Abstract
Recent advances in gene knockout techniques and the in vivo analysis of mutant mice, together with the advent of large-scale projects for systematic mouse mutagenesis and genome-wide phenotyping, have allowed the creation of platforms for the most complete and systematic analysis of gene function ever undertaken in a vertebrate. The development of high-throughput phenotyping pipelines for these and other large-scale projects allows investigators to search and integrate large amounts of directly comparable phenotype data from many mutants, on a genomic scale, to help develop and test new hypotheses about the origins of disease and the normal functions of genes in the organism. Histopathology has a venerable history in the understanding of the pathobiology of human and animal disease, and presents complementary advantages and challenges to in vivo phenotyping. In this review, we present evidence for the unique contribution that histopathology can make to a large-scale phenotyping effort, using examples from past and current programmes at Lexicon Pharmaceuticals and The Jackson Laboratory, and critically assess the role of histopathology analysis in high-throughput phenotyping pipelines.
Introduction
It was six men of Hindustan
To learning much inclined,
Who went to see the Elephant
(Though all of them were blind),
That each by observation
Might satisfy his mind…And so these men of Hindustan
Disputed loud and long,
Each in his own opinion
Exceeding stiff and strong,
Though each was partly in the right
And all were in the wrong.An extract from The Blind Men and the Elephant by John Godfrey Saxe (1816–1887)
Use of the mouse as a model organism has proved to be one of the most powerful approaches available in our efforts to understand and cure human disease (Sundberg, 1991; Peters et al., 2007; Rosenthal and Brown, 2007). Mutant mice not only provide models for major aspects of disease, serving as preclinical tools for drug discovery and efficacy testing (Zambrowicz and Sands, 2003; Zambrowicz et al., 2003a; Antony et al., 2011; Sutherland and Berns, 2011; Van Dam and De Deyn, 2011), but the phenotyping of spontaneous and genetically engineered mutant mice also generates a great deal of fundamental information about gene function that can then be used to explore the underlying pathobiology.
Through hypothesis-driven research and the use of spontaneous and induced mutations, we now have at least partial phenotypic information on just over a third of the ∼25,000 mouse genes with associated protein sequence data (see Box 1). The remainder of the genome remains closed to us, a black box that can be accurately characterised as the ‘ignorome’ – that set of protein-coding genes about which we know little apart from their genomic location and predicted exon structure. Within the ignorome lie thousands of genes whose functions hold the answer to many questions about normal physiology and disease; a major challenge of the 21st century is how we can most efficiently gain traction on this problem and create an ‘encyclopaedia’ of gene function for every gene in the genome. Over the last decade, whole-genome approaches for the discovery of gene function and new models of human disease have been developed using the mouse. These have been either phenotype driven – using ENU mutagenesis (Munroe et al., 2000; Acevedo-Arozena et al., 2008), gene trapping (Hansen et al., 2008) or insertional mutagenesis (Ivics et al., 2009) – or genotype driven through the use of targeted knockouts (Skarnes et al., 2011). All of these approaches require the use of high-throughput phenotyping pipelines (see Box 2) to characterise large numbers of animals for a wide range of phenotypic parameters; for example, the German Mouse Clinic measures more than 550 parameters in two cohorts of ten mice of each sex and genotype (Fuchs et al., 2009; Fuchs et al., 2010; Gates et al., 2010; Gailus-Durner et al., 2011). High-throughput phenotyping pipelines are central to the recently launched International Mouse Phenotyping Consortium (IMPC) project (http://www.mousephenotype.org), which has set out to phenotype knockouts for every protein coding gene in the mouse genome (Abbott, 2010). These pipelines have also been implemented in discovery strategies in the pharmaceutical industry and other major academic laboratories.
Box 1. Functional characterisation of the mouse genome.
Data from the Mouse Genome Database (Blake et al., 2011) currently (October 2011) lists 15,226 genes with mutant alleles in mice, and phenotypic information is currently available for ∼8200 of the ∼25,000 mouse genes with associated protein sequence data. At the level of protein class or Gene Ontology (GO) molecular function annotation, we only have 13,591 genes with any experimentally based functional annotations and, of the 8600 GO molecular function terms, most have very few associated gene products. Only 7000 genes have GO-biological process annotations in the mouse.
Box 2. What is a high-throughput phenotyping pipeline?
Phenotyping pipelines comprise a series of sequential assays carried out on the same cohort of mice; these assays measure a broad set of phenotypic parameters, with a high rate of throughput. The breadth of these predominantly in vivo assays is key to the pipeline approach, which is designed to detect a wide spectrum of phenotypes ranging, for example, from behaviour to dysmorphology, clinical chemistry and immune responses (Justice, 2008; Mandillo et al., 2008; Brown et al., 2009; Gates et al., 2010). The complexity of phenotypes assayed is directly proportional to the cost and inversely proportional to throughput; as assays become more labour intensive, the number of mice that can feasibly be examined decreases. This then requires the stratification of assays into primary, secondary and tertiary phenotyping efforts, whereby mice from the primary pipeline are selected for additional in-depth analysis on the basis of phenotypes uncovered in the first battery of assays.
In this Special Article, we explore the implementation and value of histopathological analysis in high-throughput mouse phenotyping pipelines, its complementarity to in vivo assays, and its unique contribution to our understanding of disease.
Histopathology as a component of high-throughput phenotyping pipelines
Histopathology is the systematic and systemic analysis of the microscopic morphological correlates of disease through examining tissues obtained from necropsy or biopsy under the microscope. Macroscopic (gross) analysis is sometimes referred to as anatomic pathology, although this term is more properly used to denote the whole discipline or medical specialty. In the current context, the pathologist examines a mouse at both micro- and macroscopic levels. Histopathology is indispensable for any high-throughput phenotype screening programme because it can identify phenotypes not detected by any other physiological or behavioural screening assays and provide contextualisation for in vivo findings. It also provides an overview of the disease processes ongoing in the whole organism and aids the interpretation of in vivo assays, such as blood cell counts, urinalysis and gait, etc. Like the elephant in the poem by John Saxe quoted above, examining only part of a phenotype, however precisely, is likely to lead to serious errors; thus, the whole phenotype must be characterised to obtain an accurate description.
Along with the development and standardisation of physiological and in vivo phenotyping methodologies over the last decade, considerable experience has been accumulated of the use of high-throughput histopathology for the characterisation of mice from systematic screening programmes in both academia and the pharmaceutical industry (Beltrandelrio et al., 2003; Sundberg and Ichiki, 2005; Yuan et al., 2009). The German Mouse Clinic, for example, has included histopathology in its pipeline since 2001 (Fuchs et al., 2009; Gailus-Durner et al., 2009; Fuchs et al., 2010), and the Toronto Centre for Phenogenomics runs histopathological analysis as an integral part of its phenotyping pipeline. Other centres have used detailed histological phenotyping for several decades as a standard first screen for defining mouse models of human diseases, using the physiological methods described above as confirmatory assays only as needed (Sundberg and Boggess, 1998; Sundberg et al., 2000; Sundberg et al., 2011). At a recent meeting in Bordeaux, France, which was organised under the auspices of EUMODIC (European Mouse Disease Clinic) and the Cancéropôle Grand Sud-Ouest, a group of pathologists and geneticists from academia and industry critically examined the advantages and problems associated with various approaches to running high-throughput histopathology phenotyping (Schofield et al., 2011); the reader is referred to this source for additional discussion and insights from a wide range of experiences.
High-throughput histopathology phenotyping in drug discovery
Evidence that the pathological processes underlying the abnormal phenotypes of knockout mice often mirror the therapeutic targets of many drugs in humans has spurred the use of knockout mice in drug discovery research (Zambrowicz and Sands, 2003). In the past decade, pharmaceutical companies have developed and utilised comprehensive high-throughput phenotype screening protocols to identify potentially useful drug targets and toxicity liabilities in genetically engineered mice. Because a primary objective of pharmaceutical companies is to discover drug targets and any related risks in the most timely and cost-effective manner possible, we believe that their approaches to phenotyping can serve as useful templates in designing a rational and cost-effective approach to high-throughput phenotyping of knockout mice in other research settings.
In order to recoup the considerable investment made in generating a new line of genetically engineered mice, comprehensive phenotyping programmes are used in pharmaceutical development to ensure that unexpected and valuable phenotypes are not missed. Experience has shown that phenotype screening programmes must be comprehensive because completely novel and unexpected phenotypes seem to appear at least as often as those predicted by specific hypotheses. The phenotyping protocols used in the pharmaceutical industry are usually performed in a centralised facility, and include assays that permit the rapid and cost-effective assessment of a wide range of physiological parameters. They are generally restricted to a carefully selected and validated battery of rapid and relatively straightforward physiological and behavioural assays.
In a programme at Lexicon Pharmaceuticals, which generated and analysed over 4650 knockout mouse lines in a high-throughput mutagenesis and phenotyping process that was designed to characterise protein functions and identify novel drug targets (Zambrowicz et al., 2003b; Zambrowicz et al., 2003a), many phenotypes were discovered that have proven useful in elucidating fundamental processes in biology and the pathogenesis of disease. Important phenotypes would have been missed if the use of histopathology in high-throughput screening had been restricted only to those lines showing early mortality or other physiological or behavioural phenotypes, emphasising the importance of histopathology in a primary screen. Even in those cases in which a phenotype was initially identified with another screening test, histopathology findings were invaluable in elucidating the underlying structural basis for physiological or behavioural phenotypes. Below, we provide some examples of the value of histopathological analysis in high-throughput screens used in evaluating knockout mouse lines at Lexicon.
Case studies from high-throughput histopathological analysis
Advantages of the systemic approach
In many knockout lines, histopathology has proved to be the most efficient and precise method to detect phenotypes in specific organ systems, including the upper respiratory tract, teeth, eyes, muscle, kidney and skin. In fact, in the Lexicon screen, histopathology was the only assay included in a comprehensive battery of tests that revealed a phenotype in many knockout lines. For example, although some tooth abnormalities are grossly visible, most reported tooth phenotypes require histopathology for full characterisation (Bei, 2009), and several knockout phenotypes were characterized by significant enamel or dentin defects that were only detected histologically (P.V., unpublished observations). Detection of tooth phenotypes by histopathology is rapid and cost effective because the cross-sections of the nose that are included in standard histopathology tissue sets include incisor and molar teeth in addition to the nasal passageways. Evaluation of these nasal sections also permits rapid diagnosis of primary ciliary dyskinesia in mice, which can also be characterised by male infertility and hydrocephalus, but most commonly presents as a clinically asymptomatic suppurative rhinosinusitis that is best detected by histological evaluation of nasal passageways (Vogel et al., 2010a).
The eye is another organ system in which histopathological evaluation provides unique advantages. Although many ocular defects can be detected by a combination of fundoscopy, angiography and intraocular pressure measurements, virtually all of these defects can be diagnosed less expensively and more definitively by histopathology (Smith et al., 2002). More importantly, the earliest lesions in many types of progressive retinal degeneration can only be detected by histopathology, so this technique is of crucial importance in high-throughput phenotyping because cost constraints generally require the use of relatively young mice. Interestingly, we also found that pigmentation defects are most easily assessed in the eye, where the close proximity of both neural-crest-derived melanocytes and pigmented neuroepithelial cells allows direct comparisons. This unique arrangement can accentuate differences in pigmentation and in fact led directly to the identification of an important hypopigmentation gene (Slc24a5) in mice that had not been found previously among any of the hundreds of pigment variants present in this organism. The effects of Slc24a5 on pigmentation had been missed previously because mutation of this gene does not have any grossly detectable effects on hair coat colour (Vogel et al., 2008). Although screening tests to detect lighter skin in mice could be developed to find additional pigmentation genes, these assays would be more time consuming and expensive than histological evaluation of eye pigmentation.
Early discovery of age-related degenerative and neoplastic phenotypes
Another important advantage provided by inclusion of histopathology in initial phenotype screens is that many phenotypes can be identified histologically before behavioural or physiological abnormalities are detectable. High-throughput phenotyping is generally performed on young mice in order to reduce husbandry costs, but it is clear that age-related phenotypes are missed when only young mice are examined, because these mice are at a stage when age-related phenotypes might not be sufficiently developed to cause metabolic, morphological or behavioural problems (Myers, 1978; Bronson, 1990; Mohr et al., 1996; Lipman, 1997).
As mentioned above, retinal degeneration can be readily detected by histopathology in young mice. In addition, we found that an increased frequency of hepatocyte mitoses detected in otherwise normal young mice is a reliable predictor of liver disease and early onset hepatic neoplasms (Read et al., 2009). Similarly, numerous ubiquitin-positive eosinophilic bodies were detected in the deep cerebellar and vestibular nuclei of young Atg4b-deficient mice [Atg4bGt(OST264114)Lex] (Read et al., 2011) long before they showed the measurable impairment of motor performance caused by knocking out this gene. In several other lines, widespread centralisation of myocyte nuclei was seen without detectable muscle weakness, high serum creatine kinase levels or prominent inflammatory infiltrates, demonstrating a muscle phenotype that was not revealed at this stage of the disease by physiological assays. Others have shown that evaluation of the diaphragm muscle can permit early detection of myopathies that would otherwise go undetected in young mice (Barton et al., 2010).
As noted previously, because histopathology encompasses evaluation of the whole animal, genetic defects can result in a characteristic pattern of lesions in multiple cell and tissue types that, when found in other lines of knockout mice, facilitate the recognition of previously unsuspected relationships between genes. Separate lines of knockout mice with similar pathological phenotypes have facilitated the discovery of important pathogenetic mechanisms, ligand-receptor pairs, cofactors or proteins located along the same metabolic pathways. For example, the frequent association in many lines of knockout mice of hydrocephalus (accumulation of cerebrospinal fluid in the cerebral ventricles) with chronic upper respiratory lesions caused by deficient ciliary clearance clearly demonstrated that dysfunctional motile cilia on ependymal cells were central to the pathogenesis of this devastating neurological condition (Vogel et al., 2010a; Vogel et al., 2010b).
In another case, finding lesions in two knockout lines of mice deficient for the enzymes that were later determined to be responsible for mucolipidosis (ML) types II and IIIC illustrates how histopathology can detect otherwise hidden phenotypes, identify genes with related functions and even elucidate fundamental pathogenetic processes (Vogel et al., 2009). With the exception of histopathology, no notable phenotypes were detected in a line of N-acetylglucosamine-1-phosphotransferase, γ subunit knockout mice (Gnptgtm1Lex/Gnptgtm1Lex) that had been subjected to comprehensive phenotypic analysis involving a wide array of tests. However, histopathological evaluation of tissues from these asymptomatic mice revealed widespread cytoplasmic vacuolisation and inclusions in secretory cells in several glandular tissues. We had previously seen a very similar disease phenotype in secretory cells in Gnptab (subunits α/β) homozygous knockout mice [GnptabGt(OST97730)Lex]. The many similarities alerted us to the fact that the genes involved in these two lines were related, and it was later found that the affected genes encode different subunits of the same enzyme complex, and that mutations in these subunits had been linked to the development of MLII and MLIII in humans. Notably, neither line of mutant mice developed the skeletal and connective tissue abnormalities or the vacuolar cytoplasmic alterations in fibrocytes or mesenchymal cells that characterise the lesions of ML in humans. Instead, the predominant lesions in both lines of mice were found in the secretory epithelial cells of several exocrine glands, including the pancreas and the parotid, submandibular salivary, nasal, lacrimal, bulbourethral and gastric glands. We initially believed that mice lacking either the α, β or γ subunits displayed clinical and pathological features that differed substantially from those reported in humans having mutations in orthologous genes, but then found a single report describing similar findings in secretory cells of the exocrine pancreas, salivary glands and gastric chief cells in patients with MLII (Elleder and Martin, 1998). The significance of these earlier findings was not fully appreciated until the pathological phenotypes in knockout mice were published (Gelfman et al., 2007; Vogel et al., 2009).
Together, these papers support a novel hypothesis that implicates secretory dysfunction rather than a classic lysosomal storage disease in the pathogenesis of the connective tissue (bone and collagen) lesions seen in human MLII and MLIII. Chondrocytes and fibroblasts are not generally thought of as secretory cells, but they do secrete large amounts of collagen and cartilage matrix during periods of rapid growth. The novel insights into the pathogenesis of these diseases in the mouse have contributed to additional investigations into the underlying pathobiology in humans (Flanagan-Steet et al., 2009; Boonen et al., 2011), and serve as an example of the advances made possible through histopathology phenotyping of seemingly normal knockout mice.
Histopathology contextualises in vivo findings
So far, we have provided examples that illustrate the indispensable role of histopathology as a first-order assay in discovering gene function. The in vivo assays in a pipeline are designed to detect a wide range of phenotypes, but each assay probes a specific aspect of the phenome and in few cases do assays provide definitive diagnostic or aetiological information about the overall disease process in the whole organism. The ancient Buddhist story referred to in Saxe’s poem is an excellent metaphor for this – that is, that each assay probes and evaluates a different part of the ‘elephant’. Unfortunately, this means that striking findings in a single assay can be misleading to investigators if taken out of context. In histopathology evaluations, the microscopic structure of virtually every organ system is considered and findings are noted in the context of the whole animal. Histopathology is therefore well equipped to provide a context for abnormal findings obtained from other physiological and behavioural assays, and can facilitate the integration of all findings to give a more complete and accurate picture of the knockout phenotype. For example, blood pressure is a physiological parameter commonly measured in high-throughput screening, with the intent to discover potential drug targets for treating hypertension. In some cases, histopathological evaluation has proven invaluable in determining whether underlying structural defects in the myocardium or other problems are responsible for the low blood pressure readings (Van Sligtenhorst et al., 2011). Similarly, drug target discovery screening for osteoporosis relies on bone density measurements provided by techniques such as microCT assay. However, histopathological evaluation of bone sections can quickly and reliably indicate whether the measured increases in bone density are likely to result in increased bone strength (as in sclerostin knockout mice) (Li et al., 2008) or increased bone fragility (as in cathepsin-K knockout mice) (Li et al., 2006). Yet another line of knockout mice showing increased bone density concurrently displayed a desirable lean phenotype by whole body dual-energy X-ray absorptiometry (DEXA) scans and body weight measurements. Histopathology confirmed the increased bone density but also revealed the presence of defects in tooth dentin accompanied by severe periodontal inflammation, which provided a contextual explanation for the lean phenotype (P.V., unpublished observations). In other cases, histopathology showed that other knockout lines found to have low body weight or lean phenotypes using physiological measurements had other precipitating causes, including protein-losing glomerulopathies or enteropathies, and malabsorption syndromes.
Cost-effectiveness of histopathology screening
One major challenge facing the field of functional genomics is the need to accurately determine the in vivo function of all genes in the most timely and cost-effective manner possible. Histopathology is not inherently a rapid or inexpensive assay, but the power of this ‘whole animal’ method of evaluation makes it an essential component of any high-throughput screening programme. The successful inclusion of histopathology in phenotyping screening programmes used in pharmaceutical drug discovery efforts proves that it is both worthwhile and possible to control the costs of histopathology evaluation, thereby reducing the numbers of missed phenotypes (false negatives). In the setting of a pharmaceutical programme, the cost effectiveness of every assay included in a testing regimen is evaluated continuously, and only those that contribute significantly to the drug development process are continued. As a result, very strong justification is required to incorporate and continue any labour-intensive or time-consuming assay as part of a high-throughput phenotyping screen. It is therefore noteworthy that histopathology phenotyping – which is both labour intensive and time consuming – was always included in the battery of primary screening assays used in evaluating knockout mice lines at both Genentech and Lexicon Pharmaceuticals.
A major cost driver in many proposed histopathology screens is the large number of knockout mice and wild-type controls often said to be needed. However, based on our experience of phenotyping over 4500 lines, it is unnecessary to include wild-type control mice with every knockout line, provided knockout mice are generated on a standard genetic background. In fact, the numbers of wild-type mice evaluated can be further reduced over time as pathologists gain experience with the lines used. The relatively low numbers of mice (two males and two females) proposed for use in an initial screen is based on our experience of performing histopathology phenotyping on 472 lines of knockout mice with targeted mutations in transmembrane and secreted proteins (Tang et al., 2010). Interestingly, over 100 of these lines had previously been evaluated by pathologists following a protocol that allowed only two knockout mice (one male and one female) in the initial screen, with provision for additional mice to be tested in cases in which suspected pathology phenotypes were found. Among these 100 lines, only one additional phenotype was detected by doubling the number of mice evaluated, and in this lone exception (Pkd1l1tm1Lex homozygous knockout mice) (Vogel et al., 2010c) the only additional phenotype noted that was missed on initial evaluation was situs inversus (reversed orientation of internal organs), which was present in less than 50% of mice lacking the gene.
On the basis of prior experience, it is likely that a rapid, accurate and cost-effective histopathological screen will require a minimum of two males and two female knockout mice; the establishment of uniform tissue collection, fixation and processing methods; and the evaluation of tissue sections by pathologists with expertise in specific organ systems. The standardisation of tissue collection, fixation and processing procedures is essential to control costs while improving efficiency and simultaneously ensuring top quality histopathological evaluations. Nevertheless, the key requirement for keeping the costs of histopathology low while keeping the quality and reliability of findings high is to have expert pathologists perform the evaluations. There are several reasons for this. An expert can very quickly recognise subtle but real lesions in a specific tissue that even an experienced pathologist might miss or need additional time to research and understand. In addition, an expert will already be familiar with and quickly identify incidental or background changes of no consequence that sometimes occur in individual mice (thereby reducing the number of false positives and time). Having inexperienced pathologists or post-docs perform pathology screening will inevitably increase the numbers of both missed (false-negative) and spurious (false-positive) phenotypes.
Training in the specialist area of mutant mouse pathology has received much discussion in recent years, with considerable concern about the availability of qualified and experienced pathologists (Barthold et al., 2007; Schofield et al., 2009). Recent investment in training programmes in France (Schofield et al., 2011) and the emergence of courses associated with the Centre for Genomic Pathology (http://ctrgenpath.net/) in the United States, together with existing programmes (Sundberg et al., 2007; Sundberg et al., 2010), are cause for optimism. These developments reflect a renewed interest among young veterinary pathologists in this new and expanding area, although further momentum needs to be built and maintained.
Histopathology and informatics
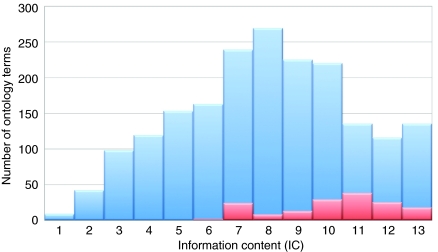
It is difficult to make a quantitative estimate of the relative value of histopathology versus in vivo phenotyping to the overall use of phenotype data. We believe that the examples discussed above more than adequately demonstrate the scientific value of such phenotyping. Recently, however, the development of the mammalian and human phenotype ontologies (Robinson et al., 2008; Smith and Eppig, 2009) – which describe phenotypes associated with mouse mutants or human diseases – has allowed us to quantitatively estimate the usefulness of different types of mouse phenotype data in discovering candidates for human diseases. The graph shown in Fig. 1 represents the information content (IC; i.e. the discriminating ability) of terms in the mammalian phenotype ontology that contain pathology information (red bars), mapped onto the total disease information in Online Mendelian Inheritance in Man (OMIM; blue bars). It is clear that the phenotype ontology terms involving histopathology occupy a position at the upper end of the IC (Shannon, 1948) spectrum compared with most in-vivo-measured phenotypes, and thus will be much more powerful for finding related phenotypes in mice and humans.
Fig. 1.
Information content (IC) of mammalian phenotype ontology terms in the OMIM dataset. Blue columns represent all phenotype ontology terms and red columns those terms that are defined using terms from the mammalian pathology ontology (MPATH). Mammalian phenotype ontology terms were used to automatically annotate all OMIM diseases by mapping between the human and mammalian phenotype ontology with PhenomeBlast software (http://bioonto.gen.cam.ac.uk:9090/PhenomeBlast-0.1/). The IC was then calculated as the probability that a given ontology term is used in the annotation of an OMIM disease. For example, abnormal circulating lipid level (MP:0003949) is a relatively undiscriminating measurement because it is found as an annotation to 430 OMIM diseases; thus, the IC for this term is low. By contrast, xanthoma (deposition of cholesterol-rich lipids under the skin; MP:0003692) has a much greater ability to discriminate between diseases because it is only found in 17 diseases in OMIM, and the IC for this term is high. Less than 10% of the phenotype ontology terms are defined using MPATH, but the spectrum of IC values shows that terms defined using MPATH are more informative and therefore more discriminating than the bulk of the phenotype ontology terms.
The use of standardised nomenclature for lesions, based on accepted ontologies such as the mammalian pathology ontology (MPATH) (Schofield et al., 2010a), should be an absolute requirement for data capture from high-throughput pathology phenotyping. Standardised nomenclature not only facilitates the retrieval of data shared through public databases, but also allows quantitative analysis of phenotyping experiments (Sundberg et al., 2011) and permits the use of this data for bioinformatic approaches that aim to discover human disease genes (Schofield et al., 2010b) by allowing the computational analysis of data from human and mouse phenotype knowledgebases (Washington et al., 2009; Hoehndorf et al., 2011).
Conclusions
Experience gained over the last decade from the use of histopathological analysis in high-throughput mouse phenotyping programmes provides insights into its power and complementarity to in vivo phenotyping assays. High-throughput histopathology is logistically feasible and can detect phenotypes that other assays are unable to identify, especially the early-onset aspects of age-related disease that do not manifest as physiological or behavioural perturbations at the age when in vivo assays are conducted. This is particularly important because most phenotyping pipelines use mice that are less than 20 weeks of age for reasons of cost.
Importantly, histopathological analysis needs to be carried out by experts to be reliable, and the availability of pathologists with sufficient experience of genetically engineered mice is frequently a concern. Acknowledgement of the importance of this expertise by funding agencies and new training programmes in some countries, as well as renewed enthusiasm for the specialty by trainees, bode well for the future, although more investment is needed. We believe, however, that the availability of experienced pathologists is unlikely to be the limiting factor in setting up high-throughput pathology programmes at the moment. In addition, the situation has been helped by the maturation of whole-slide scanning and viewing technologies, which provide a viable way to share expertise and data internationally (Potts, 2009), making the most of the global pool of expertise.
Standardisation of terminologies and the adoption of defined ontologies also enhance the ability to share and combine data, and to use computation to relate phenotypes derived from histopathological analysis to human disease. Propagation and development of these standards, together with training in basic informatics for pathologists, would add still further to the efficiency and usefulness of the approach.
When Nusslein-Vollhard and Wieschaus first conducted their saturation mutagenesis screen of Drosophila in the late 1970s, the prospect of having a phenotypic characterisation for a mutation in every gene in a mammal seemed distant to the point of being unattainable. In the case of flies, the phenotypes came first and then the sequences, with the fruit fly genome being announced complete in 2000. The complete genome sequence of the mouse was available before the large-scale mutagenesis projects began (Mouse Genome Sequencing Consortium, 2002); it is owing to the combination of having the complete genomic sequence, advances in targeted mutagenesis and the development of high-throughput technologies that we are now looking at the prospect of completion of a systematic encyclopaedia of gene function for the Mouse Genome by 2021 through the IMPC project. The inclusion of gross pathology and histopathology in the recommendations for phenotyping in this – the largest project of its kind ever undertaken – will contribute not only to our interpretation of the in vivo assays planned, but will also improve our ability to discover and understand mouse models of human disease.
Acknowledgments
The authors thank Cynthia Smith for helpful comments and providing statistics on phenotype annotation from the Mouse Genome Database.
Footnotes
FUNDING
This work was supported by the Ellison Medical Foundation [grant number AG-IA-0201-05] (to J.P.S.); the National Institutes of Health [National Institute of Aging Nathan Shock Center AG-25707, CA89713 and AR056635 (to J.P.S.); and R01 HG004838-02 (to P.N.S.)]; the Commission of the European Union [EUMODIC contract number LSHG-CT-2006-037188] (to P.N.S.); and by the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BBG0043581] (to G.V.G.).
REFERENCES
- Abbott A. (2010). Mouse project to find each gene’s role. Nature 465, 410. [DOI] [PubMed] [Google Scholar]
- Acevedo-Arozena A., Wells S., Potter P., Kelly M., Cox R. D., Brown S. D. (2008). ENU mutagenesis, a way forward to understand gene function. Annu. Rev. Genomics Hum. Genet. 9, 49–69 [DOI] [PubMed] [Google Scholar]
- Antony P. M., Diederich N. J., Balling R. (2011). Parkinson’s disease mouse models in translational research. Mamm. Genome 22, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold S. W., Borowsky A. D., Brayton C., Bronson R., Cardiff R. D., Griffey S. M., Ince T. A., Nikitin A. Y., Sundberg J. P., Valli V. E., et al. (2007). From whence will they come? A perspective on the acute shortage of pathologists in biomedical research. J. Vet. Diagn. Invest. 19, 455–456 [DOI] [PubMed] [Google Scholar]
- Barton E. R., Wang B. J., Brisson B. K., Sweeney H. L. (2010). Diaphragm displays early and progressive functional deficits in dysferlin-deficient mice. Muscle Nerve 42, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei M. (2009). Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 19, 504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrandelrio H., Kern F., Lanthorn T., Oravecz T., Piggott J., Powell D., Ramirez-Solis R., Sands A., Zambrowicz B. (2003). Saturation screening of the druggable mammalian genome. In Model Organisms in Drug Discovery (ed. Carroll P., Ftzgerald K.), pp. 251–279 Chichester, UK: John Wiley & Sons [Google Scholar]
- Blake J. A., Bult C. J., Kadin J. A., Richardson J. E., Eppig J. T. (2011). The mouse genome database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 39, D842–D848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen M., van Meel E., Oorschot V., Klumperman J., Kornfeld S. (2011). Vacuolization of mucolipidosis type II mouse exocrine gland cells represents accumulation of autolysosomes. Mol. Biol. Cell 22, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson R. T. (1990). Rate of occurrence of lesions in 20 inbred and hybrid genotypes of rats and mice sacrificed at 6 month intervals during the first years of life. In Genetic Effects on Aging II (ed. Harrison D. E.), pp. 279–358 Caldwell, NJ: The Telford Press [Google Scholar]
- Brown S. D., Wurst W., Kuhn R., Hancock J. M. (2009). The functional annotation of mammalian genomes: the challenge of phenotyping. Annu. Rev. Genet. 43, 305–333 [DOI] [PubMed] [Google Scholar]
- Elleder M., Martin J. J. (1998). Mucolipidosis type II with evidence of a novel storage site. Virchows Arch. 433, 575–578 [DOI] [PubMed] [Google Scholar]
- Flanagan-Steet H., Sias C., Steet R. (2009). Altered chondrocyte differentiation and extracellular matrix homeostasis in a zebrafish model for mucolipidosis II. Am. J. Pathol. 175, 2063–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs H., Gailus-Durner V., Adler T., Pimentel J. A., Becker L., Bolle I., Brielmeier M., Calzada-Wack J., Dalke C., Ehrhardt N., et al. (2009). The German mouse clinic: a platform for systemic phenotype analysis of mouse models. Curr. Pharm. Biotechnol. 10, 236–243 [DOI] [PubMed] [Google Scholar]
- Fuchs H., Gailus-Durner V., Adler T., Aguilar-Pimentel J. A., Becker L., Calzada-Wack J., Da Silva-Buttkus P., Neff F., Gotz A., Hans W., et al. (2010). Mouse phenotyping. Methods 53, 120–135 [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V., Fuchs H., Adler T., Aguilar Pimentel A., Becker L., Bolle I., Calzada-Wack J., Dalke C., Ehrhardt N., Ferwagner B., et al. (2009). Systemic first-line phenotyping. Methods Mol. Biol. 530, 463–509 [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V., Naton B., Adler T., Afonso L., Aguilar-Pimentel J.-A., Becker L. (2011). The German Mouse Clinic – running an open access platform. In The Mouse as a Model Organism (ed. Brakebusch T. P. C.), pp. 11–44 Berlin: Springer Verlag [Google Scholar]
- Gates H., Mallon A. M., Brown S. D. (2010). High-throughput mouse phenotyping. Methods 53, 394–404 [DOI] [PubMed] [Google Scholar]
- Gelfman C. M., Vogel P., Issa T. M., Turner C. A., Lee W. S., Kornfeld S., Rice D. S. (2007). Mice lacking alpha/beta subunits of GlcNAc-1-phosphotransferase exhibit growth retardation, retinal degeneration, and secretory cell lesions. Invest. Ophthalmol. Vis. Sci. 48, 5221–5228 [DOI] [PubMed] [Google Scholar]
- Hansen G. M., Markesich D. C., Burnett M. B., Zhu Q., Dionne K. M., Richter L. J., Finnell R. H., Sands A. T., Zambrowicz B. P., Abuin A. (2008). Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 18, 1670–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R., Schofield P. N., Gkoutos G. V. (2011). PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res. 39, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z., Li M. A., Mates L., Boeke J. D., Nagy A., Bradley A., Izsvak Z. (2009). Transposon-mediated genome manipulation in vertebrates. Nat. Methods 6, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice M. J. (2008). Removing the cloak of invisibility: phenotyping the mouse. Dis. Model. Mech. 1, 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Y., Jepsen K. J., Majeska R. J., Zhang J., Ni R., Gelb B. D., Schaffler M. B. (2006). Mice lacking cathepsin K maintain bone remodeling but develop bone fragility despite high bone mass. J. Bone Miner. Res. 21, 865–875 [DOI] [PubMed] [Google Scholar]
- Li X., Ominsky M. S., Niu Q. T., Sun N., Daugherty B., D’Agostin D., Kurahara C., Gao Y., Cao J., Gong J., et al. (2008). Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J. Bone Miner. Res. 23, 860–869 [DOI] [PubMed] [Google Scholar]
- Lipman R. (1997). Pathobiology of aging rodents: inbred and hybrid models. Exp. Gerontol. 32, 215–228 [DOI] [PubMed] [Google Scholar]
- Mandillo S., Tucci V., Holter S. M., Meziane H., Banchaabouchi M. A., Kallnik M., Lad H. V., Nolan P. M., Ouagazzal A. M., Coghill E. L., et al. (2008). Reliability, robustness, and reproducibility in mouse behavioral phenotyping: a cross-laboratory study. Physiol. Genomics 34, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr U., Dungworth D. L., Capen C. C., Carlton W. W., Sundberg J. P., Ward J. M. (1996). Pathobiology of the Aging Mouse (ed. Mohr U.). Washington, DC: ILSI Press [Google Scholar]
- Mouse Genome Sequencing Consortium (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 [DOI] [PubMed] [Google Scholar]
- Munroe R. J., Bergstrom R. A., Zheng Q. Y., Libby B., Smith R., John S. W., Schimenti K. J., Browning V. L., Schimenti J. C. (2000). Mouse mutants from chemically mutagenized embryonic stem cells. Nat. Genet. 24, 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers D. D. (1978). Review of disease patterns and life span in aging mice: genetic and environmental interactions. In Genetic Effects on Aging (ed. Bergsma D., Harrison D. E., Paul N. W.). New York: Alan R. Liss, Inc; [PubMed] [Google Scholar]
- Peters L. L., Robledo R. F., Bult C. J., Churchill G. A., Paigen B. J., Svenson K. L. (2007). The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8, 58–69 [DOI] [PubMed] [Google Scholar]
- Potts S. J. (2009). Digital pathology in drug discovery and development: multisite integration. Drug Discov. Today 14, 935–941 [DOI] [PubMed] [Google Scholar]
- Read R., Hansen G., Kramer J., Finch R., Li L., Vogel P. (2009). Ectonucleoside triphosphate diphosphohydrolase type 5 (Entpd5)-deficient mice develop progressive hepatopathy, hepatocellular tumors, and spermatogenic arrest. Vet. Pathol. 46, 491–504 [DOI] [PubMed] [Google Scholar]
- Read R., Savelieva K., Baker K., Hansen G., Vogel P. (2011). Histopathological and neurological features of Atg4b knockout mice. Vet. Pathol. 48, 486–494 [DOI] [PubMed] [Google Scholar]
- Robinson P. N., Kohler S., Bauer S., Seelow D., Horn D., Mundlos S. (2008). The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N., Brown S. (2007). The mouse ascending: perspectives for human-disease models. Nat. Cell Biol. 9, 993–999 [DOI] [PubMed] [Google Scholar]
- Schofield P. N., Brown S. D. M., Sundberg J. P., Warren M., Dubus P., Ellender M., Fiette L., Rozell B., Quintanilla-Martinez L., Raspa M., et al. (2009). PRIME importance of pathology expertise. Nat. Biotechnol. 27, 24–25 [DOI] [PubMed] [Google Scholar]
- Schofield P. N., Gruenberger M., Sundberg J. P. (2010a). Pathbase and the MPATH ontology. Community resources for mouse histopathology. Vet. Pathol. 47, 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. N., Gkoutos G. V., Gruenberger M., Sundberg J. P., Hancock J. M. (2010b). Phenotype ontologies for mouse and man: bridging the semantic gap. Dis. Model. Mech. 3, 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. N., Dubus P., Klein L., McKerlie C., Ward J. M., Sundberg J. P. (2011). Pathology of the laboratory mouse: an international workshop on challenges for high throughput phenotyping. Toxicol. Pathol. 39, 559–562 [DOI] [PubMed] [Google Scholar]
- Shannon C. E. (1948). A mathematical theory of communication. Bell System Technical J. 27, 379–423 [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Eppig J. T. (2009). The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R., John S., Nashina P., Sundberg J. (2002). Systematic Evaluation of the Mouse Eye: Anatomy, Pathology, and Biomethod (ed. Smith R. S.). Boca Raton, FL, USA: CRC Press [Google Scholar]
- Sundberg J. P. (1991). Mouse mutations: animal models and biomedical tools. Lab. Anim. 20, 40–49 [DOI] [PubMed] [Google Scholar]
- Sundberg J. P., Boggess D. (1998). Rhino-9J (hr(rh9J)): a new allele at the hairless locus. Vet. Pathol. 35, 297–299 [DOI] [PubMed] [Google Scholar]
- Sundberg J. P., Ichiki T. (2005). Genetically engineered mice handbook. Boca Raton: CRC Press [Google Scholar]
- Sundberg J. P., Boggess D., Sundberg B. A., Eilertsen K., Parimoo S., Filippi M., Stenn K. (2000). Asebia-2J (Scd1(ab2J)): a new allele and a model for scarring alopecia. Am. J. Pathol. 156, 2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg J. P., Hackman R. C., HogenEsch H., Nikitin A. Y., Ward J. M. (2007). Training mouse pathologists: five years of pathology of mouse models of human disease workshops. Toxicol. Pathol. 35, 447–448 [DOI] [PubMed] [Google Scholar]
- Sundberg J. P., Ward J. M., HogenEsch H., Nikitin A., Treuting P., Macauley J., Schofield P. (2010). Training pathologists in mouse pathology. Vet. Pathol. [Epub ahead of print] doi: 10.1177/0300985810381244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg J., Berndt A., Sundberg B., Silva K. A., Kennedy V., Bronson R., Yuan R., Paigen B., Harrison D., Schofield P. N. (2011). The mouse as a model for understanding chronic diseases of aging: the histopathologic basis of aging in inbred mice. Pathobiology Aging Age-related Dis. 1, 71719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland K. D., Berns A. (2011). Cell of origin of lung cancer. Mol. Oncol. 4, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Li L., Tang J., Li Y., Lin W. Y., Martin F., Grant D., Solloway M., Parker L., Ye W., et al. (2010). A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28, 749–755 [DOI] [PubMed] [Google Scholar]
- Van Dam D., De Deyn P. P. (2011). Animal models in the drug discovery pipeline for Alzheimer’s disease. Br. J. Pharmacol. [Epub ahead of print] doi: 10.1111/j.1476-5381.2011.01299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sligtenhorst I., Ding Z. M., Shi Z. Z., Read R. W., Hansen G., Vogel P. (2011). Cardiomyopathy in Alpha-Kinase 3 (ALPK3) deficient mice. Vet. Pathol. [Epub ahead of print] doi: 10.1177/0300985811402841 [DOI] [PubMed] [Google Scholar]
- Vogel P., Read R. W., Vance R. B., Platt K. A., Troughton K., Rice D. S. (2008). Ocular albinism and hypopigmentation defects in Slc24a5−/− mice. Vet. Pathol. 45, 264–279 [DOI] [PubMed] [Google Scholar]
- Vogel P., Payne B. J., Read R., Lee W. S., Gelfman C. M., Kornfeld S. (2009). Comparative pathology of murine mucolipidosis types II and IIIC. Vet. Pathol. 46, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P., Read R. W., Hansen G. M., Payne B. J., Small D., Sands A. T., Zambrowicz B. P. (2010a). Congenital hydrocephalus in genetically engineered mice. Vet. Pathol. [Epub ahead of print] doi: 10.1177/0300985811415708 [DOI] [PubMed] [Google Scholar]
- Vogel P., Hansen G., Fontenot G., Read R. (2010b). Tubulin tyrosine ligase-like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet. Pathol. 47, 703–712 [DOI] [PubMed] [Google Scholar]
- Vogel P., Read R., Hansen G. M., Freay L. C., Zambrowicz B. P., Sands A. T. (2010c). Situs inversus in Dpcd/Poll−/−, Nme7−/−, and Pkd1l1−/−mice. Vet. Pathol. 47, 120–131 [DOI] [PubMed] [Google Scholar]
- Washington N. L., Haendel M. A., Mungall C. J., Ashburner M., Westerfield M., Lewis S. E. (2009). Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 7, e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Tsaih S. W., Petkova S. B., de Evsikova C. M., Xing S., Marion M. A., Bogue M. A., Mills K. D., Peters L. L., Bult C. J., et al. (2009). Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell 8, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz B. P., Sands A. T. (2003). Knockouts model the 100 best-selling drugs-will they model the next 100? Nat. Rev. Drug Discov. 2, 38–51 [DOI] [PubMed] [Google Scholar]
- Zambrowicz B. P., Turner C. A., Sands A. T. (2003a). Predicting drug efficacy: knockouts model pipeline drugs of the pharmaceutical industry. Curr. Opin. Pharmacol. 3, 563–570 [DOI] [PubMed] [Google Scholar]
- Zambrowicz B. P., Abuin A., Ramirez-Solis R., Richter L. J., Piggott J., BeltrandelRio H., Buxton E. C., Edwards J., Finch R. A., Friddle C. J., et al. (2003b). Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. USA 100, 14109–14114 [DOI] [PMC free article] [PubMed] [Google Scholar]