Abstract
The cessation of progesterone (P4) production (i.e., functional regression), arguably the key event in luteolysis of the primate corpus luteum (CL), is poorly understood. Previously, we found that genes encoding proteins involved in cholesterol uptake decreased while those involved in cholesterol efflux (reverse cholesterol transport; RCT) increased in expression during spontaneous functional regression of the rhesus macaque CL, thereby potentially depleting the cholesterol reserves needed for steroidogenesis. Therefore, a comprehensive analysis of the components necessary for RCT was performed. RCT components were expressed (mRNA and/or protein) in the macaque CL including cholesterol sensors (liver × receptors α or NR1H3; and β or NR1H2), efflux proteins (ATP-binding cassette subfamilies A1 or ABCA1; and G1 or ABCG1), acceptors (apolipoproteins A1 or APOA1; and E or APOE), and plasma proteins facilitating high-density lipoprotein (HDL) formation (lecithin:cholesterol acyltransferase or LCAT; phospholipid transfer protein or PLTP). ABCA1, APOE, PLTP and NR1H3 increased, while lipoprotein receptors decreased, in expression (mRNA and/or protein) through the period of functional regression. The expression of APOA1 and APOE, as well as NR1H3, was greatest in the CL and tissues involved in regulating cholesterol homeostasis. Immunolocalization studies revealed that RCT proteins and lipoprotein receptors were expressed in large luteal cells, which possess intracellular cholesterol reserves during periods of progesterone synthesis. Lipid staining revealed changes in luteal cholesterol ester/lipid distribution that occurred following functional regression. These results indicate that decreased cholesterol uptake and increased RCT may be critical for the initiation of primate luteolysis by limiting intracellular cholesterol pools required for steroidogenesis.
Keywords: rhesus macaque, corpus luteum, functional regression, luteolysis, reverse cholesterol transport
Introduction
The CL produces P4 during the luteal phase of the menstrual cycle, and P4 is absolutely required to maintain early pregnancy in primates. However, in non-conception cycles, P4 secretion from the CL must cease to allow for the initiation of the next ovarian cycle. Therefore, understanding the factors controlling CL regression (luteolysis) is of considerable importance. Luteolysis occurs in two phases: 1) a decrease in P4 synthesis (functional regression), and 2) degradation and structural remodeling of the CL (structural regression) (Hoyer 1998; Stouffer 2003). Functional regression must occur before structural regression can proceed due to the luteoprotective effects of P4 (Hoyer 1998; Stouffer 2003). However, the molecular processes responsible for initiation of functional regression and subsequent structural regression in the primate CL are poorly defined.
Functional regression in primates is characterized by reduced sensitivity to luteinizing hormone (LH) as the CL ages (Cameron & Stouffer 1982; Eyster et al. 1985; Brannian & Stouffer 1991), and pulses of LH must increase or a more potent LH-receptor ligand (chorionic gonadotropin; CG) must be present to maintain the primate CL by the end of the normal luteal phase (Duffy et al. 1999). The mechanisms responsible for the loss of P4 synthesis in an LH-replete environment are not known. To address this void in our understanding of primate luteolysis, we previously used a microarray approach to identify global changes in gene expression occurring throughout the normal period of functional regression in the primate CL (Bogan et al. 2009). From the resultant database, it was noted that genes whose products are involved in cholesterol uptake (low-density lipoprotein receptor or LDLR; scavenger receptor type B class 1 or SCARB1) decreased in expression while ABCA1 and ABCG1 increased during the approximate time when functional regression occurred. ABCA1 and ABCG1 are two key cholesterol efflux proteins critical for RCT, a process whereby excess cholesterol is removed from cells and transported to the liver for elimination as bile or bile acids (Mahley et al. 2006). Because steroid hormones are derived from cholesterol, induction of RCT coupled with inhibiting extracellular cholesterol uptake could conceivably cause functional regression in the CL by limiting the substrate necessary for steroidogenesis.
In addition to ABCA1 and ABCG1, the RCT system requires coordination of many proteins including cholesterol sensors (NR1H3/LXRα, NR1H2/LXRβ), cholesterol acceptors (APOA1, APOE), and plasma proteins facilitating HDL formation (LCAT, PLTP, CETP). Unesterified cholesterol and phospholipids are transferred to APOA1, the major protein component of HDL, by the ABC transporters (Mahley et al. 2006). The free cholesterol bound to APOA1 is esterified by LCAT thus changing the morphology of the HDL molecule from discoidal to spherical as cholesterol esters are packaged into its hydrophobic core (Jonas 2000). The actions of LCAT allow more cholesterol to be transferred to the HDL molecule (Jonas 2000). In addition to APOA1, APOE also may be an important cholesterol acceptor in RCT as some subclasses of HDL contain APOE (Mahley et al. 2006). APOE allows for expansion of the cholesterol ester-rich core, thus increasing the cholesterol-carrying capacity of HDL molecules (Mahley et al. 2006). Additionally, APOE is an LDLR ligand and consequently allows HDL clearance via hepatic LDLR and LDLR-related proteins (Mahley 1988; Zaiou et al. 2000). Other plasma proteins involved in RCT include PLTP, which promotes the formation of pre-β HDL and thus increases the cholesterol accepting capacity of plasma (Von Eckardstein et al. 1996; Dullaart & Van Tol 2001), and CETP, which transfers cholesterol esters from HDL to APOB-containing lipoproteins (low or very-low density lipoproteins) (Mahley et al. 2006).
NR1H2 and NR1H3 are known to stimulate transcription of several important RCT genes including ABCA1, ABCG1, PLTP and CETP (Lund et al. 2003), APOE (Laffitte et al. 2001a), as well as autoregulation of NR1H3 (Laffitte et al. 2001b). They were originally identified as orphan nuclear receptors belonging to the steroid hormone receptor superfamily. They are now known to bind various oxysterols thereby serving as “cholesterol sensors” that will stimulate RCT in cases of cholesterol excess (Wojcicka et al. 2007).
To our knowledge, the presence and functionality of the RCT system in the CL has not been addressed in any species. Based on our previous genomic analysis of macaque luteal tissue obtained through the period of functional regression (Gene Expression Omnibus accession number GSE12807) (Bogan et al. 2009), the RCT system may be induced within the primate CL to facilitate the loss in P4 producing capacity at the end of non-conception cycles. To this end, the objectives of the current study are to: 1) extend our previous findings of differential mRNA expression of ABCA1 and ABCG1, as well as the lipoprotein receptors LDLR and SCARB1 (Bogan et al. 2009), by determining if their corresponding protein levels are also differentially expressed throughout the normal period of functional regression; 2) determine if additional components involved in RCT (mRNA or protein) are also expressed in the primate CL (e.g., APOA1, APOE, LCAT, PLTP, CETP, NR1H2, and NR1H3); 3) determine the luteal cell localization of RCT, as well as LDLR and SCARB1, protein expression; and 4) assess changes in cholesterol/lipid handling or storage in the primate CL during its functional regression.
Results
Levels of Lipoprotein Receptor and ABC Transporter Proteins in the Macaque CL
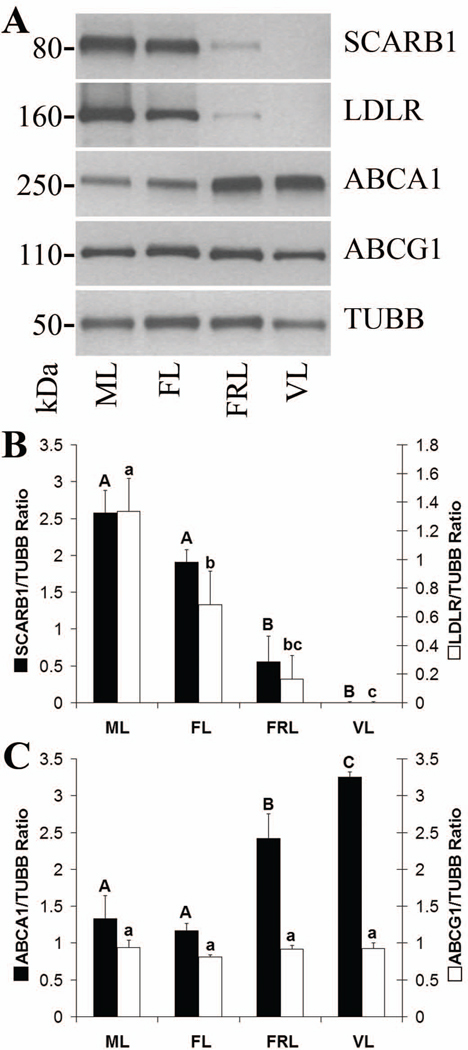
Levels of SCARB1 were not different between mid-late and functional late CL. However, there was a >3-fold decrease from functional to functionally-regressed late stage CL (p< 0.05), with SCARB1 being undetectable in very-late stage CL (Fig. 1 B). There was an approximately 2-fold decrease in LDLR from mid-late to functional late CL (p< 0.05), followed by a >4-fold decrease from functional to functionally-regressed late CL. LDLR was not detected in very-late stage CL (Fig. 1 B).
Figure 1. Lipoprotein receptor and ABC transporter protein levels throughout the period of spontaneous functional regression in the rhesus macaque CL.
Panel A contains representative Western blots for SCARB1, LDLR, ABCA1, ABCG1, and TUBB using samples pooled from CL collected at either the mid-late (ML), functional late (FL), functionally-regressed late (FRL), or very-late (VL) stages of the luteal phase. The results of densitometry analyses of individual luteal homogenates (n = 4 CL/group) for the lipoprotein receptors SCARB1 and LDLR, as well as the cholesterol efflux proteins ABCA1 and ABCG1, are presented in panels B and C, respectively. Error bars indicate one standard error of the mean (SEM). Columns with different letters of the same case are significantly different (p< 0.05).
Levels of ABCA1 protein were not different between mid-late and functional late CL, but increased >2-fold from functional to functionally-regressed late stage CL (p< 0.05), and continued to rise in the very-late stage (Fig. 1 C). There were no differences between any groups in levels of ABCG1 protein (Fig. 1 C).
Localization of Luteal SCARB1, LDLR, ABCA1, and ABCG1
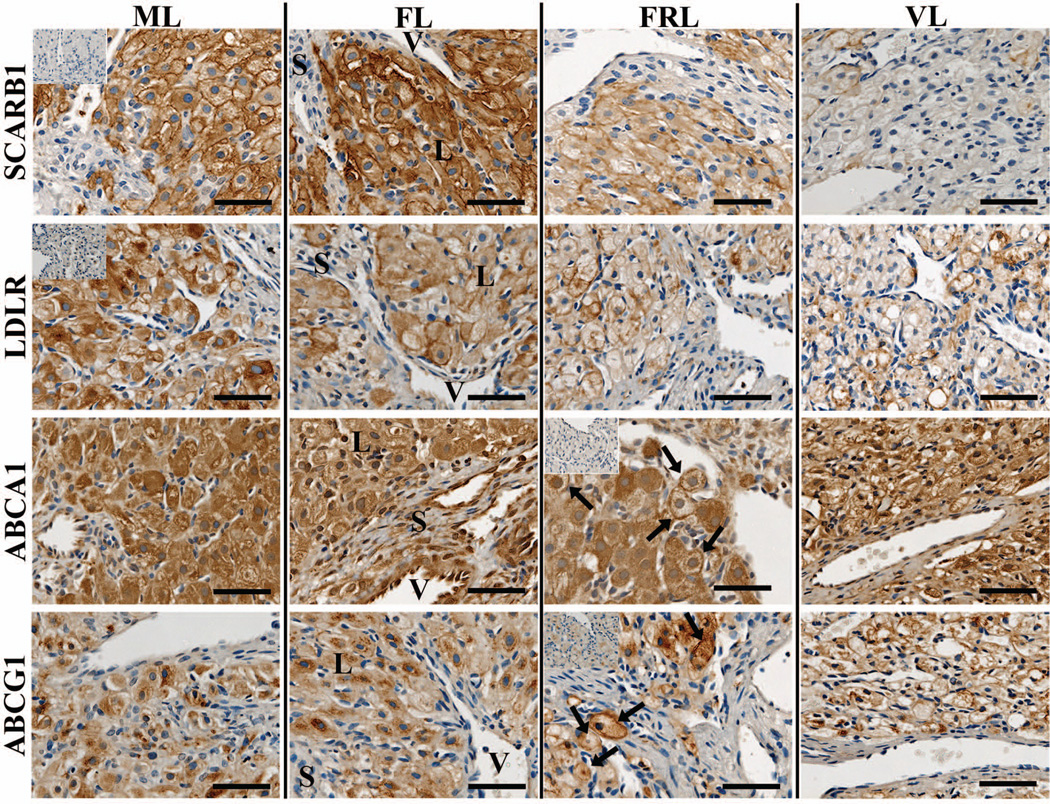
Large luteal cells stained for SCARB1 while there was minimal or no expression in small cell types such as stroma and endothelial cells (Fig. 2). Pre-absorption of the primary antibody with its immunizing peptide resulted in minimal staining. SCARB1 was evident at the plasma membrane during all stages analyzed. LDLR displayed a similar staining pattern as SCARB1 with the majority of expression occurring in large luteal cells (Fig. 2). Staining for LDLR was evident at the plasma membrane of all stages analyzed, and was absent following pre-absorption of the primary antibody with its immunizing peptide.
Figure 2. Localization of lipoprotein receptors and ABC transporters in the rhesus macaque CL throughout the period of spontaneous functional regression.
Representative photomicrographs of SCARB1, LDLR, ABCA1, and ABCG1 from the mid-late (ML), functional late (FL), functionally-regressed late (FRL), and very-late (VL) luteal stages are shown. The insets in the upper left corner of the ML samples for SCARB1 and LDLR, as well as in the FRL samples for ABCA1 and ABCG1, are sections that were processed with primary antibody preabsorbed with immunizing peptide. The approximate locations of various cell types are indicated in the FL panel for each protein and include: large luteal cells (L); small luteal or stromal cells (S); and blood vessels (V). The scale bar in the lower right hand corner of each image is 50 µm. Arrows indicate the appearance of plasma membrane localization of ABCA1 and ABCG1 in FRL stage CL.
ABCA1 expression was observed in large luteal cells, as well as in the endothelial cells and adjacent stroma lining blood vessels (Fig. 2). ABCA1 expression was diffuse throughout the cell in mid-late and functional late CL, while localization near the plasma membrane in some cells was more evident in functionally-regressed late CL. ABCG1 expression was also found in large luteal cells, while staining in small luteal cells or near the vasculature was minimal (Fig. 2). During the mid-late and functional late stages, ABCG1 expression was predominantly perinuclear with some evidence of plasma membrane staining. In functionally-regressed late CL, however, increased plasma membrane localization was observed.
CL Expression of APOA1 and APOE
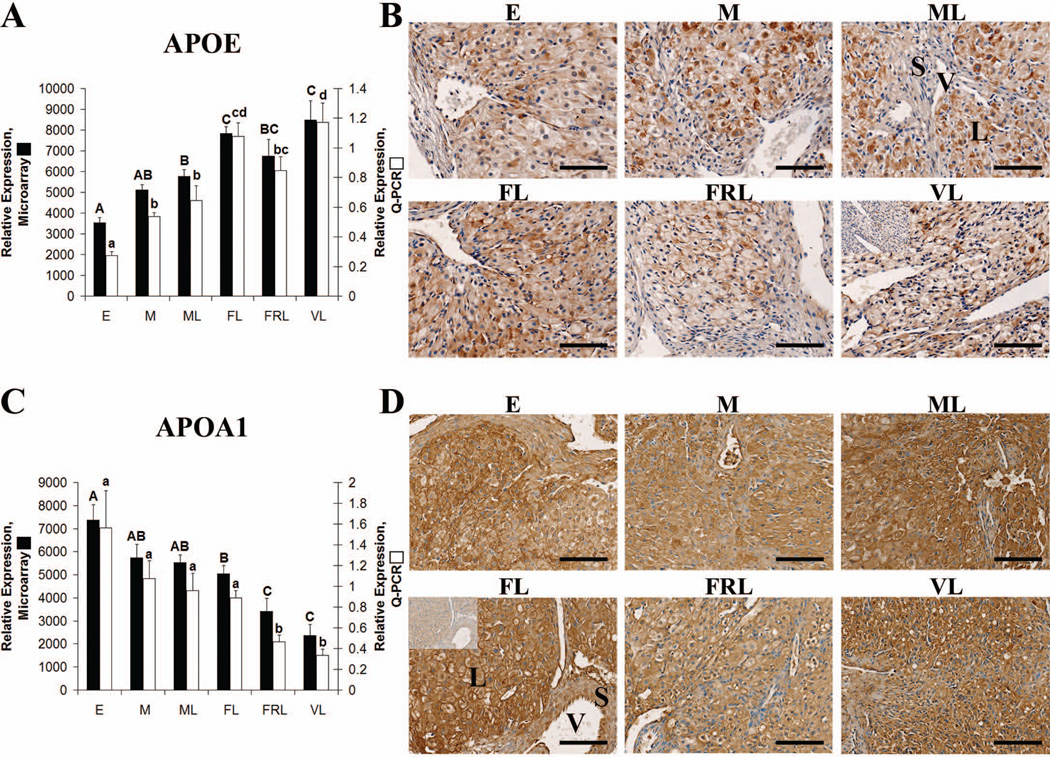
Both APOA1 and APOE mRNA were expressed in the macaque CL at all stages of the luteal phase as determined by GCOS absolute expression analysis of microarray data (Bogan et al. 2008a). APOE mRNA levels were lowest in early CL followed by an increase at each subsequent stage with the exception of a slight decrease between functional and functionally-regressed late CL. Q-PCR analysis of APOE expression yielded highly similar results, with the early CL being significantly lower than all other groups (p< 0.05) and the functional late and very-late CL being significantly higher (p< 0.05) than mid or mid-late CL (Fig. 3 A). APOA1 had a pattern of expression opposite to APOE as its mRNA levels were highest in the early luteal phase and slowly decreased thereafter with functionally-regressed late and very-late CL being significantly lower than all other stages as determined by both microarray and Q-PCR (Fig. 3 C).
Figure 3. Apolipoprotein expression in the rhesus macaque CL.
Panel A contains microarray and Q-PCR expression data for APOE during the early (E), mid (M), mid-late (ML), functional late (FL), functionally-regressed late (FRL), and very-late (VL) stages. Error bars indicate one SEM (n = 4 CL/group). Upper case letters denote significant differences (p< 0.05) for microarray data and lower case letters denote significant differences for Q-PCR data. Panel B contains representative photomicrographs for APOE IHC performed using CL obtained from the E through VL stages of the luteal phase. The inset in the upper left corner of the VL sample is from a control section that was probed with primary antibody preabsorbed with its immunizing peptide. The approximate locations of various cell types are indicated in the ML panel and include: large luteal cells (L); small luteal or stromal cells (S); and blood vessels (V). The scale bar in the lower right hand corner of each image is 100 µm. Panel C contains microarray and Q-PCR expression data for APOA1 that was analyzed in the same manner as for Panel A. Panel D contains representative photomicrographs from APOA1 IHC analysis. The details are similar to the description for panel B except the negative control inset is in the FL sample and consists of a control section probed with primary antibody preabsorbed with purified full-length human APOA1.
APOE protein was also detected in the CL at all stages of the luteal phase by IHC (Fig. 3 B). APOE localized to the large luteal cells, with some interspersed staining also noted in stroma and along vessel endothelial cells. APOE staining was primarily observed in the cytoplasm and perinuclear regions of mid and mid-late CL, with a more diffuse pattern observed in functional and functionally-regressed late sections. Strong staining for APOA1 was also detected throughout the luteal phase and in all regions of the primate CL (Fig. 3 D). Staining near the plasma membrane of large luteal cells was evident through the functionally-regressed late stage, while APOA1 was mostly detected in the cytoplasm of these cells during the very-late stage.
CL Expression of LCAT, PLTP, and CETP
LCAT and PLTP mRNA were expressed in the macaque CL at all stages of the luteal phase as determined by GCOS absolute expression analysis of microarray data (Bogan et al. 2008a). However, CETP mRNA was not detected (p> 0.065) in 20 of 24 CL, marginally detected (0.051 > p < 0.065) in 2 CL (one mid and one functional late), and detected (p< 0.05) in only 2 CL (one mid-late and one functionally-regressed late). As CETP mRNA was not detected in the majority of CL collected from all stages of the luteal phase, it does not appear that CETP is expressed by the primate CL.
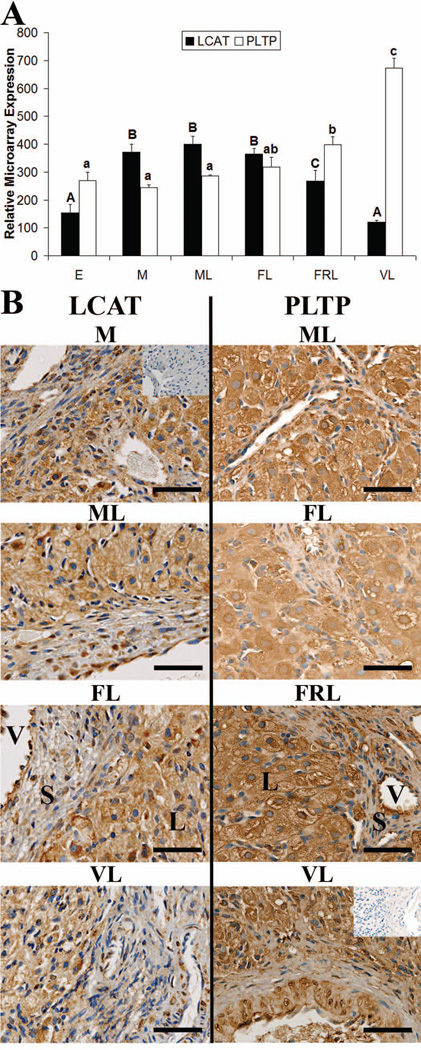
LCAT was significantly lower in early and very-late CL than all other stages (p< 0.05) (Fig. 4 A). LCAT expression did not change from mid through functional late stages, while functionally-regressed late CL had LCAT levels intermediate of functional late and very-late stages (p< 0.05). PLTP expression did not change from the early through mid-late stages, but increased from the functional late through very-late stages (Fig. 4 A). The very-late CL had significantly higher levels (p< 0.05) of PLTP than all other stages, and functionally-regressed late CL were significantly higher (p< 0.05) than early through mid-late stages.
Figure 4. Expression of proteins that promote HDL formation in the rhesus macaque CL.
Panel A contains microarray expression data for LCAT and PLTP during the early (E), mid (M), mid-late (ML), functional late (FL), functionally-regressed late (FRL), and very-late (VL) stages of the luteal phase. Error bars indicate one SEM (n = 4 CL/group). Columns with different letters of the same case are significantly different (p< 0.05). Panel B contains representative photomicrographs for LCAT and PLTP IHC analysis from throughout the luteal phase. The inset in the upper right corner of the M image (LCAT), or VL image (PLTP), are control sections where the primary antibody was excluded. The approximate locations of various cell types are indicated in the FL panel and include: large luteal cells (L); small luteal or stomal cells (S); and blood vessels (V). The scale bar in the lower right hand corner of each image is 50 µm.
LCAT protein was detected within the CL throughout the luteal phase by IHC (Fig. 4 B). Cytoplasmic staining for LCAT was noted in large luteal cells, while staining was variable in the stroma. Additionally, LCAT staining was evident in the endothelial cells lining the vasculature. PLTP protein was also present within the CL (Fig. 4 B) and displayed a similar staining pattern as LCAT with expression found primarily in large luteal cells, as well as variable staining in the stroma, and more consistent staining in the regions lining blood vessels.
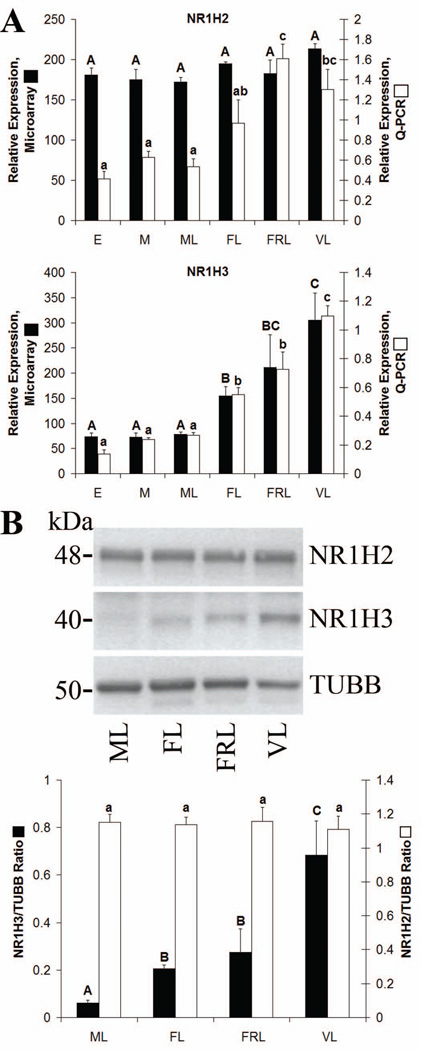
CL Expression of NR1H2 and NR1H3
Both NR1H2 and NR1H3 were expressed in the CL at all stages of the luteal phase as determined by GCOS absolute expression analysis (Bogan et al. 2008a). Levels of NR1H3 (LXRα) did not change from the early through mid-late stages, but significantly increased (p< 0.05) by the functional late stage and continued to increase through the very-late stage as determined by both microarray and Q-PCR (Fig. 5 A). Levels of NR1H2 (LXRβ) were not different between any groups as determined by microarray. However, Q-PCR analysis of NR1H2 expression indicated that while NR1H2 mRNA did not change from the early through mid-late stages, its levels increased from the mid-late through functionally-regressed late stages, with functionally-regressed late and very-late CL having significantly higher (p< 0.05) NR1H2 levels than early through mid-late stages (Fig. 5 A). These changes in NR1H2 levels detected by Q-PCR were not due to cross-amplification of NR1H3 cDNA as the primers and probe used were designed within a region unique to rhesus NR1H2.
Figure 5. NR1H2 and NR1H3 expression in the rhesus macaque CL.
Panel A contains microarray and Q-PCR expression data for NR1H2 and NR1H3 during the early (E), mid (M), mid-late (ML), functional late (FL), functionally-regressed late (FRL), and very-late (VL) stages of the luteal phase. Panel B contains Western blot data for NR1H2 and NR1H3. Images are from pooled CL collected at the ML, FL, FRL, or VL stages. The graph presents densitometry results from analysis of individual CL (n = 4 CL/group). Error bars indicate one SEM. Columns with different letters of the same case are significantly different (p< 0.05).
Western blot analysis was used to quantify levels of NR1H2 and NR1H3 protein during the mid-late through very-late stages (Fig. 5 B). There were no significant differences between stages in levels of NR1H2. Levels of NR1H3 protein increased >3-fold from the mid-late to functional late luteal phase (p< 0.05), with a further increase of >2-fold being detected in very-late relative to functionally-regressed late CL (p< 0.05).
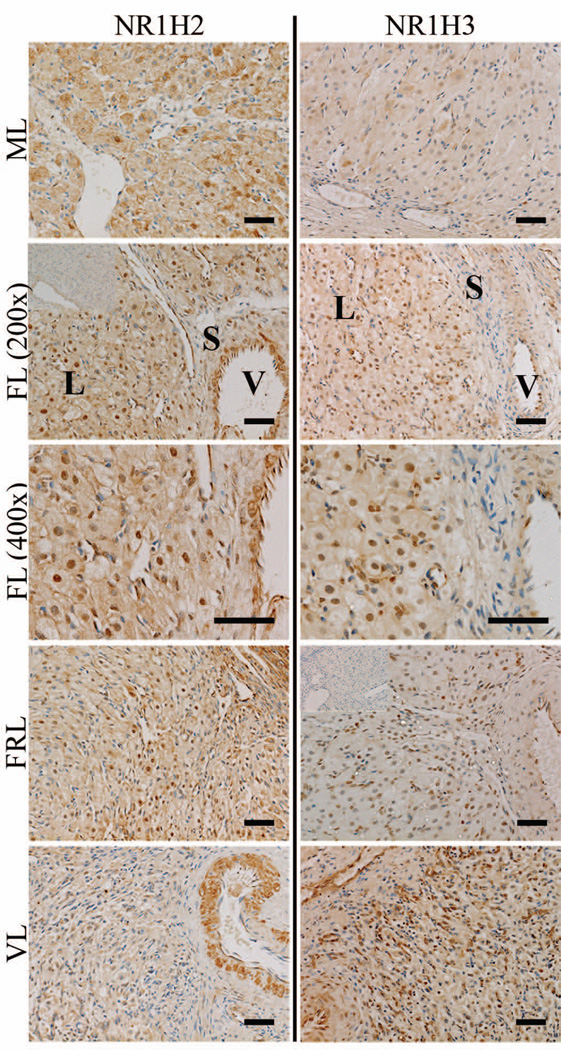
Localization of NR1H2 and NR1H3
NR1H2 protein was detected in the large luteal cells, as well as in regions adjacent to the endothelial cells lining blood vessels (Fig. 6). During the mid and mid-late stages, NR1H2 was localized primarily to the cytoplasm of large luteal cells. By the functional late stage, nuclear localization of NR1H2 became evident in large luteal cells and remained present through the functionally-regressed late stage. While NR1H2 protein was still detected in regions of the stroma adjacent to blood vessels during the very-late stage, nuclear localization in the large luteal cells was less evident.
Figure 6. Localization of NR1H2 and NR1H3 expression in the rhesus macaque CL throughout the period of spontaneous functional regression.
Representative photomicrographs of NR1H2 and NR1H3 from the mid-late (ML), functional late (FL), functionally-regressed late (FRL), and very-late (VL) stages are shown. The insets in the upper left corner of the FL 200× panel for NR1H2, and FRL panel for NR1H3, are control sections incubated with primary antibody preabsorbed with its immunizing peptide (NR1H2), or where the primary antibody was excluded (NR1H3). The approximate locations of various cell types are indicated in the FL panel for each protein and include: large luteal cells (L); small luteal or stromal cells (S); and blood vessels (V). The scale bar in the lower right hand corner of each image is 50 µm.
NR1H3 protein was detected in the same cell types as NR1H2 including the large luteal cells and regions adjacent to blood vessels (Fig. 6). Staining for NR1H3 in the CL was weak during the mid-late stage, while nuclear localization of NR1H3 was observed in large luteal cells beginning at the functional late stage. Contrary to NR1H2, strong nuclear staining of NR1H3 was still apparent in large luteal cells during the very-late stage.
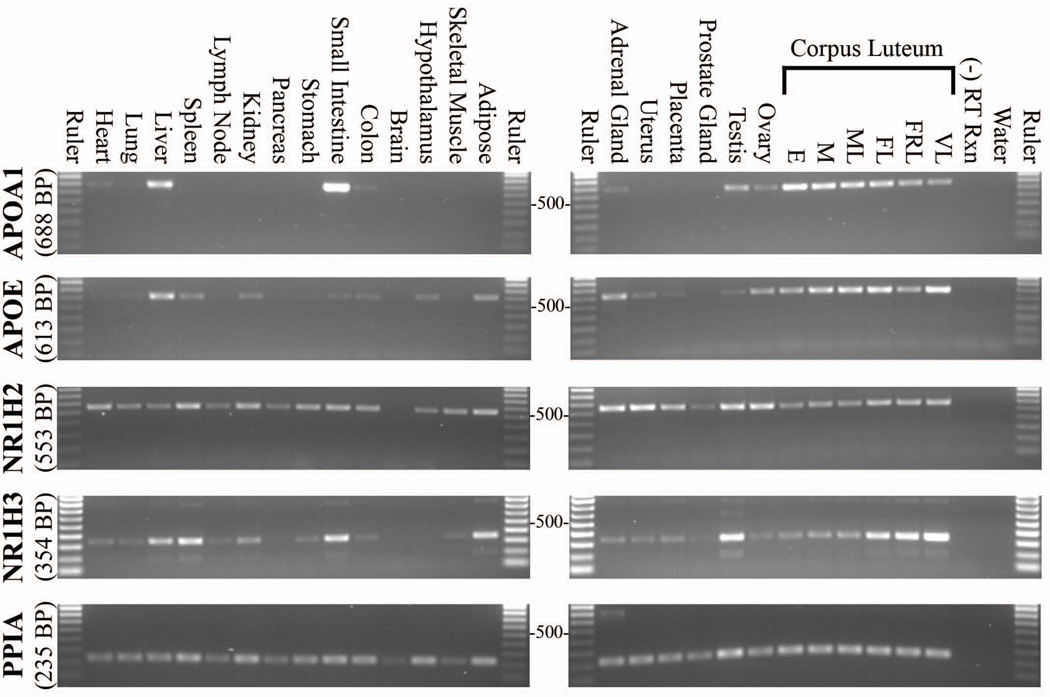
Tissue Distribution of APOA1, APOE, NR1H2 and NR1H3 mRNA
APOA1 mRNA levels were greatest in liver, small intestine, and CL, while lower levels were detected in the adrenal, testis and ovary. There was minimal or non-existent APOA1 mRNA expression in other tissues analyzed (Fig. 7). APOE mRNA was highly expressed in the CL with comparatively lower levels present in the other steroidogenic/reproductive tissues analyzed (adrenal, uterus, placenta, testis, ovary). APOE mRNA was also observed in liver, spleen, kidney, small intestine, colon, hypothalamus and adipose. NR1H2 was detected in all tissues analyzed except brain, however, the housekeeping control for brain was also weakly amplified. High cycle numbers (> 30) were needed to detect NR1H2 in most tissues. NR1H3 was expressed in all steroidogenic tissues analyzed, and was especially high in testis and late to very-late stage CL. NR1H3 was also highly expressed in liver, spleen, small intestine and adipose, while heart, lung, lymph node, kidney, stomach, colon, and skeletal muscle displayed lower mRNA levels.
Figure 7. Tissue distribution of APOA1, APOE, NR1H2, and NR1H3 gene expression in the rhesus macaque.
Agarose gel images of PCR amplification products are shown. Gene-specific primers were used to amplify cDNA collected from various rhesus macaque tissues. The number of cycles used in each image is: APOA1 = 25, APOE = 24, NR1H2 = 34, NR1H3 = 26; and PPIA = 26. These cycle numbers correspond approximately with the mid-linear range of amplification for each gene as determined from pooled luteal cDNA. A 100 base-pair (BP) ruler is included with each image, and the 500 BP band is marked. Expected product size for each gene is indicated.
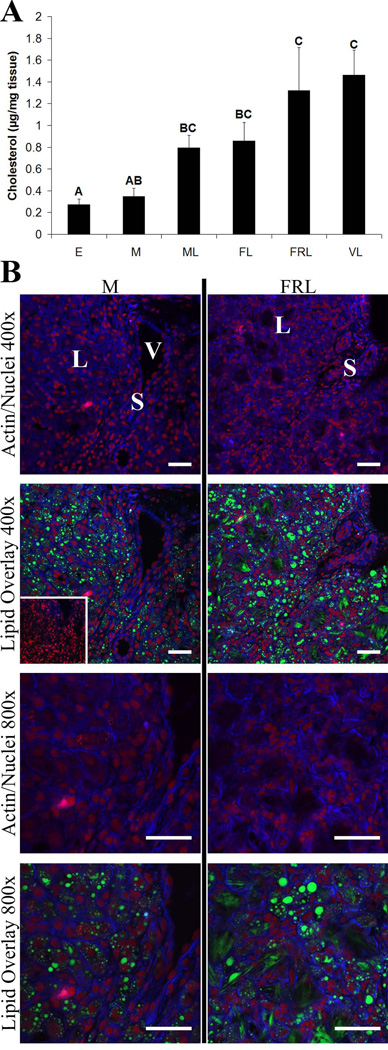
Cholesterol Levels and Localization
Cholesterol levels in luteal homogenates increased throughout the luteal phase with functionally-regressed and very-late stages having significantly higher concentrations of cholesterol (p< 0.05) than early or mid CL (Fig. 8 A). During the mid luteal phase, lipid droplets were abundant throughout large luteal cells. However, functionally-regressed late CL were associated with the formation of much larger lipid droplets or aggregates possibly residing in extracellular regions (Fig. 8 B).
Figure 8. Rhesus macaque luteal cholesterol levels and cholesterol ester/lipid droplet localization prior to and following spontaneous functional regression.
Panel A contains normalized cholesterol concentrations in CL extracts from the early (E), mid (M), mid-late (ML), functional late (FL), functionally-regressed late (FRL), and very-late (VL) stages of the luteal phase. Error bars indicate one SEM. Columns with different letters of the same case are significantly different (p< 0.05). Representative lipid droplet staining and localization results, as determined by confocal microscopy, are presented in panel B for the mid and functionally-regressed late stages. Actin is represented by blue fluorescence, nuclei are red, and neutral lipids including cholesterol esters are green. The inset in the mid 400× lipid overlay is a parallel section that was extracted with methanol to remove all lipids (background control). The approximate locations of various cell types are indicated in the first image for each stage and include: large luteal cells (L); small luteal or stromal cells (S); and blood vessels (V). The scale bar in the lower right hand corner of each image is 50 µm.
Discussion
The initial finding that stimulated further research into the RCT system in the CL was based on the divergent patterns of expression between the ABC transporters and lipoprotein receptors during normal functional regression of the primate CL (Bogan et al. 2009). In the current study, the corresponding protein levels were found to parallel previous mRNA results for ABCA1, SCARB1, and LDLR, but not ABCG1. However, ABCG1 expression was maintained throughout the periods covering functional and structural regression. When considered together, there is clearly an increase in the ratio of ABC transporters to lipoprotein receptors in the primate CL near the time of functional regression. ABCA1 is a full-size ABC transporter (i.e. containing two ATP binding cassettes and two transmembrane domain regions linked together) (Kaminski et al. 2006), which is involved in HDL metabolism by transferring unesterified cholesterol and phospholipids to lipid-free or lipid-poor APOA1 or APOE (Mahley et al. 2006). ABCG1 is a half-size ABC transporter which has been reported to cooperate with ABCA1 in cholesterol export by transferring cholesterol to nascent or pre-β HDL particles, but not lipid-free APOA1 (Gelissen et al. 2006). In murine macrophages, ABCA1 and ABCG1 are additive with regard to cholesterol efflux (Wang et al. 2007).
Expression of ABCA1, ABCG1, SCARB1, and LDLR was localized primarily to the large luteal cells. These large cells have intracellular cholesterol reserves during the luteal phase that serve as a substrate for P4 synthesis (Niswender et al. 2000; Stouffer 2006). The high steroidogenic output of the CL, estimated at up to 40 mg of P4 per day in humans (Lipsett 1978), relies primarily on blood-borne cholesterol as de novo synthesis is only important under severely lipid-deprived states (Gwynne & Strauss 1982; Niswender et al. 2000; Stouffer 2006). Thus, the balance between cholesterol uptake and efflux is likely the primary factor regulating levels of intracellular cholesterol stores. Staining for SCARB1, LDLR, and ABCG1 within the non-steroidogenic stroma was rarely observed. However, ABCA1 was frequently also expressed in regions adjacent to blood vessels. As expected for cell surface receptors, staining for SCARB1 and LDLR was observed near the plasma membrane at all stages analyzed, although SCARB1 had more pronounced membrane localization. This is likely due to differences in the lipid uptake mechanism between the two receptors as the LDLR works by receptor-mediated endocytosis which may result in its distribution between the cytoplasm and plasma membrane, whereas SCARB1 works via a selective uptake pathway not involving internalization of the receptor (Glass et al. 1983; Reaven et al. 1984). In contrast, ABCA1 and ABCG1 were primarily expressed in the cytoplasmic and perinuclear regions, with plasma membrane staining observed more frequently in functionally-regressed late CL. It has been reported that ABCG1 localization is redistributed from intracellular sites to the plasma membrane following treatment with NR1H2/NR1H3 agonists (Wang et al. 2006). Additionally, ABCA1 may be important for intracellular cholesterol movement. ABCA1 expression has been found in early and late endosomes and lysosomes, the golgi complex, as well as the plasma membrane with trafficking of ABCA1 occurring between these sites (Orso et al. 2000; Neufeld et al. 2001). Certain ABCA1 mutants found in Tangier’s disease (familial HDL-deficiency syndrome) fail to redistribute to the plasma membrane (Tanaka et al. 2003), and mice with targeted inactivation of ABCA1 have impaired cellular lipid trafficking via the trans-golgi secretory pathway (Orso et al. 2000). Thus, ABCA1 and ABCG1 redistribution within the cell may be important for RCT and may represent another mechanism whereby RCT is regulated in addition to changes in their expression.
Because apolipoproteins are produced in the liver or other tissues and secreted into the bloodstream, synthesis of apolipoproteins within the CL is not a prerequisite for RCT to occur. However, the overall level of cholesterol efflux may be limited by the amount of apolipoprotein acceptors (Santamarina-Fojo et al. 2001). As the CL is a highly steroidogenic tissue with large cholesterol reserves (Niswender et al. 2000; Stouffer 2006), intraluteal synthesis of apolipoprotein acceptors may be necessary to support the large amounts of cholesterol that would need to be disposed of during functional regression. Both APOA1 and APOE mRNA were expressed throughout the luteal phase, and relative microarray expression units of both genes ranged from approximately 2300 up to 8500, levels several-fold higher than any other RCT genes analyzed in the current study. This indicates that the primate CL may synthesize large amounts of lipoproteins that could serve as cholesterol acceptors during RCT. APOA1 expression has been primarily attributed to the liver and small intestine (Yokoyama 2006), which is consistent with our SQ-PCR data. We also detected strong APOA1 expression in the CL, as well as appreciable levels in other steroidogenic tissues including testis, ovary, and adrenal gland. APOE was more widely expressed than APOA1, although the CL had APOE levels higher or comparable to tissues richest in APOE such as the liver and adipose. While both APOA1 and APOE were expressed at high levels throughout the luteal phase, their patterns of expression were nearly opposite as APOA1 steadily decreased while APOE increased from the early through very-late stages. This indicates that the two genes are under different transcriptional control in the CL, and rising levels of APOE may compensate for decreasing APOA1.
APOA1 and APOE proteins were detected in the CL by IHC. Because these are circulating proteins, their presence in the CL may be a result of both intraluteal and extraluteal sources, although intracellular staining is likely due to intraluteal synthesis. In steroidogenic tissues, the uptake of cholesterol esters from HDL occurs without internalization of the corresponding lipoproteins (Glass et al. 1983; Reaven et al. 1984) such that non-luteal sources of APOA1 and APOE would likely remain extracellular. APOE was found in large luteal cells, in scattered regions of the stroma, and in the lining of blood vessels. APOE was predominantly localized intracellularly, especially in the perinuclear region during the mid and mid-late stages. By the late stage (functional or functionally-regressed), APOE appeared more diffuse throughout the cytoplasm. Intracellular and total luteal staining for APOE was noticeably reduced by the very-late stage as compared to the early through functional late stages, which is in contrast to its mRNA levels. Besides the fact that IHC is not a quantitative procedure, there could be a number of reasons for this apparent discrepancy including: 1) APOE mRNA levels do not match its protein levels, 2) the extraluteal contribution to APOE staining is unknown and may be lower in the functionally-regressed late to very-late stages due to decreased lipoprotein receptor expression, and 3) intraluteal APOE protein synthesis parallels mRNA levels and differences in staining intensity are due to changes in the rate of APOE secretion from luteal cells. The latter possibility is arguably the most-probable explanation as the staining pattern observed is consistent with intraluteal synthesis and storage of APOE during periods of high P4 production (mid and mid-late), followed by an enhanced secretion of APOE during functional and structural regression. Strong staining for APOA1 was observed throughout the luteal phase, with a large contribution possibly coming from extraluteal sources. Staining along the plasma membrane was noticed from the early through functionally-regressed late stages, possibly due in large part to extracellular APOA1 bound to SCARB1. However, during the very-late stage when SCARB1 is virtually absent, APOA1 localization to the plasma membrane was less pronounced while cytoplasmic APOA1 staining remained strong, consistent with intraluteal synthesis.
Similar to the apolipoproteins, luteal expression of LCAT and PLTP is not absolutely required for RCT to occur as they can arrive via the bloodstream from extraluteal sources. However, intraluteal synthesis of these proteins may enhance the efficiency of the RCT system in the CL. Both LCAT and PLTP mRNA were expressed in the CL throughout the luteal phase. Furthermore, both proteins were detected in the CL by IHC. Intracellular staining was found in large luteal cells. The non-steroidogenic stroma was mostly devoid of LCAT and PLTP except for small patches of staining. Also, staining was present in regions immediately adjacent to blood vessels. This pattern of expression is consistent with intraluteal synthesis of LCAT and PLTP by large luteal cells, followed by their subsequent secretion. LCAT is functionally-dependant on APOA1, and to a lesser extent APOE, as LCAT specifically interacts with and is activated by these apolipoproteins (Jonas 2000).
NR1H2 and NR1H3 are key regulators of the RCT system. In mice, NR1H3 is highly expressed in the liver, intestine, and adipose; but was reported to be present at low or non-detectable levels in steroidogenic tissues such as the adrenal, testis and ovary (Repa & Mangelsdorf 2000). Based on our SQ-PCR data, it appears that in the macaque NR1H3 is also highly expressed in the liver, small intestine, and adipose as was reported in mice. High levels of NR1H3 were also noted in the macaque spleen, testis and CL. In contrast, NR1H2 is ubiquitously expressed in mice (Repa & Mangelsdorf 2000), which is consistent with our findings in the macaque whereby NR1H2 mRNA levels were similar in most tissues. In the macaque CL, there was a significant increase in NR1H3 mRNA and protein prior to the drop in serum P4 levels (mid-late to functional late). Both NR1H2 and NR1H3 were expressed within the same cell types of the CL. Their expression within large luteal cells overlaps with the localization of other RCT proteins analyzed in the current study. Genes reported to be transcriptional targets of NR1H2 and NR1H3 include: ABCA1, ABCG1, PLTP, and CETP (Lund et al. 2003), NR1H3 (Laffitte et al. 2001b), as well as APOE (Laffitte et al. 2001a). With the exception of CETP which was not expressed in the macaque CL, our previous (Bogan et al. 2009) and current findings are consistent with an activation of NR1H2 and NR1H3 in the primate CL near the time of functional regression based on increased ABCA1, ABCG1, PLTP, NR1H3, and APOE mRNA levels.
This report is the first to detail the presence of components necessary for a functional RCT system in the CL of any species. The only other report on the RCT system that is relevant to the CL came from studies utilizing luteinizing granulosa cells obtained from women undergoing in vitro fertilization (Drouineaud et al. 2007). In that study, synthetic NR1H2/NR1H3 agonists inhibited P4 synthesis while stimulating mRNA expression of ABCA1, ABCG1, APOE, PLTP, and NR1H3 (Drouineaud et al. 2007). In the present study, expression of all components necessary for a functional RCT system was found within the macaque CL, and particularly within large luteal cells, including cholesterol sensors (NR1H2, NR1H3), cholesterol efflux proteins (ABCA1, ABCG1), cholesterol acceptors (APOA1, APOE), as well as plasma proteins critical for HDL maturation (LCAT, PLTP). The mRNA and/or protein levels of known NR1H2/NR1H3 target genes underwent dynamic changes near the time of functional regression in the macaque CL consistent with an induction of these receptors and consequently RCT during this period. It seems logical that an induction of RCT activity in the CL could deplete cholesterol stores and subsequently inhibit P4 synthesis. Surprisingly, however, cholesterol concentrations in luteal homogenates increased throughout the luteal phase including the period of functional regression. Analysis of lipid droplets within luteal tissue indicated that clear differences in cholesterol ester/lipid storage or localization had occurred between fully functional (mid CL) and functionally-regressed late CL. Following functional regression, droplets that were noticeably larger compared to mid CL were present, with some appearing to reside in the extracellular space. Thus, cholesterol appears to be sequestered from the steroidogenic machinery in the form of these large droplets, but is not necessarily completely removed or cleared from the tissue. Collectively, these results indicate that the RCT system may be induced in the primate CL near the time of functional regression, and consequently, may play an integral role in the cessation of P4 synthesis and ultimately luteolysis. Future studies are needed to further define the functional importance of the RCT system in the primate CL.
Materials and Methods
CL Collection
All animal protocols were approved by the Oregon National Primate Research Center’s Institutional Animal Care and Use Committee. Regularly cycling female rhesus macaques between 5 and 10 years of age were used for these studies. The care and handling of rhesus macaques was performed as described previously (Wolf et al. 1990). CL collected between days 10–12 (mid-late stage), 14–16 (late stage), or 18–19 (very-late stage) after the midcycle LH surge were used for analysis as these stages encompass the periods immediately preceding functional regression (mid-late stage), during functional regression (late stage), and structural regression (very-late stage, menses). The late stage CL were further divided into two groups (n = 4 CL/group) based on serum concentrations of P4 with CL from animals maintaining high serum levels of P4 (5.4 ± 1.2 ng/ml) considered functional late stage CL, and CL from animals having low serum levels of P4 (0.4 ± 0.1 ng/ml) considered to be functionally-regressed late stage CL as described previously (Bogan et al. 2009). In some cases, CL collected between days 3–5 (early stage, developing CL), and 7–8 (mid stage, fully functional CL) were also included in the analyses.
Messenger RNA Quantification by Microarray and Taqman Quantitative Real-Time PCR (Q-PCR) Analyses
Microarray expression data were obtained using raw fluorescence intensity files (.cel files) from two previously published microarray experiments (Bogan et al. 2009; Bogan et al. 2008a) that are accessible through the NCBI GEO database (accession numbers GSE10367 and GSE12807; http://www.ncbi.nlm.nih.gov/geo/). The raw expression intensities corresponding to early, mid, mid-late, and very-late stages (Bogan et al. 2008a), as well as functional and functionally-regressed late stages (Bogan et al. 2009) were normalized by the robust multichip average (RMA) algorithm (Irizarry et al. 2003) using Affymetrix Expression Console version 1.0 software. Probe sets corresponding to APOA1, APOE, LCAT, PLTP, CETP, NR1H2 and NR1H3 were located (http://www.affymetrix.com/analysis/index.affx) and used to isolate their expression data from the newly generated microarray expression database.
To determine whether or not a gene was expressed in the macaque CL, a database containing gene chip operating system (GCOS) absolute expression analysis data was referenced. This database was developed by the Affymetrix Microarray Core of the Oregon Health & Science University Gene Microarray Shared Resource. For each individual GeneChip genome array hybridization performed as part of two previous microarray projects (Bogan et al. 2009; Bogan et al. 2008a), a GCOS absolute expression analysis was performed on non-normalized data with parameters α1 and α2 set to 0.05 and 0.065 (Affymetrix defaults), respectively. These parameters determine whether a transcript is detected and therefore present (P) (p ≤ 0.05), marginally detected (p = 0.051 to 0.065), or not detected and thus considered to not be expressed within the CL (p > 0.065).
Q-PCR was performed as described previously (Bogan et al. 2008a) with the exception that target genes were normalized to mitochondrial ribosomal protein S10 (MRPS10). Primer and probe sequences used for Q-PCR analysis are provided in Table 1.
Table 1.
Q-PCR forward and reverse primer, as well as MGB probe sequences.
| Gene Symbol |
Forward Primer (5’-3’) | Reverse Primer (5’-3’) | Probe (5’-3’): 6FAM-Sequence-MGBNFQ MRPS10 only: VIC-Sequence-MGBNFQ |
|---|---|---|---|
| APOA1 | ACTCAAAGACAGCGGCAAAGA | GTCCCAGTTGTCCAGGAGCTT | CCTTCAAACTGGGACACATA |
| APOE | CTGGGTGCAGACACTGTCTGA | GCTGTTCCTCCAGTTCCGATT | TCAACTCCTTCATGGTCTCA |
| NR1H2 | GGGAACTGGACTTTCGTCTCAA | GAAACATTTGCTGGGTGAATGTC | TCTCTAGAATGGTCAACCCAG |
| NR1H3 | TGTGCACGAATGACTGTTCTTTT | CCCTTCTCAGTCTGTTCCACTTC | ATCCGGCCCAGAAAACAAAA |
| MRPS10 | AATGTGCCCAACCTTCATGTC | TCCAGGCAAACTGTTCCTTCA | TGAAGGCCATGCAGTCTCTCAAGTCCC |
| SQ-PCR forward and reverse primer sequences, and annealing temperature used in PCR reaction. | |||
|
Gene Symbol |
Forward Primer (5’-3’) | Reverse Primer (5’-3’) | Annealing Temperature (°C) |
| APOA1 | CCACGGCCCTTCAGGATGAA | TCAGATGCTCGCTGGCCTTG | 64° |
| APOE | GTCGCTTTTGGGATTACCTG | TGCTCCTTCACCTCGTCCAG | 53° |
| NR1H2 | TACAGGACAAGAAGCTACCG | ACATGCTCAATCAGGTTCAG | 55° |
| NR1H3 | CAGAGAGGAAGCCAGGATGC | CACTTGCGAAGCCGACACTC | 62° |
| PPIA | CCAGGGTTTATGTGTCAGGG | TGCCTTCTTTCACTTTGCCA | 56° |
Semi-quantitative PCR (SQ-PCR) was performed using cDNA generated from various rhesus macaque tissues. Reverse-transcription PCR was performed using 1 µg RNA, and the cDNA was diluted to a 100 µl final volume as described elsewhere (Bogan et al. 2008a). Primers were designed using Vector NTI software (Informax, Inc., Bethesda, MD) to amplify a region of 200–700 base pairs in length, and were contained within regions of each gene that are homologous among all known transcript variants. Primer sequences and annealing temperatures used in the PCR reactions are provided in Table 1. PCR reactions utilized 6 µl of cDNA and 400 nM of gene-specific primers in a 30 µl reaction. The reactions were stopped at 3 different cycle numbers to allow collection of 10 µl aliquots. After addition of 5× nucleic acid loading buffer (Bio-Rad, Hercules, CA), 5 µl of PCR product (10 µl for NR1H2) was resolved by 1% agarose (w/v) gel electrophoresis. To estimate the linear range of amplification, PCR was performed using CL cDNA, and the reactions were terminated at 24, 27, 30, 33, and 36 cycles. Based on these results (data not shown), SQ-PCR was performed on cDNA from 21 different tissues, and 3 different cycle numbers were attempted for each gene corresponding to mid-linear range, just prior to saturation, and post-saturation. Cyclophilin A (PPIA) was used as a housekeeping control.
Western Blot Analysis
Rabbit monoclonal antibodies produced against human antigens corresponding to SCARB1 (clone EP1556Y, catalog no. 1971), LDLR (clone EP1553Y, catalog no. 1956), and ABCG1 (clone EP1366Y, catalog no. 1945) were purchased from Epitomics, Inc. (Burlingame, CA). Mouse monoclonal antibodies produced against human antigens were used for ABCA1 (clone AB.H10, catalog no. sc-58219, Santa Cruz Biotechnology; Santa Cruz, CA) and NR1H3 (clone PPZ0412, catalog no. PP-PPZ0412, R&D Systems; Minneapolis, MN). A rabbit polyclonal antibody produced against human NR1H2 was purchased from Aviva Systems Biology (San Diego, CA; catalog no. ARP38906).
Either 12 µg (ABCA1), 20 µg (SCARB1, NR1H2), 30 µg (ABCG1, LDLR), or 50 µg (NR1H3) of protein were loaded into individual wells. Antibodies were used at the following concentrations: anti-ABCA1, 0.2 µg/ml; anti-ABCG1, 0.017 µg/ml; anti- LDLR, 0.14 µg/ml; anti-SCARB1, 0.027 µg/ml; anti-NR1H2, 1 µg/ml; and anti-NR1H3, 1.25 µg/ml. Protein levels from individual CL (n = 4 CL/stage) were normalized to β-tubulin (TUBB), and Western blot procedures and subsequent densitometry analysis were performed as described previously (Bogan et al. 2008a; Bogan et al. 2008b).
Immunohistochemistry (IHC)
The anti-ABCG1, LDLR, SCARB1, NR1H2 and NR1H3 antibodies used in Western blot analysis were also used for IHC. For ABCA1, a rabbit polyclonal antibody (catalog no. NB400-105) produced against human ABCA1 was purchased from Novus Biologicals, Inc. (Littleton, CO). A rabbit polyclonal antibody against human APOE (catalog no. 1831) and rabbit monoclonal antibody against human LCAT (clone EPR1384Y, catalog no. 2013) were purchased from Epitomics, Inc. A chicken polyclonal antibody produced against human APOA1 was purchased from GenWay Biotech, Inc. (San Diego, CA; catalog no. 15-288-20069F), and a mouse monoclonal antibody produced against human PLTP was from GeneTex, Inc. (Irvine, CA; clone 2F3-G4, catalog no. GTX90539) Antibodies were used at the following concentrations: anti-ABCA1, 3 µg/ml; anti-ABCG1, 0.33 µg/ml; anti-LDLR, 0.875 µg/ml; anti-SCARB1, 0.27 µg/ml; anti-APOE, 0.46 µg/ml; anti-APOA1, 2 µg/ml; anti-LCAT, 0.19 µg/ml; anti-PLTP, 5 µg/ml; anti-NR1H2, 3 µg/ml; and anti-NR1H3, 20 µg/ml. Specificity was determined by probing a CL section with primary antibody that had been pre-absorbed with its immunizing peptide if available (anti-ABCA1, ABCG1, LDLR, SCARB1, APOE, APOA1, NR1H2), or the primary antibody was excluded if no peptide was available for pre-absorption (anti-LCAT, PLTP and NR1H3). CL sections from 2–3 animals per stage were analyzed for each antibody. IHC analysis and image capture were performed as described previously (Bogan et al. 2008b).
Cholesterol Quantification and Localization
Cholesterol was extracted according to a published protocol with minor modifications (McDonald et al. 2007). Briefly, frozen CL were homogenized in 3 ml of PBS followed by addition of 6.6 ml of a chloroform/methanol (1:1 v:v) solution, and were vortexed for 2 minutes. Samples were centrifuged at 1400 × g for 5 minutes, and the lower organic layer was transferred to a new tube. Samples were dried under an air stream in a 37° C water bath, and re-suspended in 2 ml toluene. The lipid extracts were run over a 100 mg silica solid-phase-extraction (SPE) column (Biotage AB, Charlottesville, VA) pre-conditioned with 2 ml of hexane, and washed with 1 ml of hexane. Cholesterol was eluted with 8 ml of 0.5% (v:v) isopropanol in hexane, dried down, and re-suspended in 500 µl of methanol. Cholesterol concentrations were determined using a fluorometric assay kit (Cayman Chemical Co., Ann Arbor, MI) following the manufacturer’s recommendations with slight modifications. A final volume of 1 µl of all unknowns was used for the assay, and the methanol concentration was held constant at 1% in all unknowns and standards. Total cholesterol content was normalized to tissue weight.
For lipid droplet analysis, fresh frozen CL collected at either the mid or functionally-regressed late stages were cut in 10 µm slices. Sections were fixed with 4% paraformaldehyde and 1% calcium chloride (w:v) in TBS for 10 minutes at room temperature, and washed in 3 changes of PBS. Sections were blocked for 30 minutes at room temperature with Image-iT Signal Enhancer (Invitrogen Co., Carlsbad, CA). Sections were stained with spectrally-distinct fluorophores for actin (phalloidin-Alexa Fluor 350 conjugate, Invitrogen), nuclei (TO-PRO-3 iodide, Invitrogen), and neutral lipids including cholesterol esters (LipidTox, Invitrogen) following the manufacturer’s recommendations. A negative control for neutral lipid fluorescence was included consisting of a section extracted with methanol for 15 minutes at room temperature. Slides were imaged using a Leica TCS SP spectral confocal imaging system.
Statistical Analyses
Normalized microarray, Q-PCR, and Western blot data were log-transformed if necessary to stabilize variance. Data were analyzed using one-way ANOVA followed by pairwise comparisons with the Student-Newman-Keuls (SNK) test, and differences were considered statistically significant at p< 0.05.
Acknowledgments
The authors would like to thank Dr. Ov Slayden who provided the microscope and camera for capturing digital photomicrographs, as well as technical assistance on their use. Thanks to Dr. Marina Peluffo for assistance with confocal microscopy image capture, and Ms. Melinda Murphy for general laboratory technical assistance. We would also like to thank the following core facilities at the Oregon National Primate Research Center: Molecular and Cellular Biology Core (Drs. Eliot Spindel and Yibing Ja), and the Imaging and Morphology Core (Dr. Anda Cornea and Ms. Barbra Mason).
Funding
This work was supported by the following National Institutes of Health grants: U54 HD55744 (JDH), R01 HD42000 (JDH), RR00163 (JDH), and T32 training grant HD007133 (RLB).
Footnotes
Publisher's Disclaimer: Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in Reproduction, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the journal accepts no responsibility for any errors or omissions it may contain. The definitive version is available at doi:10.1530/REP-09-0005, 2009, Society for Reproduction and Fertility.
Declaration of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD. Systematic determination of differential gene expression in the primate corpus luteum during the luteal phase of the menstrual cycle. Molecular Endocrinology. 2008a;22:1260–1273. doi: 10.1210/me.2007-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD. Prostaglandin Synthesis, Metabolism, and Signaling Potential in the Rhesus Macaque Corpus Luteum Throughout the Luteal Phase of the Menstrual Cycle. Endocrinology. 2008b;149:5861–5871. doi: 10.1210/en.2008-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Hennebold JD. Dynamic Changes in Gene Expression That Occur During the Period of Spontaneous Functional Regression in the Rhesus Macaque Corpus Luteum. Endocrinology. 2009;150:1521–1529. doi: 10.1210/en.2008-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannian JD, Stouffer RL. Progesterone production by monkey luteal cell subpopulations at different stages of the menstrual cycle: changes in agonist responsiveness. Biology of Reproduction. 1991;44:141–149. doi: 10.1095/biolreprod44.1.141. [DOI] [PubMed] [Google Scholar]
- Cameron JL, Stouffer RL. Gonadotropin receptors of the primate corpus luteum. II. Changes in available luteinizing hormone- and chorionic gonadotropin-binding sites in macaque luteal membranes during the nonfertile menstrual cycle. Endocrinology. 1982;110:2068–2073. doi: 10.1210/endo-110-6-2068. [DOI] [PubMed] [Google Scholar]
- Drouineaud V, Sagot P, Garrido C, Logette E, Deckert V, Gambert P, Jimenez C, Staels B, Lagrost L, Masson D. Inhibition of progesterone production in human luteinized granulosa cells treated with LXR agonists. Molecular Human Reproducton. 2007;13:373–379. doi: 10.1093/molehr/gam019. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Stewart DR, Stouffer RL. Titrating luteinizing hormone replacement to sustain the structure and function of the corpus luteum after gonadotropin-releasing hormone antagonist treatment in rhesus monkeys. The Journal of Clinical Endocrinology and Metabolism. 1999;84:342–349. doi: 10.1210/jcem.84.1.5362. [DOI] [PubMed] [Google Scholar]
- Dullaart RP, Van Tol A. Role of phospholipid transfer protein and prebeta-high density lipoproteins in maintaining cholesterol efflux from Fu5AH cells to plasma from insulin-resistant subjects. Scandanavian Journal of Clinical and Laboratory Investigation. 2001;61:69–74. doi: 10.1080/00365510151068027. [DOI] [PubMed] [Google Scholar]
- Eyster KM, Ottobre JS, Stouffer RL. Adenylate cyclase in the corpus luteum of the rhesus monkey. III. Changes in basal and gonadotropin-sensitive activities during the luteal phase of the menstrual cycle. Endocrinology. 1985;117:1571–1577. doi: 10.1210/endo-117-4-1571. [DOI] [PubMed] [Google Scholar]
- Gelissen IC, Harris M, Rye KA, Quinn C, Brown AJ, Kockx M, Cartland S, Packianathan M, Kritharides L, Jessup W. ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:534–540. doi: 10.1161/01.ATV.0000200082.58536.e1. [DOI] [PubMed] [Google Scholar]
- Glass C, Pittman RC, Weinstein DB, Steinberg D. Dissociation of tissue uptake of cholesterol ester from that of apoprotein A-I of rat plasma high density lipoprotein: selective delivery of cholesterol ester to liver, adrenal, and gonad. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:5435–5439. doi: 10.1073/pnas.80.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwynne JT, Strauss JF., 3rd The role of lipoproteins in steroidogenesis and cholesterol metabolism in steroidogenic glands. Endocrine Reviews. 1982;3:299–329. doi: 10.1210/edrv-3-3-299. [DOI] [PubMed] [Google Scholar]
- Hoyer PB. Regulation of luteal regression: the ewe as a model. Journal of the Society for Gynecologic Investigation. 1998;5:49–57. doi: 10.1016/S1071-5576(97)00105-6. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jonas A. Lecithin cholesterol acyltransferase. Biochimica et Biophysica Acta. 2000;1529:245–256. doi: 10.1016/s1388-1981(00)00153-0. [DOI] [PubMed] [Google Scholar]
- Kaminski WE, Piehler A, Wenzel JJ. ABC A-subfamily transporters: structure, function and disease. Biochimica et Biophysica Acta. 2006;1762:510–524. doi: 10.1016/j.bbadis.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Laffitte BA, Repa JJ, Joseph SB, Wilpitz DC, Kast HR, Mangelsdorf DJ, Tontonoz P. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proceedings of the National Academy of Sciences of the United States of America. 2001a;98:507–512. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Joseph SB, Walczak R, Pei L, Wilpitz DC, Collins JL, Tontonoz P. Autoregulation of the human liver X receptor alpha promoter. Molecular and Cellular Biology. 2001b;21:7558–7568. doi: 10.1128/MCB.21.22.7558-7568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsett MB. Steroid Hormones. In: Yen SC, Jaffe RB, editors. Reproductive Endocrinology. Philadelphia: WB Saunders Press; 1978. pp. 80–92. [Google Scholar]
- Lund EG, Menke JG, Sparrow CP. Liver X receptor agonists as potential therapeutic agents for dyslipidemia and atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003;23:1169–1177. doi: 10.1161/01.ATV.0000056743.42348.59. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Huang Y, Weisgraber KH. Putting cholesterol in its place: apoE and reverse cholesterol transport. The Journal of Clinical Investigation. 2006;116:1226–1229. doi: 10.1172/JCI28632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JG, Thompson BM, McCrum EC, Russell DW. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods in Enzymology. 2007;432:145–170. doi: 10.1016/S0076-6879(07)32006-5. [DOI] [PubMed] [Google Scholar]
- Neufeld EB, Remaley AT, Demosky SJ, Stonik JA, Cooney AM, Comly M, Dwyer NK, Zhang M, Blanchette-Mackie J, Santamarina-Fojo S, et al. Cellular localization and trafficking of the human ABCA1 transporter. The Journal of Biological Chemistry. 2001;276:27584–27590. doi: 10.1074/jbc.M103264200. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Stulnig TM, Lin CY, Yeo AL, Nowotny P, Liu ET, Steffensen KR. Liver X receptors regulate adrenal steroidogenesis and hypothalamic-pituitary-adrenal feedback. Molecular Endocrinology. 2007;21:126–137. doi: 10.1210/me.2006-0187. [DOI] [PubMed] [Google Scholar]
- Niswender GD, Juengel JL, Silva PJ, Rollyson MK, McIntush EW. Mechanisms controlling the function and life span of the corpus luteum. Physiological Reviews. 2000;80:1–29. doi: 10.1152/physrev.2000.80.1.1. [DOI] [PubMed] [Google Scholar]
- Orso E, Broccardo C, Kaminski WE, Bottcher A, Liebisch G, Drobnik W, Gotz A, Chambenoit O, Diederich W, Langmann T, et al. Transport of lipids from golgi to plasma membrane is defective in tangier disease patients and Abc1-deficient mice. Nature Genetics. 2000;24:192–196. doi: 10.1038/72869. [DOI] [PubMed] [Google Scholar]
- Reaven E, Chen YD, Spicher M, Azhar S. Morphological evidence that high density lipoproteins are not internalized by steroid-producing cells during in situ organ perfusion. The Journal of Clinical Investigation. 1984;74:1384–1397. doi: 10.1172/JCI111549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annual Review of Cell and Developmental Biology. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- Santamarina-Fojo S, Remaley AT, Neufeld EB, Brewer HB., Jr Regulation and intracellular trafficking of the ABCA1 transporter. Journal of Lipid Research. 2001;42:1339–1345. [PubMed] [Google Scholar]
- Stouffer RL. Progesterone as a mediator of gonadotrophin action in the corpus luteum: beyond steroidogenesis. Human Reproduction Update. 2003;9:99–117. doi: 10.1093/humupd/dmg016. [DOI] [PubMed] [Google Scholar]
- Stouffer RL. Structure, function, and regulation of the corpus luteum. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. Vol. 3. New York: Raven Press; 2006. pp. 475–526. [Google Scholar]
- Tanaka AR, Abe-Dohmae S, Ohnishi T, Aoki R, Morinaga G, Okuhira K, Ikeda Y, Kano F, Matsuo M, Kioka N, et al. Effects of mutations of ABCA1 in the first extracellular domain on subcellular trafficking and ATP binding/hydrolysis. The Journal of Biological Chemistry. 2003;278:8815–8819. doi: 10.1074/jbc.M206885200. [DOI] [PubMed] [Google Scholar]
- Von Eckardstein A, Jauhiainen M, Huang Y, Metso J, Langer C, Pussinen P, Wu S, Ehnholm C, Assmann G. Phospholipid transfer protein mediated conversion of high density lipoproteins generates pre beta 1-HDL. Biochimica et Biophysica Acta. 1996;1301:255–262. doi: 10.1016/0005-2760(96)00050-1. [DOI] [PubMed] [Google Scholar]
- Wang N, Ranalletta M, Matsuura F, Peng F, Tall AR. LXR-induced redistribution of ABCG1 to plasma membrane in macrophages enhances cholesterol mass efflux to HDL. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:1310–1316. doi: 10.1161/01.ATV.0000218998.75963.02. [DOI] [PubMed] [Google Scholar]
- Wang X, Collins HL, Ranalletta M, Fuki IV, Billheimer JT, Rothblat GH, Tall AR, Rader DJ. Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. The Journal of Clinical Investigation. 2007;117:2216–2224. doi: 10.1172/JCI32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcicka G, Jamroz-Wisniewska A, Horoszewicz K, Beltowski J. Liver X receptors (LXRs). Part I: structure, function, regulation of activity, and role in lipid metabolism. Postȩpy Higieny I Medycyny Doświadczalnej (Online) 2007;61:736–759. [PubMed] [Google Scholar]
- Wolf DP, Thomson JA, Zelinski-Wooten MB, Stouffer RL. In vitro fertilization-embryo transfer in nonhuman primates: the technique and its applications. Molecular Reproduction and Development. 1990;27:261–280. doi: 10.1002/mrd.1080270313. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Assembly of high-density lipoprotein. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:20–27. doi: 10.1161/01.ATV.0000195789.39418.e8. [DOI] [PubMed] [Google Scholar]
- Zaiou M, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH, Segall ML, Phillips MC, Lund-Katz S. Apolipoprotein E;-low density lipoprotein receptor interaction. Influences of basic residue and amphipathic alpha-helix organization in the ligand. Journal of Lipid Research. 2000;41:1087–1095. [PubMed] [Google Scholar]