made. In addition to usual workup for CKD, investigations included interrogation of genomic DNA for sequence variation in UMOD, Western blotting of urine from available pretransplant patients, and calculation of fractional excretion of uric acid. All participants gave informed consent (NRES 08/H0306/62).
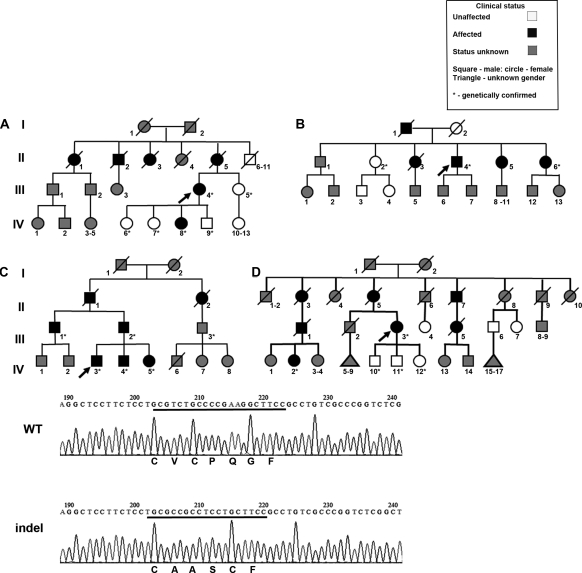
Figure 2.
Genetic features of study subjects. (Upper panel) Pedigrees of the four kindreds (A through D) showing extensive history of renal disease. The arrows denote index cases. (Lower panel) The upper sequence shows wild-type UMOD, and the lower sequence shows the indel mutation identified in all four families, with the predicted amino acids indicated below each sequence. The indel sequence alteration results in the replacement of five amino acids (including a cysteine residue) in the second calcium-binding EGF-like domain with four novel residues (also including a cysteine).
Table 1.
Clinical and biochemical features of index cases and affected family members
| Age (years)a | Gender | Presentation | Initial Ultrasound Findings | FEUr (%) | Age at RRT (years) | Age at Tx (years) | Age at Death (years) | Genotype | |
|---|---|---|---|---|---|---|---|---|---|
| Index cases | |||||||||
| AIII.4 | 47 | F | HT, CKD, polyuria, polydipsia | “Small kidneys” | 54 | 55 | Indel | ||
| BII.4 | 43 | M | HT, CKD3, proteinuria | “Small kidneys” | 48 | 50 | Indel | ||
| CIV.3 | 35 | M | Asymptomatic, normal biochemistryb | “Normal” (at 30 years) | 9.2 | Indel | |||
| DIII.3 | 46 | F | HT, CKD4, recurrent UTI | “Normal IVU” (at 46 years) | 52 | Indel | |||
| Family members | |||||||||
| AII.1 | 50 | F | HT, “chronic nephritis,” CVAc | 50 | NK | ||||
| AII.2 | NK | M | HT, CKD, LVF | 68 | NK | ||||
| AII.3 | 45 | F | “Died of renal failure” | 45 | NK | ||||
| AII.5 | 61 | F | HT, CKD5 | Small kidneys (“like walnuts”) | 61 | 63 | 63 | NK | |
| AIV.8 | 28 | F | Asymptomatic, urate 0.37 mmol/L,d otherwise normal biochemistryb | 8.0 | Indel | ||||
| BI.1 | 49 | M | “Died of renal failure” | 49 | NK | ||||
| BII.1 | 51 | M | HT, CKD4, proteinuria (NB has diabetes) | Left 10.5 cm, right 10 cm | NK | ||||
| BII.3 | 38 | F | HT, CKD5 | 38 | 48 | 51 | NK | ||
| BII.5 | 41 | F | HT, CKD | 43 | NK | ||||
| BII.6 | 39 | F | CKD, polydipsia, proteinuria, recurrent UTI | Left 8.3 cm, right 8.5 cm | 41 | 44 | Indel | ||
| CII.1 | NK | M | Malignant HT, CKD, anaemia | 47 | NK | ||||
| CII.2 | NK | F | HT, “chronic pyelonephritis” MI | NK | 46 | NK | |||
| CIII.1 | 57 | M | HT, CKD3, polyuria, gout | Left 9.5 cm, right 10.5 cm | 67 | 68 | Indel | ||
| CIII.2 | 60 | M | HT, CKD 4, proteinuria | Horseshoe kidney | 65 | Indel | |||
| CIV.4 | 34 | M | HT, CKD3 | Left 11.8 cm, right 10.8 cm | Indel | ||||
| CIV.5 | 30 | F | Asymptomatic, normal biochemistryb | Both 10.7 cm | 13.5 | Indel | |||
| DII.3 | NK | F | “Uremic seizures” | 52 | NK | ||||
| DII.5 | NK | F | “Uremia” | 59 | NK | ||||
| DII.7 | NK | M | “Uremia” | 47 | NK | ||||
| DI.8 | 49 | F | Malignant HT, “uremia” | 49 | NK | ||||
| DIII.1 | 50 | M | HT, CKD4 | 53 | 63 | NK | |||
| DIII.5 | NK | F | HT, CKD | 59 | 68 | NK | |||
| DIV.2 | NK | F | HT, CKD | 49 | Indel |
CKD, chronic kidney disease; CVA, cerebrovascular accident; FEUr, fractional excretion of urate, which is equal to [(urine urate × serum creatinine)/(serum urate × urine creatinine)] × 100%; HT, hypertension; IVU, intravenous urogram; LVF, left ventricular failure; MI, myocardial infarction; NK, not known; RRT, renal replacement therapy; Tx, transplant; UTI, urinary tract infection.
Age at first presentation.
Serum sodium, potassium, estimated GFR, venous bicarbonate, corrected calcium, phosphate, urate.
Clinical information in quotation marks is from historical documents or patient recollection.
Normal range, 0.15 to 0.35 mmol/L.
Mutation Screening and Genotyping
The East Anglian Regional Genetics Service performed direct sequencing of UMOD according to a UKGTN-approved protocol (http://www.cuh.org.uk/addenbrookes/services/clinical/genetics/genetics_index.html). Intragenic single nucleotide polymorphisms were typed by direct sequencing. Polymorphic markers closely flanking UMOD were PCR-amplified using a fluorescent reporter primer, and products were electrophoresed according to standard methods.
Expression Constructs and Site-directed Mutagenesis
Full-length UMOD cDNA was amplified from normal human kidney using gene-specific primers and cloned into the mammalian expression vector pcDNA3.1. Site-directed mutagenesis was performed using mutagenic oligonucleotides and the QuikChange kit (Stratagene), and all constructs were sequence-verified before use.
Cell Culture, Transient Transfection, and Secretion Assays
HEK293 cells were cultured in DMEM containing 5 mM glucose and 10% FCS at 37°C in 5% CO2/95% air. 2 × 105 cells were plated into each well of a six-well plate and transfected using Lipofectamine Plus (Invitrogen) according to the manufacturer's recommendations. At specified time intervals between 4 and 72 hours post-transfection, 150μl aliquots of media were removed and analyzed by Western blot. At 72 hours, the cells were lysed with radioimmuno precipitation assay buffer for analysis by Western blotting. All of the experiments were carried out in triplicate for each UMOD isoform. Endo-β-acetylglucosaminidase (Endo H) and peptide-N-glycosidase (PNGase F) treatment of cell lysates was carried out according to the manufacturer's instructions (New England Biolabs).
Western Blot Analysis
Proteins were separated by gel electrophoresis in 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membrane (Immobilon-P). The membranes were blocked overnight in PBS containing 0.1% Tween 20 (PBST) and 5% nonfat dry milk and washed three times in PBST. The membranes were then incubated at room temperature for 1 hour with primary antibody: sheep anti-human uromodulin (1:1000, R & D Systems), anti-β-actin (1:1000), or anti-green fluorescent protein (1:2000; Abcam) in PBST containing 1% BSA, washed three times in PBST, and then incubated at room temperature with the appropriate horseradish peroxidase-labeled secondary antibody. The membranes were washed again and visualized with enhanced chemiluminescence (Amersham). Protein bands were quantified using ImageJ (v1.43U; National Institutes of Health) densitometry software. Statistical analyses were performed using GraphPad Prism 5.0. Nonparametric data were analyzed using the Mann-Whitney U-test, and the results were considered statistically significant at P < 0.05.
Urine Samples
Urine was collected into sterile receptacles containing 0.02% sodium azide and stored at −80°C until required. For Western blotting, thawed samples were diluted in 5 mM Tris-Cl (pH 8.0) to achieve equivalence of urinary creatinine, denatured by the addition of an equal volume of protein loading buffer containing β-mercaptoethanol, and boiled before separation on a 10% SDS-polyacrylamide gel.
Immunofluorescence and Immunohistochemistry
Transfected HEK293 cells grown on coverslips coated with poly-L-lysine (100 μg/ml) were fixed with 4% paraformaldehyde for 10 minutes, permeabilized with 0.2% Triton X-100 for 8 minutes, and washed with Hanks' balanced salt solution containing 5% FCS. Fixed cells were incubated with sheep anti-uromodulin antibody at 1:1000 dilution and mouse anti-PD1 (Abcam) at 1:300 overnight at 4°C. The cells were washed three times in Hanks' balanced salt solution containing 5% FCS and incubated with AlexaFluor 488 donkey anti-sheep and AlexaFluor 546 donkey anti-mouse antisera (Invitrogen) at 1:1000 dilution for 2 hours at room temperature.
Formalin-fixed, paraffin-embedded sections were deparaffinized in xylene, rehydrated, and permeabilized with proteinase K (50 μg/ml). Sections were blocked with 3% BSA/TBST for 15 minutes and incubated with sheep anti-uromodulin antibody (1:2000) in a humidified atmosphere for 24 hours at 4°C. Sections were washed in TBST and incubated for 2 hours with a fluorescently labeled anti-sheep antibody (1:1000) (Invitrogen). All of the sections were washed and mounted using Vectashield mounting medium, and all of the images were captured using a Zeiss LSM510 META confocal microscope.
Results
UMOD Kindreds
The four kindreds reported here all had an extensive family history of renal disease, with hypertension as a prominent feature. We were able to obtain variable amounts of data on 30 individuals, of whom only one (CIII.1; Figure 2) has had clinical gout. Macroscopic hematuria was absent from the cohort. Two women (BII.6 and DIII.3) reported recurrent urinary tract infections.
Kindred A.
The 61-year-old proband (AIII.4; Figure 2) presented elsewhere at age 47 with polydipsia and polyuria. Investigations revealed bilateral small kidneys and impaired excretory function. She developed end-stage renal failure at age 54 and received a living unrelated kidney transplant the following year. Her deceased mother and three deceased maternal relatives were all described as having either CKD or “chronic nephritis” (Figure 2 and Table 1). Her mother had presented immediately predialysis at age 61 and underwent renal transplantation 2 years later. All of the proband's children were asymptomatic and, after appropriate counseling, elected to undergo diagnostic testing.
Kindred B.
The proband, a 48-year-old man (BII.4; Figure 2), was already receiving renal replacement therapy. Initial investigations carried out elsewhere 5 years earlier had revealed hypertension, CKD, and bilateral small kidneys. A renal biopsy had not been performed. In light of his family history, he was concerned whether his children should be offered any screening. His father and three of his five siblings had renal disease that had progressed to end-stage renal failure.
Kindred C.
The 35-year-old index case (CIV.3) presented because of a family history of renal disease affecting his brother, father, paternal uncle, grandfather, and great aunt (Figure 2). He was normotensive with no urinary dipstick abnormality and had normal renal imaging and function. Because his affection status was uncertain, initial UMOD sequencing was performed on his clinically affected brother (CIV.4).
Kindred D.
The proband (DIII.3) was a 78-year-old woman who had received a renal transplant in 1984 at age 52, having presented at age 46 with hypertension and progressive CKD. Her mother died at age 59 from uremia, and there were six other maternal relatives with a diagnosis of renal disease or uremia (Figure 2 and Table 1). She was concerned about the implications for her own offspring.
In the 26 individuals where age of ESRD/death was recorded, the median was 50.5 years (range, 38 to 68 years).
Identification of UMOD Mutation
In all of these kindreds, the family history was compatible with an adult-onset autosomal dominant nephropathy. In the absence of other diagnostic markers, UMOD testing was offered as a first-line diagnostic test, because the presentations with CKD and small kidneys were suspicious for UMOD-associated kidney disease (UAKD). The same indel mutation (c.278_289delTCTGCCCCGAAGinsCCGCCTCCT; p.V93_G97del/ins AASC) was identified in all four probands and in all clinically affected individuals who were available for testing. Predictive and diagnostic testing was subsequently offered to other family members who requested it, after formal nephrology and genetics review. Evidence for a founder effect was sought by haplotype analysis of available polymorphic microsatellite markers and intragenic single nucleotide polymorphisms. No shared common haplotype was found in all four families (data not shown).
The Indel Sequence Change Predicts a Structural Change in Uromodulin
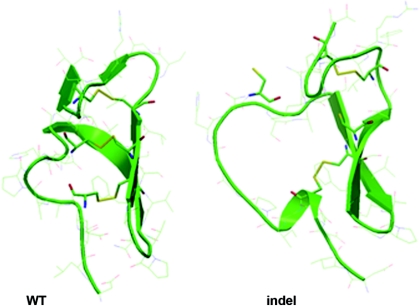
The replacement of c.278_289 TCTGCCCCGAAG with CCGCCTCCT results in the replacement of five amino acids including a cysteine residue in the second uromodulin calcium-binding EGF-like domain, with four novel residues also including a cysteine residue (Figure 2, lower panel). Structural alignments using Phyre (16) and nFOLD3 (17) confirmed the presence of an EGF-like domain from residues 65 to 107 (Uniprot reference P07911). This region contains six highly conserved cysteine residues that are predicted to disulfide-bond in a 1–3, 2–4, 5–6 pattern. Figure 3 (left panel) demonstrates a model of this domain on the basis of the corresponding structure of a fibrillin-1 EGF-like domain (Protein Data Bank code 1emo), with the three disulfide bridges maintaining a compact fold of two major β sheets and N- and C-terminal loops. A similar model for the indel mutant reveals a loss of disulfide bridging at the fifth cysteine, even although it is replaced with a new cysteine, with resultant unfolding of the N-terminal region (Figure 3, right panel).
Figure 3.
Structural models of wild-type (WT) and indel uromodulin isoforms. Shown are models of part of the WT and indel uromodulin isoform, on the basis of the solved EGF-like domain of fibrillin-1 (Protein Data Bank code 1emo). The indel isoform loses disulfide bridging at the fifth cysteine, with consequent unfolding of the N-terminal region.
The Indel Mutation Results in Trafficking Defects
To assess the effect of the indel mutation on uromodulin processing and trafficking, equal amounts of expression constructs containing wild-type (WT), indel, C150S UMOD, or empty vector were transfected into HEK293 cells. The C150S substitution has previously been shown to result in marked ER retention of mutant uromodulin (11). Transfection efficiency among the different expression constructs was verified by cotransfection with an enhanced green fluorescent protein-expressing plasmid.
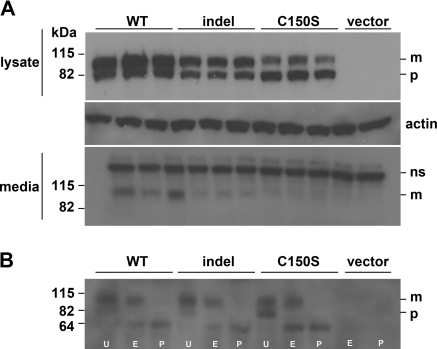
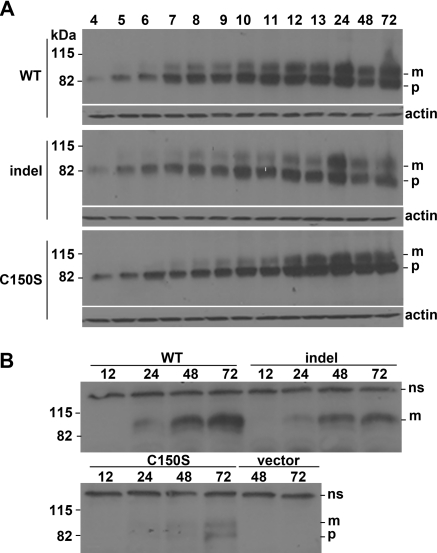
The media and cell lysates were initially collected at 72 hours post-transfection and were analyzed by Western blotting. Each experiment was carried out in triplicate and independently repeated three times. Transfection efficiencies for wild type, indel, and C150S were not different at 33, 34, and 31%, respectively. Comparison of total cell lysates (Figure 4A) shows that both the 82-kD precursor (ER resident) and the 100-kD mature, glycosylated uromodulin protein could be detected in all transfected cells. Densitometric analysis confirmed twice as much mature protein as precursor in wild-type transfected cells, compared with 1.5 and 0.4 times for indel and C150S transfected cells, respectively (P = 0.041 and 0.003 versus WT). The mature form of uromodulin was also readily detected in the media of wild type–expressing cells but was approximately halved in the indel-expressing cells (Figure 4A). As expected from previous observations, very little of the C150S mature form was detectable in media (11).
Figure 4.
Western blot analyses of wild-type and mutant uromodulin expressed in HEK293 cells at 72 hours post-transfection. (A) Lysates of HEK293 cells transfected with wild-type (WT), indel, C150S, or vector constructs. Uromodulin is seen as an 82-kD precursor (p) and a 100 kD mature (m) protein. The middle panel shows β-actin as loading control. The lower panel shows media of these same cells. Indel transfected cells secrete less mature uromodulin (m) than do WT cells at this time-point. ns, nonspecific band. (B) Cleavage pattern of cell lysates treated with Endo H, PNGase F, or no enzyme confirms that the immature form (p) is ER-resident and predominates in C150S, as shown by Endo H digestion.
To assess glycosylation of the uromodulin isoforms, total cell extracts from wild-type and mutant isoforms of uromodulin-expressing cells were treated with Endo H or PNGase F and changes visualized by Western blot. Endo H cleaves high mannose oligosaccharides of ER-resident glycoproteins and PNGase F cleaves all types of N-linked glycosylation. The 82-kD (or ER resident) WT uromodulin is cleaved by Endo H to a 60-kD deglycosylated form, whereas PNGase F cleaves both the 100- and 82-kD forms to the 60-kD form. Figure 4B shows that the indel behaved as did WT in this respect, whereas C150S displayed a previously reported intracellular processing defect (11).
To study the time course of the observed behavior of mutant uromodulin, glycosylation and secretion were analyzed at a series of intermediate time-points from 4 to 72 hours post-transfection. The 82-kD immature protein was detectable from 4 hours in cell lysates of all constructs (Figure 5A). Mature (100 kD) protein was first detectable at 5 hours in wild-type and indel but not C150S transfected cells. By 10 hours, there was a greater amount of the mature 100-kD WT protein compared with the same time point in both the indel and C150S, irrespective of comparable quantities of the immature form at this and later time points. However, by 24 hours, the ratio of immature to mature protein was comparable in both WT and indel transfected cells. These temporal differences indicate delayed processing of the misfolded indel uromodulin.
Figure 5.
Western blot analyses showing maturation of wild-type and mutant uromodulin over time. (A) Detection of uromodulin in total cell lysates from HEK293 cells transfected with WT, indel, or C150S UMOD, 4 to 72 hours post-transfection. The lower panels show β-actin loading controls. (B) Detection of uromodulin in the same media at 12, 24, 48, and 72 hours. Less mature (m) uromodulin can be detected in the indel transfected than in WT media at 24, 48, and 72 hours. The immature 82-kD precursor (p) is prominent in the C150S transfected cells. ns, nonspecific band. Both panels are from the same gel.
Analysis of media from these transfected cells (Figure 5B) showed that secreted WT uromodulin could be detected at 24 hours, whereas no mutant protein could be detected from either the indel or C150S transfected cells at this time. The levels of indel uromodulin were lower compared with WT at 48 and 72 hours, whereas levels of C150S were lowest at all time points. Detection of the ER-resident immature 82-kD protein in the media of C150S transfected cells at the 72-hour time point suggested decreased cell viability.
Localization and Trafficking Defects of the Indel Isoform
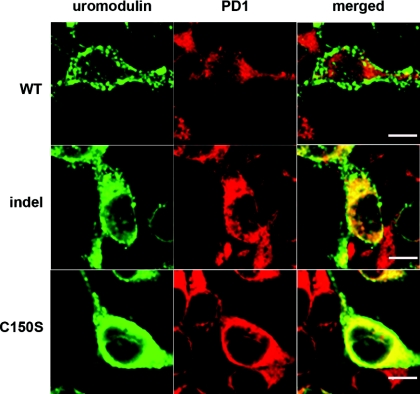
We next assessed whether the delay in protein secretion displayed by the indel protein in vitro was mirrored by ER retention, as reported for C150S. As shown in Figure 6, immunofluorescence microscopy of transiently transfected cells 48 hours after transfection revealed little colocalization of WT uromodulin with the ER marker, PD1. In contrast, both the indel and C150S mutant isoforms clearly colocalized with PD1, C150S to a greater extent. Taken together with the Western blot experiments, these data are suggestive of an intermediate phenotype for the indel protein.
Figure 6.
Cellular distribution of wild-type (WT) and mutant uromodulin proteins expressed in HEK293 cells, 48 hours post-transfection. Cells transfected with WT, indel, or C150S UMOD were costained with anti-uromodulin (left panels, green) and anti-PD1 (middle panels, red) antibodies. (right panels) The merged images clearly show colocalization of uromodulin and the ER-resident protein PD1, in both indel- and C150S-expressing cells. Bar, 5 μm.
Histology and Urine Biochemistry
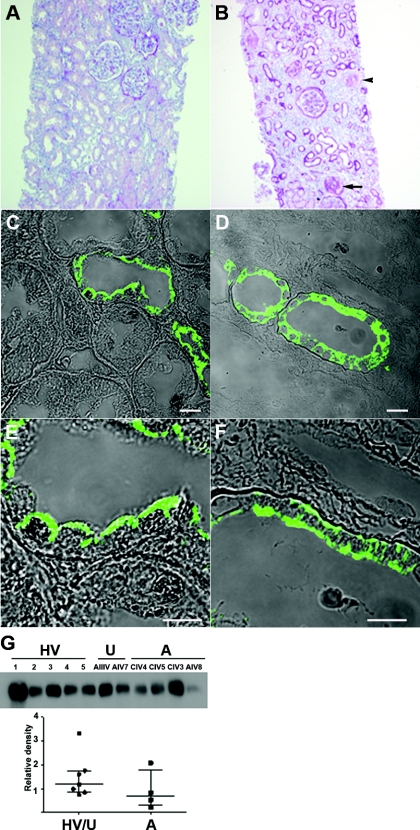
Of the three individuals from this cohort who underwent renal biopsy, only one had tissue available for review (Figure 2, patient CIV.4), which revealed streaky tubulointerstitial atrophy and fibrosis, with secondary focal segmental glomerulosclerosis (Figure 7B). Mild nonspecific lymphocytic inflammation was observed in atrophic foci but not elsewhere. Small arteries showed prominent myointimal thickening. An earlier review elsewhere reported the absence of immune deposits on fluorescence or electron microscopy, the latter revealing very focal foot process effacement only and no glomerular basement membrane abnormalities. Uromodulin localization was determined in residual tissue from this biopsy and compared with normal kidney (Figure 7, C through F). Similar to other reports (9,18), apical staining was seen in the latter (Figure 7, C and E), whereas in the indel biopsy, uromodulin was located throughout the cell (Figure 7, D and F), suggesting cellular retention and/or disorganized trafficking.
Figure 7.
Renal histology and urine biochemistry. (A) Normal renal cortical histology. (B) Renal biopsy of patient CIV.4, demonstrating parenchymal tubular atrophy and interstitial fibrosis, with associated glomerulosclerosis (arrow). Small arteries show prominent myointimal thickening (arrowhead). (C through F) Uromodulin expression is abnormal in the patient (D and F) compared with normal kidney (C and E). Bar, 10 μm. (G) Western blot of urine from healthy volunteers (HV: 1 to 5), unaffected relatives (U: AIII.V and AIV.7), and affected individuals with preserved renal function (A: CIV.4, CIV.5, CIV.3, and AIV.8). The amounts of uromodulin were quantified by densitometry, protein loading having been corrected for urinary creatinine.
Western blotting of urine from the four available pretransplant affected individuals not on allopurinol (AIV.8, CIV.3, CIV.4, and CIV.5; amounts corrected for urine creatinine) revealed variable levels trending toward a reduction in uromodulin compared with a group of unaffected relatives and healthy volunteers (Figure 7G; P = 0.25). Notably, the fractional excretion of uric acid was within normal limits in the available affected subjects (Table 1).
Discussion
In this report we have functionally studied a recurrent but previously uncharacterized uromodulin mutation identified in four autosomal dominant families seen in a single center renal clinic, all of UK origin. This cohort contains 12% of the index cases we have genetically screened (three other non-indel UMOD mutations have also been found). A founder effect appears unlikely, because closely linked markers were not haploidentical in all four families. Mechanisms for such recurrent sequence alterations are poorly understood.
Clinical severity increased with age but notably, gout was not a prominent feature in any of these families, which was also noted in the only other clinical report featuring this sequence change (19). Relatively late presentation was also described there, similar to our median age to ESRD or death. We have shown that the trafficking defects, ER retention, and reduction in the amount of protein secreted both in vitro and in vivo by indel uromodulin is intermediately severe in comparison with the C150S mutant or indeed the majority of other reported mutations involving cysteine residues (19–22). This predicts the observed milder clinical course and perhaps the predominant absence of gout in our cohort. Some of our patients are currently devoid of clinical features despite carrying the mutation, which is entirely consistent with the experience of the majority of the older cases, where late but rapid deterioration of renal function occurred.
Pathogenicity of the indel is predicted because it disrupts the structure of the second EGF-like domain. Pathogenic UMOD mutations are mostly clustered in the N-terminal region containing three EGF-like domains and eight conserved cysteine residues. EGF domains are protein folds widely distributed in nature, which are often involved in cell adhesion and numerous protein-protein interactions. Mutations in other EGF domains are responsible for disorders such as Marfan syndrome (MIM 154700) and familial hypercholesterolemia (MIM 143890). The replacement of one cysteine with another in the indel mutation, even in a closely related position, is not able to maintain structural or functional integrity of uromodulin (Figure 3). The indel amino acid change also lies adjacent to the proposed Ca2+-binding site within this region (23), suggesting that calcium binding within EGF-like 2 could be disrupted.
Uromodulin has recently been shown to interact with and regulate the renal outer medullary potassium channel ROMK2, which is also present in the loop of Henle (24). In that study, a variety of point mutant uromodulin proteins still possessed the ability to interact with ROMK2 in vitro but failed in vivo to increase ROMK2's surface expression and current amplitude. Also, given that ROMK2 functionally interacts with the apical sodium potassium chloride cotransporter (NKCC2), one might speculate that uromodulin is indirectly able to influence this transporter. Hypokalemia might therefore have been predicted in our cohort, but none was observed.
Of note, UMOD C147W transgenic (18) and Nkcc2−/− mice (25) share a number of similarities including a urine concentrating defect, which has been anecdotally reported in children with UAKD (20). Interestingly, polydipisia and/or polyuria have been reported within our cohort (Table 1). Another similarity between our cohort and UMOD transgenic strains is the absence of hyperuricemia, which of itself cannot therefore be a major contributor to CKD (18,26).
Because fractional excretion of urate has been previously proposed as a biochemical screening tool for UAKD (27,28), it is also notable that in the available genetically confirmed, but as yet clinically unaffected subgroup of our cohort, this parameter was not reduced. In these cases, it cannot be accounted for by CKD. Occasional normal levels were in fact reported in one study (10), and further evaluation in larger groups would be useful. This subgroup also had normal serum potassium, bicarbonate, calcium, and phosphate levels.
Although UAKD is judged to be rare, its true prevalence is not known, particularly among CKD patients presenting with already-reduced renal size. Our approach is proactively to screen UMOD in such individuals, particularly if they present with a positive family history of renal disease, no underlying diagnosis, and/or notable interstitial fibrosis on renal biopsy (the latter a feature here and in other UMOD reports) (9,18,23). This strategy has yielded important results both for these patients and their families and for those considering living related kidney donation, where establishing a negative genetic diagnosis in a potential donor will be important given the relatively late emergence of the phenotype. We therefore propose that genetic screening should be more widely applied. Allied to this, identification of mutations in additional genes (to explain causation in the approximately 50% of patients unaccounted for by UMOD) is an important goal.
With anecdotal evidence that it is useful, it has been proposed that allopurinol, a urate-lowering agent, should be offered to all those affected by UAKD, to slow progression (29,30). There is a need for proper, controlled trials of this and/or other potential therapies, to which end accurate genotypic as well as phenotypic characterization are important. The cohort reported here provides a unique opportunity to contribute to this debate, because it includes currently clinically unaffected gene carriers.
The absence of validated preventive therapies makes decisions regarding screening of clinically unaffected but at-risk relatives more difficult. It is clearly an issue for patients including some within this cohort. Whether or not genetic screening is pursued, we suggest that at-risk relatives should remain under some form of continuing surveillance for hypertension, CKD, and other cardiovascular risk factors.
Disclosures
None.
Acknowledgments
We thank Drs. Vicky Bardsley, David Wright, and Seema Qamar for their assistance. This work was supported by the National Institute for Health Research Cambridge Biomedical Research Centre and the Wellcome Trust.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1. Hateboer N, Gumbs C, Teare MD, Coles GA, Griffiths D, Ravine D, Futreal PA, Rahman N: Confirmation of a gene locus for medullary cystic kidney disease (MCKD2) on chromosome 16p12. Kidney Int 60: 1233–1239, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, Shirts B, Xu L, Zhu H, Barmada MM, Bleyer AJ: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zivná M, Hulková H, Matignon M, Hodanová K, Vylet'al P, Kalbácová M, Baresová V, Sikora J, Blazková H, Zivný J, Ivánek R, Stránecký V, Sovová J, Claes K, Lerut E, Fryns J-P, Hart PS, Hart TC, Adams JN, Pawtowski A, Clemessy M, Gasc J-M, Gübler M-C, Antignac C, Elleder M, Kapp K, Grimbert P, Bleyer AJ, Kmoch S: Dominant renin gene mutations associated with early-onset hyperuricemia, anemia, and chronic kidney failure. Am J Hum Genet 85: 204–213, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bingham C, Ellard S, van't Hoff WG, Simmonds HA, Marinaki AM, Badman MK, Winocour PH, Stride A, Lockwood CR, Nicholls AJ, Owen KR, Spyer G, Pearson ER, Hattersley AT: Atypical familial juvenile hyperuricemic nephropathy associated with a hepatocyte nuclear factor-1β gene mutation. Kidney Int 63: 1645–1651, 2003 [DOI] [PubMed] [Google Scholar]
- 5. Piret S, Danoy P, Dahan K, Reed A, Pryce K, Wong W, Torres R, Puig J, Müller T, Kotanko P, Lhotta K, Devuyst O, Brown M, Thakker R: Genome-wide study of familial juvenile hyperuricaemic (gouty) nephropathy (FJHN) indicates a new locus, FJHN3, linked to chromosome 2p22.1-p21. Hum Genet 129: 51–58, 2011 [DOI] [PubMed] [Google Scholar]
- 6. Parsons CL, Bautista SL, Stein PC, Zupkas P: Cyto-injury factors in urine: A possible mechanism for the development of interstitial cystitis. J Urol 164: 1381–1384, 2000 [PubMed] [Google Scholar]
- 7. Orskov I, Ferencz A, Orskov F: Tamm-Horsfall protein or uromucoid is the normal urinary slime that traps type 1 fimbriated Escherichia coli. Lancet 315: 887, 1980 [DOI] [PubMed] [Google Scholar]
- 8. Mo L, Huang H-Y, Zhu X-H, Shapiro E, Hasty DL, Wu X-R: Tamm-Horsfall protein is a critical renal defense factor protecting against calcium oxalate crystal formation. Kidney Int 66: 1159–1166, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Zaucke F, Boehnlein JM, Steffens S, Polishchuk RS, Rampoldi L, Fischer A, Pasch A, Boehm CWA, Baasner A, Attanasio M, Hoppe B, Hopfer H, Beck BB, Sayer JA, Hildebrandt F, Wolf MTF: Uromodulin is expressed in renal primary cilia and UMOD mutations result in decreased ciliary uromodulin expression. Hum Mol Genet 19: 1985–1997, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gersch MS, Sautin YY, Gersch CM, Henderson G, Bankir L, Johnson RJ: Does Tamm-Horsfall protein-uric acid binding play a significant role in urate homeostasis? Nephrol Dial Transplant 21: 2938–2942, 2006 [DOI] [PubMed] [Google Scholar]
- 11. Bernascone I, Vavassori S, Di Pentima A, Santambrogio S, Lamorte G, Amoroso A, Scolari F, Ghiggeri GM, Casari G, Polishchuk R, Rampoldi L: Defective intracellular trafficking of uromodulin mutant isoforms. Traffic 7: 1567–1579, 2006 [DOI] [PubMed] [Google Scholar]
- 12. Jennings P, Aydin S, Kotanko P, Lechner J, Lhotta K, Williams S, Thakker RV, Pfaller W: Membrane targeting and secretion of mutant uromodulin in familial juvenile hyperuricemic nephropathy. J Am Soc Nephrol 18: 264–273, 2007 [DOI] [PubMed] [Google Scholar]
- 13. Williams SE, Reed AAC, Galvanovskis J, Antignac C, Goodship T, Karet FE, Kotanko P, Lhotta K, Morinière V, Williams P, Wong W, Rorsman P, Thakker RV: Uromodulin mutations causing familial juvenile hyperuricaemic nephropathy lead to protein maturation defects and retention in the endoplasmic reticulum. Hum Mol Genet 18: 2963–2974, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Köttgen A, Hwang S-J, Larson MG, Van Eyk JE, Fu Q, Benjamin EJ, Dehghan A, Glazer NL, Kao WHL, Harris TB, Gudnason V, Shlipak MG, Yang Q, Coresh J, Levy D, Fox CS: Uromodulin levels associate with a common UMOD variant and risk for Incident CKD. J Am Soc Nephrol 21: 337–344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rampoldi L, Caridi G, Santon D, Boaretto F, Bernascone I, Lamorte G, Tardanico R, Dagnino M, Colussi G, Scolari F, Ghiggeri GM, Amoroso A, Casari G: Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet 12: 3369–3384, 2003 [DOI] [PubMed] [Google Scholar]
- 16. Phyre, version 0.2. http://www.sbg.bio.ic.ac.uk/∼phyre/ Accessed September 26, 2011
- 17. The nFOLD3 Protein Fold Recognition Server. http://www.reading.ac.uk/bioinf/nFOLD/ Accessed September 26, 2011
- 18. Bernascone I, Janas S, Ikehata M, Trudu M, Corbelli A, Schaeffer C, Rastaldi MP, Devuyst O, Rampoldi L: A transgenic mouse model for uromodulin-associated kidney diseases shows specific tubulo-interstitial damage, urinary concentrating defect and renal failure. Hum Mol Genet 19: 2998–3010, 2010 [DOI] [PubMed] [Google Scholar]
- 19. Wolf MTF, Mucha BE, Attanasio M, Zalewski I, Karle SM, Neumann HPH, Rahman N, Bader B, Baldamus CA, Otto E, Witzgall R, Fuchshuber A, Hildebrandt F: Mutations of the uromodulin gene in MCKD type 2 patients cluster in exon 4, which encodes three EGF-like domains. Kidney Int 64: 1580–1587, 2003 [DOI] [PubMed] [Google Scholar]
- 20. Wolf MTF, Beck BB, Zaucke F, Kunze A, Misselwitz J, Ruley J, Ronda T, Fischer A, Eifinger F, Licht C, Otto E, Hoppe B, Hildebrandt F: The uromodulin C744G mutation causes MCKD2 and FJHN in children and adults and may be due to a possible founder effect. Kidney Int 71: 574–581, 2007 [DOI] [PubMed] [Google Scholar]
- 21. Dahan K, Devuyst O, Smaers M, Vertommen D, Loute G, Poux J-M, Viron B, Jacquot C, Gagnadoux M-F, Chauveau D, Büchler M, Cochat P, Cosyns J-P, Mougenot B, Rider MH, Antignac C, Verellen-Dumoulin C, Pirson Y: A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol 14: 2883–2893, 2003 [DOI] [PubMed] [Google Scholar]
- 22. Lens XM, Banet JF, Outeda P, Barrio-Lucía V: A novel pattern of mutation in uromodulin disorders: Autosomal dominant medullary cystic kidney disease type 2, familial juvenile hyperuricemic nephropathy, and autosomal dominant glomerulocystic kidney disease. Am J Kid Dis 46: 52–57, 2005 [DOI] [PubMed] [Google Scholar]
- 23. Turner JJO, Stacey JM, Harding B, Kotanko P, Lhotta K, Puig JG, Roberts I, Torres RJ, Thakker RV: Uromodulin mutations cause familial juvenile hyperuricemic nephropathy. J Clin Endocrinol Metab 88: 1398–1401, 2003 [DOI] [PubMed] [Google Scholar]
- 24. Renigunta A, Renigunta V, Saritas T, Decher N, Mutig K, Waldegger S: Tamm-Horsfall glycoprotein interacts with renal outer medullary potassium channel ROMK2 and regulates its function. J Biol Chem 286: 2224–2235, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kemter E, Rathkolb B, Bankir L, Schrewe A, Hans W, Landbrecht C, Klaften M, Ivandic B, Fuchs H, Gailus-Durner V, Hrabé de Angelis M, Wolf E, Wanke R, Aigner B: Mutation of the Na+-K+-2Cl- cotransporter NKCC2 in mice is associated with severe polyuria and a urea-selective concentrating defect without hyperreninemia. Am J Physiol Renal Physiol 298: F1405–F1415, 2010 [DOI] [PubMed] [Google Scholar]
- 26. Kemter E, Rathkolb B, Rozman J, Hans W, Schrewe A, Landbrecht C, Klaften M, Ivandic B, Fuchs H, Gailus-Durner V, Klingenspor M, de Angelis MH, Wolf E, Wanke R, Aigner B: Novel missense mutation of uromodulin in mice causes renal dysfunction with alterations in urea handling, energy, and bone metabolism. Am J Physiol Renal Physiol 297: F1391–F1398, 2009 [DOI] [PubMed] [Google Scholar]
- 27. Simmonds HA, McBride MB, Hatfield PJ, Graham R, Mccaskey J, Jackson M: Polynesian women are also at risk for hyperuricaemia and gout because of a genetic defect in renal urate handling. Br J Rheumatol 33: 932–937, 1994 [DOI] [PubMed] [Google Scholar]
- 28. McBride MB, Rigden S, Haycock GB, Dalton N, Van't Hoff W, Rees L, Venkat Raman G, Moro F, Ogg CS, Cameron JS, Simmonds HA: Presymptomatic detection of familial juvenile hyperuricaemic nephropathy in children. Pediatr Nephrol 12: 357–364, 1998 [DOI] [PubMed] [Google Scholar]
- 29. Fairbanks LD, Cameron JS, Venkat-Raman G, Rigden SPA, Rees L, Van't Hoff W, Mansell M, Pattison J, Goldsmith DJA, Simmonds HA: Early treatment with allopurinol in familial juvenile hyerpuricaemic nephropathy (FJHN) ameliorates the long-term progression of renal disease. Quart J Med 95: 597–607, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Simmonds HA, CJ, Goldsmith DJ, Fairbanks LD, Raman GV: Familial juvenile hyperuricaemic nephropathy is not such a rare genetic metabolic purine disease in Britain. Nucleoside Nucleotide Nucleic Acids 25: 1071–1075, 2006 [DOI] [PubMed] [Google Scholar]