SUMMARY
Back pain related to intervertebral disc degeneration is the most common musculoskeletal problem, with a lifetime prevalence of 82%. The lack of effective treatment for this widespread problem is directly related to our limited understanding of disc development, maintenance and degeneration. The aim of this study was to determine the developmental origins of nucleus pulposus cells within the intervertebral disc using a novel notochord-specific Cre mouse. To trace the fate of notochordal cells within the intervertebral disc, we derived a notochord-specific Cre mouse line by targeting the homeobox gene Noto. Expression of this gene is restricted to the node and the posterior notochord during gastrulation [embryonic day 7.5 (E7.5)-E12.5]. The Noto-cre mice were crossed with a conditional lacZ reporter for visualization of notochord fate in whole-mount embryos. We performed lineage-tracing experiments to examine the contribution of the notochord to spinal development from E12.5 through to skeletally mature mice (9 months). Fate mapping studies demonstrated that, following elongation and formation of the primitive axial skeleton, the notochord gives rise to the nucleus pulposus in fully formed intervertebral discs. Cellular localization of β-galactosidase (encoded by lacZ) and cytokeratin-8 demonstrated that both notochordal cells and chondrocyte-like nucleus pulposus cells are derived from the embryonic notochord. These studies establish conclusively that notochordal cells act as embryonic precursors to all cells found within the nucleus pulposus of the mature intervertebral disc. This suggests that notochordal cells might serve as tissue-specific progenitor cells within the disc and establishes the Noto-cre mouse as a unique tool to interrogate the contribution of notochordal cells to both intervertebral disc development and disc degeneration.
INTRODUCTION
Back pain accounts for more than half of all musculoskeletal disabilities, making it one of the most prevalent and costly medical conditions (Murphy and Volinn, 1999; Goetzel et al., 2003; Ramage-Morin and Gilmour, 2010). The most common cause of back pain is disc degeneration, the etiology of which is poorly defined, making it difficult to distinguish from the physiological processes of growth, aging or adaptive remodeling (Adams and Roughley, 2006). Disc degeneration has perhaps best been defined as an aberrant, cell-mediated response to progressive structural failure (Adams and Roughley, 2006): a cascade beginning with changes to the cellular microenvironment within the disc that progresses over decades resulting in structural breakdown and functional impairment (Smith et al., 2011). Degeneration of the intervertebral disc (IVD) is characterized by increased extracellular matrix breakdown, abnormal (fibrotic) matrix synthesis, inflammation, and in-growth of nociceptive nerves and blood vessels into a typically aneural and avascular tissue. Although the exact cause(s) of disc degeneration are unclear, initiation and progression of the degenerative cascade involve multiple interdependent factors, including altered mechanical loading (Ohshima et al., 1995), reduction in nutrient supply (Urban et al., 2004), altered cellular composition (Aguiar et al., 1999) and hereditary factors (Kawaguchi et al., 1999; Videman et al., 2001; Battie and Videman, 2006). Currently, there is no effective disease-modifying treatment for individuals with this disease who develop persistent pain.
The IVD is a specialized connective tissue structure consisting of three distinct yet interdependent tissues: the outer fibrillar annulus fibrosus (AF), the central viscous nucleus pulposus (NP) and the cartilage end-plates (CEPs; which anchor the discs to the adjacent vertebral body bones). The NP is composed of a proteoglycan and water gel held together loosely by an irregular network of type II collagen and elastin fibers. The major proteoglycan of the disc is aggrecan, which, owing to its highly anionic glycosaminoglycan content, provides the osmotic properties enabling the NP to maintain height and turgor against compressive loads (Watanabe et al., 1998; Adams and Roughley, 2006; Setton and Chen, 2006). The NP contains two distinct cell types: large clusters of notochordal cells and smaller, more disperse cartilage-like cells (Roberts, 2002). Interestingly, the loss of notochordal cells is associated with the onset of disc degeneration, suggesting that these cells are required for the maintenance of the NP (Aguiar et al., 1999; Boos et al., 2002; Hunter et al., 2003).
Although the role of the notochord during gastrulation has been extensively studied (Beddington and Robertson, 1999), the function of the notochord during IVD formation remains relatively unknown. In the early mouse embryo, the node is formed at embryonic day 7.5 (E7.5) and is the source of cells that will form the rod-like structure of the notochord (Yamanaka et al., 2007). The notochord is an important source of signals for patterning of the developing embryo, specifically axis determination in both dorso-ventral and left-right planes (Johnson et al., 1994; Beddington and Robertson, 1999; Beckers et al., 2007). The vertebral column is formed by aggregation of the somatic mesenchyme around the notochord, forming a continuous perichordal tube that undergoes segmentation at E13.5 to form condensed and non-condensed regions that give rise to the vertebrae and IVDs, respectively (Dalgleish, 1985; Urban et al., 2000). Within the IVD, notochordal cells proliferate and accumulate a gelatinous, glycosaminoglycan-rich extracellular matrix, which separates the original cell mass into a network of small cell clusters. The NP becomes surrounded by the mesenchyme-derived AF (Christ and Wilting, 1992; Eloy-Trinquet and Nicolas, 2002) (Fig. 1).
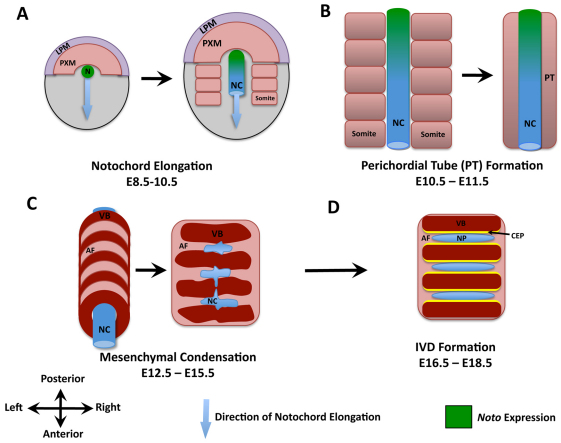
Fig. 1.
Schematic representation of the main stages in axial skeletogenesis. (A) Formation of the node (N) and elongation of the notochord (NC). LPM, lateral plate mesoderm; PXM, paraxial mesoderm. (B) Aggregation of the somatic mesenchyme around the notochord leads to formation of a continuous perichordal tube (PT). Localization of Noto expression at these time points is indicated in green. (C) Condensation of the axial mesenchyme leads to spine segmentation and perichordal disc formation. AF, annulus fibrosis; VB, vertebral bodies. (D) Formation of intervertebral discs (IVDs) is associated with the disappearance of the notochord within the vertebral bodies, and its expansion within the IVD. CEP, cartilage end-plates; NP, nucleus pulposus.
Beginning as early as the second decade of life in humans, notochordal cells are no longer detected within the NP, which is populated instead by smaller more disperse cartilage-like cells (Chelberg et al., 1995; Zhou et al., 2008). Currently, identification of these two cell types is restricted to cell morphology analysis and use of a limited number of notochord-specific markers (Fujita et al., 2005; Gilson et al., 2010; Minogue et al., 2010). There have historically been two conflicting hypotheses regarding the origin of the cartilage-like cells of the NP. It was originally suggested that these cells were of mesenchymal origin, a consequence of the migration of cells to the NP from the surrounding CEP (Vujovic et al., 2006). During tissue formation, notochordal cells were believed to direct mesenchymal cell migration, stimulate matrix synthesis and, upon completion of their role, undergo apoptosis or necrosis (Trout et al., 1982; Kim et al., 2003). Alternatively, it has been suggested that notochordal cells are progenitors for all NP cells, and undergo terminal differentiation to give rise to chondrocyte-like cells (Pazzaglia et al., 1989; Boos et al., 2002; Liebscher et al., 2010). In these two scenarios, notochordal cells play distinct roles, that of organizer and tissue-specific progenitor, respectively. Although recent studies sought to address this longstanding debate and suggested that NP cells are derived from the embryonic notochord, technical limitations prevented a definitive conclusion regarding cell fate (Choi et al., 2008).
To clearly define the developmental origins of the NP, we have targeted the gene encoding the notochord-specific transcription factor Noto to generate a mouse line expressing Cre recombinase specifically in the notochord. Noto is a highly conserved homeobox transcription factor whose expression is restricted to the organizer node and the nascent notochord during gastrulation and axis elongation (E7.5–E12.5); here, it regulates node morphogenesis, notochord ciliogenesis and left-right patterning (Fig. 1A,B) (Abdelkhalek et al., 2004; Yamanaka et al., 2007; Zizic Mitrecic et al., 2010). Expression of Noto is detected at E7.5 within the ventral node, with the onset of primitive streak formation. Between E8.0 and E9.0, Noto is expressed in the node and newly formed notochord but not in the anterior node or mature notochord. Noto expression is restricted to the notochordal plate and posterior notochord until E12.5, after which its expression is no longer detected (Abdelkhalek et al., 2004). Using this novel notochord-specific Cre mouse, we provide evidence that all cells present within the NP, from tissue formation through to skeletal maturity, are of notochord origin.
RESULTS
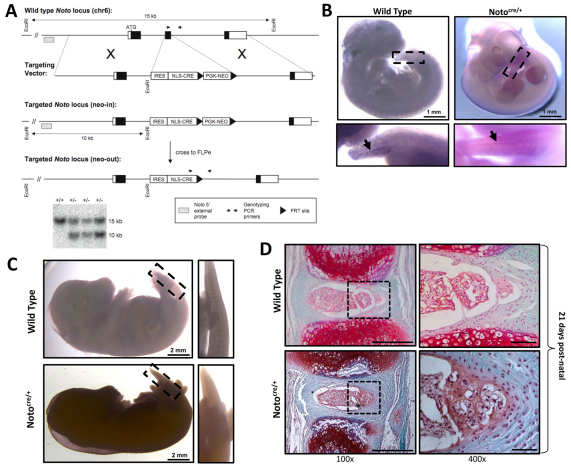
The Noto-cre allele was created by knocking the site-specific recombinase gene cre into exon 2 of the Noto locus (Fig. 2A). Using this strategy, cre is expressed in all Noto-expressing cells, specifically in the node and posterior notochord between E7.5 and E12.5 (Yamanaka et al., 2007), and in all of their descendents. To ensure that the temporal regulation of Noto expression was not altered in our transgenic strain, in situ hybridization was performed. In both Notocre/+ and wild-type littermate controls (Noto+/+), Noto expression was detected in the posterior notochord at E11.5 (Fig. 2B) and was undetectable by E15.5 (Fig. 2C).
Fig. 2.
Generation of the notochord-specific Cre mouse line. (A) Targeting strategy used to generate the Notocre/+ line. An internal ribosome entry site-nuclear localization signal-Cre recombinase (IRES-NLS-CRE) cassette replaced exon 2 of the Noto locus. Positively targeted ES cell clones were confirmed by Southern blot using an external 5′ probe; the wild-type allele is 15 kb and the targeted allele is 10 kb. Representative positive ‘neo-in’ clones are shown. The positions of genotyping PCR primers for wild-type and ‘neo-out’ mice are also shown. (B) Targeting of the Noto locus does not affect its temporal regulation, as demonstrated by whole mount in situ hybridization at E11.5. Noto expression is detected in both Notocre/+ and wild-type (Noto+/+) littermate control embryos, localized to the posterior node in the tail region (insert and arrows). (C) Noto expression is downregulated after E12.5, confirmed by in situ hybridization at E15.5 showing no detectable Noto expression. (D) Heterozygous inactivation of Noto does not disrupt notochord formation or IVD development. IVD formation and tissue architecture was examined in Notocre/+ mice and wild-type (Noto+/+) littermate controls using Safranin-O/Fast Green staining on paraffin embedded sections at P21. Enlarged view of the NP and inner AF tissues are shown in the right-hand box. Scale bars: 500 μm for 100× images and 50 μm for 400× images.
Previous studies demonstrated that homozygous disruption of the Noto gene leads to embryonic or early postnatal lethality, but heterozygous animals are viable with no reported phenotype (Abdelkhalek et al., 2004; Beckers et al., 2007; Yamanaka et al., 2007). To ensure that heterozygous inactivation of Noto did not alter notochord formation or IVD development, spine segments from Notocre/+ mice were assessed by histology at postnatal day 21 (P21) and compared with wild-type littermate controls (Noto+/+). Examination of tissue architecture and joint formation by Safranin-O/Fast Green staining demonstrated no alterations in notochord structures or IVD formation in Notocre/+ mice (Fig. 2D). Therefore, all subsequent studies were conducted using the heterozygous Notocre/+ mouse to interrogate IVD formation.
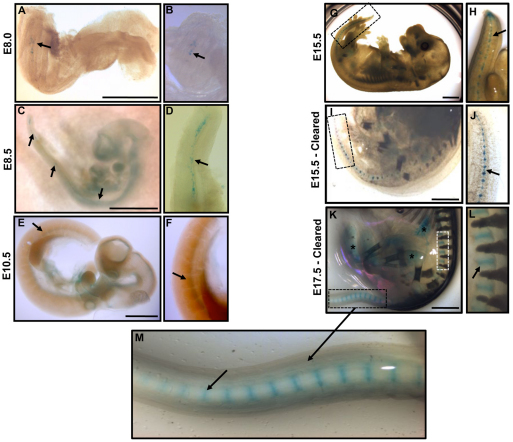
Through mating with the conditional Rosa26 (R26)-lacZ reporter [Gt(ROSA)26Sortm1so] (Soriano, 1999), fate mapping studies were conducted. Embryos carrying both the Noto-cre allele and the R26-lacZ reporter showed specific β-galactosidase expression in a subset of node and notochordal cells at E8.0, the stage at which the node is reported to undergo elongation to form the notochord (Fig. 3A,B). The mouse notochord has previously been reported to form in a non-sequential pattern with multiple organizing centers (Yamanaka et al., 2007). A discontinuous pattern of Noto expression was observed at E8.5, in which different centers of notochord formation were seen along the rostro-caudal axis (Fig. 3C,D). These centers ultimately connected to form the solid rod-like notochord by E10.5 (Fig. 3E,F).
Fig. 3.
Whole-mount examination of β-galactosidase staining in Notocre/+;R26R/+ embryos. (A–M) Staining pattern of β-galactosidase, demonstrating the localization of notochord-derived cells throughout embryonic development (stages indicated). Higher-magnification view of β-galactosidase expression at each respective time point is presented in panels to the right. Arrows denote positive β-galactosidase staining at each embryonic stage. (I–M) β-galactosidase expression in E15.5 and E17.5 embryos, rendered clear to enable tissue visualization, shows expression within IVDs in the developing spine. Asterisks (*) indicate non-specific background staining; specificity of Cre expression was confirmed by comparing background staining in Noto+/+ littermate controls (data not shown). Scale bars: 1 mm.
The segmentation of the notochord can be seen at E15.5 and E17.5 (Fig. 3G–M). Owing to opacity of embryos at these later developmental stages, the staining protocol was modified to include an embryo clearing step that renders the embryo translucent while leaving the β-galactosidase stain intact (Schatz et al., 2005). β-galactosidase expression within the IVD was observed in the upper thoracic region of the embryo, flanked by developing vertebrae (Fig. 3L). The formation of the NP through notochord condensation and segmentation was seen throughout the embryonic time points, originating caudally and proceeding rostrally. This was evident in the tail region at E15.5, where the nuclei pulposi remain connected via the remaining notochord (Fig. 3J, arrow).
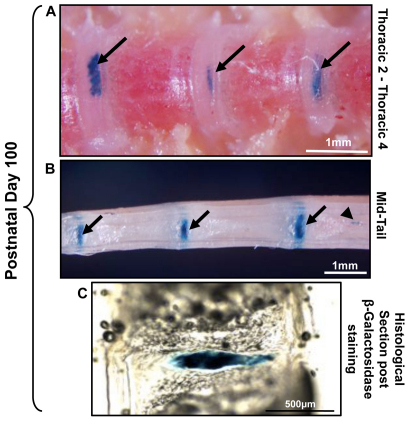
At postnatal time points, β-galactosidase expression was detected throughout the thoracic spine to caudal tip of the tail (Fig. 4). Small clusters of notochordal cells were detected that were not incorporated into the NP of the IVD but seemed instead to form notochord remnants (also known as intraosseous benign notochordal cell tumors) within the vertebral bone (Fig. 4B). These notochord remnants were indiscriminately localized throughout the spine at postnatal time points, and were present in >90% of spines examined. In order to more accurately localize β-galactosidase expression to specific cellular components of the IVD, tissues were examined by histology. Cryosections were performed on skeletally mature mice (P100) in order to preserve β-galactosidase staining. β-galactosidase was localized specifically to the NP, with no stain present in the surrounding AF or CEPs (Fig. 4C).
Fig. 4.
β-galactosidase staining at postnatal time points in Notocre/+;R26R/+ mice. (A,B) P100 skeletally mature murine spine;β-galactosidase staining was present from the tail to the upper thoracic spine. Arrows indicate positive β-galactosidase staining in the IVD; arrowhead denotes notochord remnant cells within the vertebral bone, which go on to form intraosseous benign notochordal cell tumors. (C) At P100, β-galactosidase staining was evident throughout the NP, whereas the AF was negative. Representative 30 μm sagittal cryosection of an IVD stained for β-galactosidase is shown (originally from tail region in B).
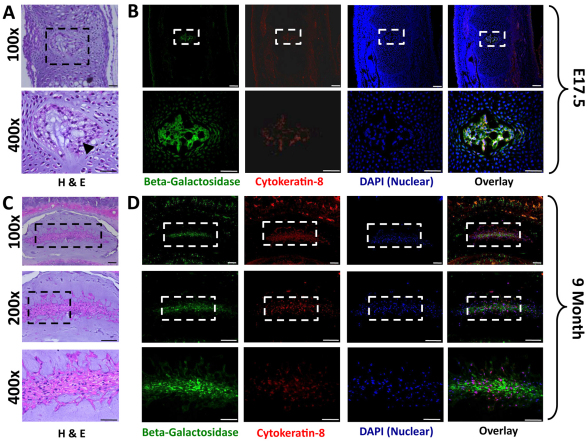
To examine β-galactosidase expression within specific cell types of the NP, paraffin-embedded IVD samples were examined by immunohistochemistry for expression of β-galactosidase and cytokeratin-8, a commonly used marker of notochordal cells (Gilson et al., 2010) (Fig. 5). Within the newly formed IVD (E15.5), cells demonstrated a physaliferous appearance characteristic of notochordal cells and expressed both β-galactosidase and cytokeratin-8 (Fig. 5A,B). In postnatal life, the NP underwent a dramatic change in cellular composition and became progressively populated by cartilage-like NP cells which seemed to be smaller than notochordal cells and were surrounded by pericellular extracellular matrix (Fig. 5C). We therefore examined the NP at 9 months to determine the contribution of the notochord to this heterogeneous population. At this stage, all cells were positive for β-galactosidase, while only a subset of cells maintained cytokeratin-8 colocalization (Fig. 5D). The detection of β-galactosidase in the chondrocyte-like NP cells demonstrated that they are derived from the embryonic notochord.
Fig. 5.
Colocalization of β-galactosidase and cytokeratin-8 in the NP. (A) Representative hematoxylin and eosin (H&E)-stained sections of IVDs at E17.5 (tail region). Cells within the NP demonstrate characteristic physaliferous morphology associated with notochordal cells (arrowhead), and are surrounded by embryonic mesenchyme. (B) Immunolocalization of β-galactosidase (green) and cytokeratin-8 (red) expression. No cytokeratin-8-negative cells were detected at this stage, suggesting that all cells maintain a notochord phenotype. Lower panels are a magnified view of each respective panel above. (C) Representative H&E-stained sections of a 9-month-old mouse lumbar disc demonstrates the heterogeneous cellular composition of the mature NP. (D) Colocalization of β-galactosidase (green) and cytokeratin-8 (red) in serial sections demonstrates that, although all cells are β-galactosidase positive, only a subset of cells maintain cytokeratin-8 expression (100×, 200× and 400× scale bars are 100 μm, 100 μm and 50 μm, respectively).
DISCUSSION
Our ability to demonstrate the fate and function of notochordal cells within the IVD has thus far been limited. A key tool for understanding cell function in model organisms is the Cre-loxP system for conditional mutagenesis (Lewandoski, 2001). This approach can be extremely informative when carefully selected Cre recombinase lines are paired with loxP-flanked conditional reporter genes. Although notochord-expressing Cre mouse lines have been generated using elements from the Foxa2 and Nodal genes, Cre expression was also detected in the ventral neural tube, or floor plate, and the definitive endoderm in these lines (Stein and Kessel, 1995; Uetzmann et al., 2008). By contrast, expression of the homeobox gene Noto is restricted to the node and the posterior notochord during gastrulation (E7.5–E12.5) and is not detected in the notochord or derivative structures after this stage (von Dassow et al., 1993; Stein and Kessel, 1995; Talbot et al., 1995). Furthermore, Noto-GFP knock-in permitted analysis of notochord formation and morphogenesis during gastrulation (Yamanaka et al., 2007). We therefore targeted the Noto locus to generate a Cre knock-in mouse (Noto-cre) and provide considerable evidence that the notochord acts as the embryonic precursor of all cells found within the mature NP. We demonstrate that all cells within the NP, both the large notochordal cells as well as the smaller chondrocyte-like NP cells, are of notochordal origin.
Our results are consistent with a previous study by Choi and colleagues that used an inducible Shh-creERT2 system to mark the notochord during development (Choi et al., 2008). The Shh-creERT2 system does have certain limitations; for example, Shh is expressed in the notochord as well as in many other cell types in the early embryo (Bouldin et al., 2010; Miller et al., 2001), making it difficult to definitively track lineages at later stages. Also, the tamoxifen that is injected into pregnant mice for induction of creERT2 in utero can persist for up to 48 hours, which could potentially lead to Cre induction in any Shh-expressing cells during that time window. Our model demonstrates that Noto-expressing notochordal cells give rise to all cells within the mature NP. Consistent with these findings, recent studies examining the gene expression profile of specific IVD cell populations in bovine samples reported a common gene expression signature in notochordal and NP cells that was not detected in articular chondrocytes (including brachyury and cytokeratins 8, 18 and 19), and suggested that this resulted from a common lineage (Minogue et al., 2010). Furthermore, studies using rabbit-derived cells demonstrated the ability of notochordal cells to undergo directed in vitro differentiation, producing cells that are morphologically and phenotypically similar to chondrocyte-like NP cells (Kim et al., 2009).
Initially, the floating head (flh) gene was identified in zebrafish and was found to encode a homeodomain protein that was essential for notochord formation along the body axis (Talbot et al., 1995). Orthologs of flh were found in Xenopus and chicks, and were named Not genes. The mammalian equivalent was identified with the report of an autosomal recessive mutation in mice that disrupted development of the caudal notochord (Abdelkhalek et al., 2004). The Not gene was renamed Noto in mammals and has been shown to be involved in axis patterning, notochord ciliogenesis, and node and tail bud morphogenesis (Beckers et al., 2007; Mitrecic et al., 2010). It was the highly specific nature of Noto that made it an ideal tool to track the notochord, and it is for that reason that we generated a the Noto-cre mouse. Given that the Noto transcript is detected only in the node and posterior notochord (Yamanaka et al., 2007), we initially suspected that Noto-cre expression was going to be limited to the posterior region of the spine. Our results, however, show that Noto-cre is detected in the tail but also throughout the axial spine to the upper thoracic region, thereby demonstrating the potential of this mouse to explore the phenotype and function of notochordal cells during development and in postnatal life, including the generation of notochord-specific gene knockouts.
By mating Noto-cre mice with the conditional R26-lacZ reporter mouse, these studies have traced cells of notochord origin during the dynamic process of notochord formation, elongation, condensation and eventual formation of the NP within the IVD. We demonstrate that notochord segmentation and NP formation proceeds in the rostral-to-caudal direction, as can be seen in the tail region of E15.5 and E17.5 mice. At these stages, notochord-derived cells are localized within fully formed IVDs in the thoracic region, while in the tail the notochord remains connected to the condensed regions that will form the NP.
Interestingly, during the early stage of notochord formation and elongation (E8.0), cre expression did not seem to be uniform throughout the notochord, and was instead detected in distinct cell clusters or centers. This heterogeneous pattern of notochord formation has previously been reported using Noto-driven reporters (i.e. NotoeGFP) (Abdelkhalek et al., 2004; Yamanaka et al., 2007) as well as other node and notochord enhancer transgenics (Nishizaki et al., 2001; Sawada et al., 2005). In the current study, this patchy expression was however resolved at later stages of development (E17.5), when β-galactosidase expression was detected throughout the notochord, suggesting that the heterogeneous nature of the early notochord is not an artifact of our system. We suggest instead that the heterogeneity often observed in the notochord is biologically significant. It might be that mesodermal cells can be incorporated into the notochord and then differentiate via interaction with their neighbors. This has been observed in transplantation of zebrafish Noto (flh) mutant cells to wild-type hosts – these cells lack a gene that is crucial for notochord formation, yet they can still incorporate and contribute to the notochord (Amacher and Kimmel, 1998). Future studies will be needed to determine the exact cause of heterogeneous expression in various notochord transgenic lines.
Speculations regarding the developmental origins of the heterogeneous population of cells that constitute the mature NP have argued that the cartilage-like cells originate either from the notochord, or from the surrounding mesenchyme following migration of cells to the NP from the adjacent CEPs or inner AF. We present evidence that notochordal cells are the precursor cells of the NP, and undergo differentiation to generate chondrocyte-like cells during postnatal development. This transition might be induced by a variety of factors present in the extracellular environment of the IVD. Most notably, with aging the nutrient supply to the already avascular and hypoxic NP becomes increasingly limited owing to calcification of the end-plates (Urban et al., 2004). The resulting changes in oxygen, glucose and lactic acid concentration within the NP might contribute to terminal notochordal cell differentiation to chondrocyte-like cells.
The lack of effective biological treatment for the widespread clinical problem of IVD degeneration is directly related to our limited understanding of the mechanisms regulating the processes of disc development, maintenance and degeneration. This gap in our knowledge is due to an incomplete understanding of the specific phenotypes of IVD cell types and the relative importance of the individual growth factors, secreted molecules and matrix components that constitute the unique extracellular environment of the IVD. Given its role in maintaining the physiological function of the IVD, a more complete understanding of notochord and NP biology is of vital importance. The described notochord-specific Cre mouse model will be a valuable tool in future studies to interrogate the function of candidate regulatory molecules during the processes of IVD development, homeostasis and degeneration, with the eventual goal of identifying candidate biological treatments for disc degeneration.
In summary, this report provides evidence demonstrating the notochord origin of all cells within the NP and demonstrates the potential of the Noto-cre mouse in understanding IVD biology.
METHODS
Generation of the notochord-specific Noto-cre mouse strain
All experiments were performed under Institutional Animal Care and Use Committee guidelines at The University of Western Ontario and Toronto Centre for Phenogenomics. To generate the Noto-cre knock-in mouse, the Noto targeting vector was amplified using long-range PCR (Roche HiFi Taq polymerase) from a BAC clone that contained the Noto locus (RP23-417O15; library derived from female C57BL/6J tissues). PCR primers added EcoRI and NotI sites to the 5′ and 3′ homology arms, respectively (sequence provided in Table 1). The 3′ homology arm PCR product (∼3 kb) was ligated into a pICN vector, and confirmed by restriction digest and sequencing. The pICN vector contains an IRES-NLS-CRE cassette (generously provided by Andras Nagy, Samuel Lunenfeld Research Institute, Toronto, Canada) arranged as follows: EcoRI-IRES-NLS-CRE-FRT-PGK-NEO-FRT-NotI/SacI. The 5′ homology arm PCR product (∼5 kb) was digested with EcoR1, ligated into the pICN-Noto-3′ vector, and confirmed by restriction digest and sequencing.
Table 1.
Primers and PCR conditions for Notocre/+ and lacZ reporter mice
The final pNoto-5′-ICN-Noto-3′ plasmid was linearized and electroporated in G4 embryonic stem (ES) cells (George et al., 2007) using standard protocols. After neomycin selection, ∼170 colonies were screened by Southern blot. A 5′ external probe was isolated from a plasmid generously shared by Anja Beckers and Achim Gossler (Abdelkhalek et al., 2004) and used to confirm positively targeted clones (Fig. 2A). Ten of ∼120 clones were positive, giving a targeting efficiency of ∼8%. Two independent positive ES cell clones were used to produce founder chimeras. Both clones produced chimeras and were crossed to CD1 (ICR) for germline transmission. Positive F1 mice were confirmed by Southern blot, and crossed to a FLPe deleter strain [MGI Strain: B6;SJL-Tg(ACTFLPe)9205Dym/J] for recombination between FRT sites and removal of PGK-NEO.
Genotyping of mice
Mice were genotyped by PCR analysis using ear clipping DNA samples and Noto-cre knock-in was confirmed by PCR genotyping using the Noto wild-type (WT) and ‘neo-out’ primers (location indicated in Fig. 2A; sequences provided in Table 1). WT primers produce a 329 bp band and ‘neo-out’ primers produce a 415 bp band using the PCR program: for denaturing, 98°C for 30 seconds; for annealing, 60°C for 30 seconds; for extension, 72°C for 30 seconds for 39 cycles. Targeting of the R26 locus in conditional lacZ reporter mice, Gt(ROSA)26Sortm1sor, was confirmed by genotyping using previously characterized primers (Soriano, 1999) (Table 1). WT primers produce a 650 bp band and primers specific for the targeted locus produce a 340 bp band using the PCR conditions: for denaturing, 94°C for 30 seconds; for annealing, 62°C for 60 seconds; for extension, 72°C for 60 seconds for 34 cycles.
Whole-mount in situ hybridization
Whole-mount in situ hybridization on E11.5 and E15.5 embryos was performed as previously described (Lickert et al., 2002). Briefly, embryos were fixed at the time of collection in 4% paraformaldehyde overnight at 4°C. For hybridization, embryos were rehydrated through a MeOH/PBT [phosphate buffered saline (PBS) plus 0.1% Tween-20 (Sigma)] series into PBT and incubated with proteinase K (10 μg/ml in PBT) for 8 minutes at room temperature. Digestion was stopped by washing with 2 mg/ml glycine in PBT, and embryos were post-fixed in 4% paraformaldehyde with 0.2% glutaraldehyde in PBT, washed in PBT and hybridized overnight at 70°C with 1 μg/ml of digoxigenin-labeled riboprobe (Roche) in hybridization solution [50% formamide, 5× SSC (pH 4.5), 1% SDS, 50 μg/ml yeast tRNA, and 50 μg/ml heparin]. Embryos were washed three times in hybridization solution for 30 minutes at 70°C, rinsed three times in TNT [10 mM Tris-HCl (pH 7.5), 0.5 M NaCl and 0.1% Tween-20] for 5 minutes each at room temperature and incubated for 1 hour with 100 μg/ml RNase A in TNT. After three washes in 50% formamide, 2× SSC (pH 4.5), 0.5 M NaCl and 0.1% Tween-20 for 30 minutes at 65°C, followed by three washes in MAB [100 mM maleic acid (pH 7.5), 150 mM NaCl, 2 mM levamisole and 0.1% Tween-20] for 5 minutes at room temperature, samples were blocked for 2 hours in 10% sheep serum in MAB/2% Roche blocking agent. Embryos were incubated overnight at 4°C with anti-digoxigenin alkaline phosphatase-coupled antibody (1:5000 diluted in MAB/2% Roche block/1% sheep serum plus 0.5 mg/ml mouse embryo powder). Following MAB washes, the embryos were washed three times in NTMT [100 mM Tris-HCl (pH 9.5), 50 mM MgCl2, 100 mM NaCl, and 0.1% Tween-20] for 5 minutes each, and stained with BM Purple (Roche) at room temperature. Gene expression patterns are representative of three or more stage-matched embryos. Imaging was done on a Leica stereo light microscope with Leica Application Suite software.
Lineage tracing and β-galactosidase staining
Notocre/+ mice were mated with R26 conditional lacZ reporter mice [Gt(ROSA)26Sortm1sor (Jackson Labs)], so that β-galactosidase is produced upon Cre-mediated excision of STOP sequence upstream of lacZ. For timed matings, insemination was confirmed by vaginal sperm plug morning after coitus, which was counted as E0.5. Pregnant females were sacrificed at stated embryonic time points. Genotypic analysis of embryos was done by PCR of DNA extract, using primer sets indicated in Table 1. Whole-mount β-galactosidase staining was carried out using standard protocols (Hogan, 1994). Briefly, embryos were fixed for 30 minutes (E<13.5) or 2 hours (E>13.5) in 0.2% glutaraldehyde/1% formaldehyde/0.02% Nonidet P-40 in PBS at room temperature. Following three washes in PBS (10 minutes each at room temperature), the embryos were incubated in X-gal staining solution [5 mM K3Fe(CN)6/5 mM K4Fe(CN)6/2 mM MgCl2/1 mg/ml X-gal in PBS] overnight at room temperature on rocker. Embryos were then washed with PBS 3×10 minutes at room temperature and kept in PBS overnight to allow stain to develop. Whole-mount embryos were imaged with a Nikon SMZ 1500 stereo microscope.
For visualization of β-galactosidase staining in embryos >E15, a clearing step was performed as previously described (Schatz et al., 2005). Briefly, following β-galactosidase staining embryos were cleared by a series of solutions containing decreasing KOH and increasing glycerol concentrations: 100:0, 80:20, 50:50, 20:80 and 0:100%, respectively, for 3 days each.
For analysis at P100, spine microdissection was necessary to allow for adequate stain penetration. Following β-galactosidase staining as outlined above, tissues were post-fixed in 4% formaldehyde overnight at room temperature, embedded in OCT (VWR) and sagittal sections were cut at a thickness of 10 μm.
Histology
Whole embryos or postnatal spinal columns were isolated at indicated stages and fixed in 4% paraformaldehyde overnight at 4°C, dehydrated in a graded series of ethanol, cleared in xylene and embedded in paraffin. Serial sections of paraffin-embedded samples sectioned sagittally at a thickness of 5 μm were stained with 0.1% Safranin-O, 0.02% Fast Green or Harris’ hematoxylin (Sigma) and Eosin-Y (Sigma).
Immunohistochemistry
Paraffin-embedded sections were cut at a thickness of 5 μm using a microtome (Leica) and collected on Superfrost Plus slides (Fisher Scientific). Sections were then de-waxed in xylene and rehydrated by successive immersion in descending concentrations of alcohol. Samples were blocked for non-specific binding by incubating sections with the corresponding species-specific serum albumin (1%) in PBS with 0.1% Tween-20 (Sigma) for 1 hour (Sigma) and then incubated with primary antibody directed against β-galactosidase (Abcam 1:500) and cytokeratin-8 (Abcam 1:200), and then placed in a humidified chamber overnight at 4°C. After washes with PBST and incubation of goat anti-rabbit FITC and goat anti-mouse Texas Red (Jackson ImmunoResearch Laboratories; 1:500) for 90 minutes, mounting was performed with VECTASHIELD Mounting Medium with DAPI (Burlingame, CA). Images were captured with a Leica DMRA2 fluorescence microscope and processed with Northern Eclipse (Empix) software.
TRANSLATIONAL IMPACT.
Clinical issue
Low back pain is a common musculoskeletal problem attributed to degeneration of the intervertebral disc (IVD) and, more specifically, the inner nucleus pulposus (NP). The lack of effective treatments is directly related to our limited understanding of the mechanisms regulating the processes of IVD development, maintenance and degeneration. To reverse the pathological process of IVD degeneration, it has been suggested that the loss of cell viability and alterations in cellular phenotype in the NP should be targeted. This approach, however, requires characterization of the developmental origins of cell types within the NP and a better understanding of the cellular phenotype of the distinct cell types responsible for maintenance of the IVD.
Results
To trace the developmental origins of NP cells within the IVD, the authors derive a notochord-specific Cre mouse line by targeting the homeobox transcription factor Noto, a gene that is expressed exclusively in the node and the posterior notochord during gastrulation. By crossing this Noto-cre mouse with a conditional lacZ reporter mouse, they are able to perform lineage tracing experiments to examine the contribution of the notochord to spinal development. The results show that, following formation of the axial skeleton, the notochord gives rise to the NP in the fully formed IVD. Localization of β-galactosidase, encoded by lacZ, demonstrates that all cells in the NP are derived from the embryonic notochord.
Implications and future directions
This study demonstrates that notochordal cells serve as tissue-specific stem cells within the IVD, capable of differentiating to form the NP. Further understanding of the mechanisms regulating the transition from notochord to NP cells might aid in the development of methods to induce the regeneration of NP tissue. The Noto-cre mouse described here will be a valuable tool for interrogating the function of molecules that regulate the function of notochordal cells within the IVD, with the eventual goal of identifying candidate biological treatments for disc degeneration.
Acknowledgments
The authors thank Jorge Cabezas for his expert animal care at Mount Sinai Hospital and the Toronto Centre for Phenogenomics, as well as Dawn Bryce and Tom Chrones at The University of Western Ontario for their technical advice.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for intellectual content, and all authors approved the final version to be published. C.A.S. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors were involved in study conception and design, and analysis and interpretation of the data. M.R.M., O.J.T. and C.A.S. were involved in data acquisition.
FUNDING
This work was supported by the Canadian Institutes of Health Research (CIHR) [MOP-77803] (to J.R.); the Natural Sciences and Engineering Research Council of Canada [RGPIN-371546] (to C.A.S.); the Canadian Arthritis Network [09-PGP-BIO-07] (to C.A.S); the CIHR Joint Motion Program in Musculoskeletal Health Training and Leadership (to M.R.M.); the CIHR (Doctoral Award) (to O.J.T.); the Canadian Arthritis Network (to C.A.S.); and The Arthritis Society Scholar Award (to C.A.S.).
REFERENCES
- Abdelkhalek H. B., Beckers A., Schuster-Gossler K., Pavlova M. N., Burkhardt H., Lickert H., Rossant J., Reinhardt R., Schalkwyk L. C., Muller I., et al. (2004). The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 18, 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. A., Roughley P. J. (2006). What is intervertebral disc degeneration, and what causes it? Spine 31, 2151–2161 [DOI] [PubMed] [Google Scholar]
- Aguiar D. J., Johnson S. L., Oegema T. R. (1999). Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp. Cell Res. 246, 129–137 [DOI] [PubMed] [Google Scholar]
- Amacher S. L., Kimmel C. B. (1998). Promoting notochord fate and repressing muscle development in zebrafish axial mesoderm. Development 125, 1397–1406 [DOI] [PubMed] [Google Scholar]
- Battie M. C., Videman T. (2006). Lumbar disc degeneration: epidemiology and genetics. J. Bone Joint Surg. Am. 88 (Suppl. 2), 3–9 [DOI] [PubMed] [Google Scholar]
- Beckers A., Alten L., Viebahn C., Andre P., Gossler A. (2007). The mouse homeobox gene Noto regulates node morphogenesis, notochordal ciliogenesis, and left right patterning. Proc. Natl. Acad. Sci. USA 104, 15765–15770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington R. S., Robertson E. J. (1999). Axis development and early asymmetry in mammals. Cell 96, 195–209 [DOI] [PubMed] [Google Scholar]
- Boos N., Weissbach S., Rohrbach H., Weiler C., Spratt K. F., Nerlich A. G. (2002). Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine 27, 2631–2644 [DOI] [PubMed] [Google Scholar]
- Bouldin C. M., Gritli-Linde A., Ahn S., Harfe B. D. (2010). Shh pathway activation is present and required within the vertebrate limb bud apical ectodermal ridge for normal autopod patterning. Proc. Natl. Acad. Sci. USA 107, 5489–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelberg M. K., Banks G. M., Geiger D. F., Oegema T. R., Jr (1995). Identification of heterogeneous cell populations in normal human intervertebral disc. J. Anat. 186, 43–53 [PMC free article] [PubMed] [Google Scholar]
- Choi K. S., Cohn M. J., Harfe B. D. (2008). Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev. Dyn. 237, 3953–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B., Wilting J. (1992). From somites to vertebral column. Ann. Anat. 174, 23–32 [DOI] [PubMed] [Google Scholar]
- Dalgleish A. E. (1985). A study of the development of thoracic vertebrae in the mouse assisted by autoradiography. Acta Anat. 122, 91–98 [DOI] [PubMed] [Google Scholar]
- Eloy-Trinquet S., Nicolas J. F. (2002). Clonal separation and regionalisation during formation of the medial and lateral myotomes in the mouse embryo. Development 129, 111–122 [DOI] [PubMed] [Google Scholar]
- Fujita N., Miyamoto T., Imai J., Hosogane N., Suzuki T., Yagi M., Morita K., Ninomiya K., Miyamoto K., Takaishi H., et al. (2005). CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem. Biophys. Res. Commun. 338, 1890–1896 [DOI] [PubMed] [Google Scholar]
- George S. H., Gertsenstein M., Vintersten K., Korets-Smith E., Murphy J., Stevens M. E., Haigh J. J., Nagy A. (2007). Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl. Acad. Sci. USA 104, 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson A., Dreger M., Urban J. P. (2010). Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res. Ther. 12, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzel R. Z., Hawkins K., Ozminkowski R. J., Wang S. (2003). The health and productivity cost burden of the “top 10” physical and mental health conditions affecting six large U.S. employers in 1999. J. Occup. Environ. Med. 45, 5–14 [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington R., Costantini F., Lacey E. (1994). Manipulating The Mouse Embryo: A Laboratory Manual. New York: Cold Spring Harbor Press [Google Scholar]
- Hunter C. J., Matyas J. R., Duncan N. A. (2003). The notochordal cell in the nucleus pulposus: a review in the context of tissue engineering. Tissue Eng. 9, 667–677 [DOI] [PubMed] [Google Scholar]
- Johnson R. L., Laufer E., Riddle R. D., Tabin C. (1994). Ectopic expression of sonic hedgehog alters dorsal-ventral patterning of somites. Cell 79, 1165–1173 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Osada R., Kanamori M., Ishihara H., Ohmori K., Matsui H., Kimura T. (1999). Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine 24, 2456–2460 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Deasy B. M., Seo H. Y., Studer R. K., Vo N. V., Georgescu H. I., Sowa G. A., Kang J. D. (2009). Differentiation of intervertebral notochordal cells through live automated cell imaging system in vitro. Spine 34, 2486–2493 [DOI] [PubMed] [Google Scholar]
- Kim K. W., Lim T. H., Kim J. G., Jeong S. T., Masuda K., An H. S. (2003). The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine 28, 982–990 [DOI] [PubMed] [Google Scholar]
- Lewandoski M. (2001). Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2, 743–755 [DOI] [PubMed] [Google Scholar]
- Lickert H., Kutsch S., Kanzler B., Tamai Y., Taketo M. M., Kemler R. (2002). Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev. Cell 3, 171–181 [DOI] [PubMed] [Google Scholar]
- Liebscher T., Haefeli M., Wuertz K., Nerlich A. G., Boos N. (2010). Age-related variation in cell density of human lumbar intervertebral disc. Spine 36, 153–159 [DOI] [PubMed] [Google Scholar]
- Miller L. A., Wert S. E., Whitsett J. A. (2001). Immunolocalization of sonic hedgehog (Shh) in developing mouse lung. J. Histochem. Cytochem. 49, 1593–1604 [DOI] [PubMed] [Google Scholar]
- Minogue B. M., Richardson S. M., Zeef L. A., Freemont A. J., Hoyland J. A. (2010). Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 12, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrecic M. Z., Mitrecic D., Pochet R., Kostovic-Knezevic L., Gajovic S. (2010). The mouse gene Noto is expressed in the tail bud and essential for its morphogenesis. Cells Tissues Organs 192, 85–92 [DOI] [PubMed] [Google Scholar]
- Murphy P. L., Volinn E. (1999). Is occupational low back pain on the rise? Spine 24, 691–697 [DOI] [PubMed] [Google Scholar]
- Nishizaki Y., Shimazu K., Kondoh H., Sasaki H. (2001). Identification of essential sequence motifs in the node/notochord enhancer of Foxa2 (Hnf3beta) gene that are conserved across vertebrate species. Mech. Dev. 102, 57–66 [DOI] [PubMed] [Google Scholar]
- Ohshima H., Urban J. P., Bergel D. H. (1995). Effect of static load on matrix synthesis rates in the intervertebral disc measured in vitro by a new perfusion technique. J. Orthop. Res. 13, 22–29 [DOI] [PubMed] [Google Scholar]
- Pazzaglia U. E., Salisbury J. R., Byers P. D. (1989). Development and involution of the notochord in the human spine. J. R. Soc. Med. 82, 413–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage-Morin P. L., Gilmour H. (2010). Chronic pain at ages 12 to 44. Health Rep. 21, 53–61 [PubMed] [Google Scholar]
- Roberts S. (2002). Disc morphology in health and disease. Biochem. Soc. Trans. 30, 864–869 [DOI] [PubMed] [Google Scholar]
- Sawada A., Nishizaki Y., Sato H., Yada Y., Nakayama R., Yamamoto S., Nishioka N., Kondoh H., Sasaki H. (2005). Tead proteins activate the Foxa2 enhancer in the node in cooperation with a second factor. Development 132, 4719–4729 [DOI] [PubMed] [Google Scholar]
- Schatz O., Golenser E., Ben-Arie N. (2005). Clearing and photography of whole mount X-gal stained mouse embryos. Biotechniques 39, 650, 652,, 654 passim. [DOI] [PubMed] [Google Scholar]
- Setton L. A., Chen J. (2006). Mechanobiology of the intervertebral disc and relevance to disc degeneration. J. Bone Joint Surg. Am. 88 Suppl. 2, 52–57 [DOI] [PubMed] [Google Scholar]
- Smith L. J., Nerurkar N. L., Choi K. S., Harfe B. D., Elliott D. M. (2011). Degeneration and regeneration of the intervertebral disc: lessons from development. Dis. Model. Mech. 4, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- Stein S., Kessel M. (1995). A homeobox gene involved in node, notochord and neural plate formation of chick embryos. Mech. Dev. 49, 37–48 [DOI] [PubMed] [Google Scholar]
- Talbot W. S., Trevarrow B., Halpern M. E., Melby A. E., Farr G., Postlethwait J. H., Jowett T., Kimmel C. B., Kimelman D. (1995). A homeobox gene essential for zebrafish notochord development. Nature 378, 150–157 [DOI] [PubMed] [Google Scholar]
- Trout J. J., Buckwalter J. A., Moore K. C. (1982). Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat. Rec. 204, 307–314 [DOI] [PubMed] [Google Scholar]
- Uetzmann L., Burtscher I., Lickert H. (2008). A mouse line expressing Foxa2-driven Cre recombinase in node, notochord, floorplate, and endoderm. Genesis 46, 515–522 [DOI] [PubMed] [Google Scholar]
- Urban J. P., Smith S., Fairbank J. C. (2004). Nutrition of the intervertebral disc. Spine 29, 2700–2709 [DOI] [PubMed] [Google Scholar]
- Urban J. P. G., Roberts S., Ralphs J. R. (2000). The nucleus of the intervertebral disc from development to degeneration. Am. Zool. 40, 53–61 [Google Scholar]
- Videman T., Gibbons L. E., Battie M. C., Maravilla K., Vanninen E., Leppavuori J., Kaprio J., Peltonen L. (2001). The relative roles of intragenic polymorphisms of the vitamin d receptor gene in lumbar spine degeneration and bone density. Spine 26, E7–E12 [DOI] [PubMed] [Google Scholar]
- von Dassow G., Schmidt J. E., Kimelman D. (1993). Induction of the Xenopus organizer: expression and regulation of Xnot, a novel FGF and activin-regulated homeo box gene. Genes Dev. 7, 355–366 [DOI] [PubMed] [Google Scholar]
- Vujovic S., Henderson S., Presneau N., Odell E., Jacques T. S., Tirabosco R., Boshoff C., Flanagan A. M. (2006). Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J. Pathol. 209, 157–165 [DOI] [PubMed] [Google Scholar]
- Watanabe H., Yamada Y., Kimata K. (1998). Roles of aggrecan, a large chondroitin sulfate proteoglycan, in cartilage structure and function. J. Biochem. 124, 687–693 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Tamplin O. J., Beckers A., Gossler A., Rossant J. (2007). Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev. Cell 13, 884–896 [DOI] [PubMed] [Google Scholar]
- Zhou G. Q., Yang F., Leung V. V. L., Cheung K. M. C. (2008). Molecular and cellular biology of the intervertebral disc and the use of animal models. Curr. Orthop. 22, 267–273 [Google Scholar]
- Zizic Mitrecic M., Mitrecic D., Pochet R., Kostovic-Knezevic L., Gajovic S. (2010). The mouse gene Noto is expressed in the tail bud and essential for its morphogenesis. Cells Tissues Organs 192, 85–92 [DOI] [PubMed] [Google Scholar]