Abstract
EAT-2A and EAT-2B are single SH2-domain proteins, which bind to phosphorylated tyrosines of SLAM family receptors in murine NK cells. Whilst EAT-2 is a positive regulator in human cells, a negative regulatory role was attributed to the adapter in NK cells derived from EAT-2A-deficient 129Sv mice. To evaluate whether the genetic background or the presence of a selection marker in the mutant mice could influence the regulatory mode of these adapters, we generated EAT-2A-, EAT-2B- and EAT-2A/B-deficient mice using C57BL/6 (B6) ES cells. We found that NK cells from EAT-2A- and EAT-2A/B-deficient mice were unable to kill tumor cells in a CD244 or CD84 dependent manner. Furthermore, EAT-2A/B positively regulate phosphorylation of Vav-1, which is known to be implicated in NK cell killing. Thus, like in humans, the EAT-2 adapters act as positive regulators of SLAM family receptor specific NK cell functions in B6 mice.
Introduction
A large body of evidence supports the notion that NK cells participate in the defense against infections, in the regulation of immune responses and the surveillance of stressed or cancer cells (1). Effector functions of NK cells are regulated by the coordinated interaction of activating and inhibitory receptors (1). Ligation of activating receptors on the surface of NK cells results in cytokine production, cytolysis and migration, which is inhibited by the triggering of inhibitory receptors. Well-defined inhibitory receptors include the MHC Class I-recognizing members of the murine Ly49 family, human killer cell immunoglobulin-like receptors (KIRs) and CD94/NKG2 in both species (1–3). The inhibitory receptors mediate their effects through one or more immunoreceptor tyrosine-based inhibitory motifs (ITIM) in their cytoplasmic domains. Established human and mouse NK cell activating receptors are NKG2D, NKRP1, CD16, DNAM1, activating human KIRs and activating murine Ly49. As several activating NK cell receptors do not contain cytoplasmic domains, they associate with and signal through adapter molecules such as DAP12, FcR-γ and CD3ζ, which contain the immunoreceptor tyrosine-based activation motif (ITAM) (1, 4).
In recent years, there has been accumulating evidence implicating the SLAM family of receptors (SLAMF1–9) and their specific intracellular adapters in immune regulation (5, 6). SLAMF receptors, which are expressed on hematopoietic cells (6) are self-ligand adhesion molecules with the exception of CD244 and its ligand CD48. After receptor ligation, the tyrosines present on their intracellular domain are phosphorylated permitting the association to the SAP family of adaptors: SAP, EAT-2A and, in rodents, EAT-2B (ERT (9)). These adapters are essentially composed of an SH2 domain and a short C-terminal tail, and are able to trigger biochemical signals that seem crucial for the SLAM-dependent and SLAM-independent functions (5, 6). In human NK cells, SAP and EAT-2 mediate the cytotoxic function of CD244, CD319 and CD352 (6). SAP positively regulates mouse NK cell functions, which are initiated by the SLAMF receptors. However, EAT-2A and EAT-2B play a dual role regulating the function of the SLAMF receptors in NK cells derived from a 129Sv background (7–9).
Because extensive polymorphisms as well as differences in expression have been found in the SLAMF locus between 129Sv and B6 mouse strains (6, 10), we set out to test the hypothesis that the strain background in which the EAT-2A/B knockout mice are generated influences the positive or negative regulatory function of a receptor. To this end, we targeted B6 ES cells to generate novel EAT-2A-, EAT-2B-, EAT-2A/B and EAT-2A/B × SAP-deficient mice, as well as CD244-deficient mice without selection cassettes on a B6 background. We find that EAT-2A and EAT-2B positively regulate cytotoxicity mediated by CD244 and CD84 in B6 mouse NK cells.
Materials and Methods
Generation of EAT-2A-, EAT-2B- and EAT-2A/B-deficient B6 mice
A B6 BAC clone containing the EAT-2A and EAT-2B genes was used to construct a targeting vector with a Neomycin resistance cassette flanked by two LoxP sites. EAT-2A or EAT-2B targeted ES cell clones generated by standard methods were injected into blastocysts, and the chimeric mice were crossed with B6 mice. In order to delete the Neomycin resistance gene from the targeted locus, EAT-2A and EAT-B heterozygous mice were crossed with B6 Cre-deleter mice (11) (Fig. S1 and S2).
To generate EAT-2A/B double deficient mice, we used a modified EAT-2B targeting vector to retarget the previously generated EAT-2A mutant ES clone (Fig. S3). Co-integration of the two targeting vectors on the same chromosome was assessed by in vitro transfecting targeted ES-cell-clones with a Cre recombinase expression vector. Deletion of the whole EAT-2 locus was confirmed by PCR (Fig. S3). To delete Neomycin and Hygromycin resistance genes from EAT-2A/B targeted loci, homozygous EAT2A/B−/− mice were bred with B6 Cre-deleter mice (11).
NK cell isolation
Splenocytes harvested from wt or mutant B6 mice were processed in phosphate-buffered saline (PBS) with 2% fetal calf serum. After red blood cell lysis, NK cells were isolated from spleen cells using magnetic microbeads according to the manufacturer's recommendations (Miltenyi Biotec). Purified NK cells (>92% NK1.1 positive) were cultured in DMEM medium supplemented with 1000 units recombinant human IL-2 (Biolegend) for 7 days, as described (14).
Cell lines
The cell lines RMAS / CD48+ or RMAS / CD48− (H-2blo), P815/CD48+ or P815/CD48− (H-2d), B16, YB2/0, CHO and YAC-1 were cultured in supplemented DMEM medium, as described (13). To generate CD84+ stable cells, CD84 cDNA was cloned into the pcDNA3.1 expression vector that was then stably transfected into P815 or B16 tumor cells.
Cytotoxicity assays
Specific lysis of targets was determined by using a standard 4h [51Cr]-release assay in 96-well U-bottom plates as previously described (12). Alternative non-radioactive cytotoxicity assay was used to quantitatively measure dehydrogenase (LDH) that is released upon cell lysis (CytoTox 96, Promega). Redirected killing assays using P815 targets were performed as previously described (13).
In vivo tumor clearance assay
Tumor clearance assays were performed, as previously described (14). Briefly, the target cells were labeled with 5μM CFSE (Molecular Probes) at 37°C for 10 min. CFSE-labeled target cells were washed three times with cell culture media. 3×106 target cells were injected i.p. in 300μl PBS. The mice were sacrificed and peritoneal cells were recovered after 18 hours. The residual target cells were counted by FACS.
Immunoprecipitation and Western blot analysis
Approximately 10–20 × 106 IL-2-generated LAK cells were labeled with anti-CD244 mAb for 30 min on ice. Anti-mouse antibody was used for crosslinking at 37°C for 15 min. Cells were lysed and CD244 or Vav-1 was immunoprecipitated. Immunoprecipitation and western blot analysis were performed as previously described (13)
Results and Discussion
Impaired in vivo killing by EAT-2A/B−/− and SAP−/− NK cells using CD244− and CD84–specific target cells
To determine whether the genetic background could play a role in the function of EAT-2A and EAT-2B, EAT-2A and EAT-2B deficient mice were generated from B6 derived ES cells (Bruce4) in which the first exon of EAT-2A gene or EAT-2B gene was replaced by the LoxP-flanked selection markers neomycin and/or hygromycin. After breeding the mutant mice with the Cre-deleter transgenic mouse (11) none of the mutant mice contained the selection cassettes (Fig. S1, S2 and S3). EAT-2A, EAT-2B or both transcripts were not detected by RT-PCR in NK cells from the resulting mutant mouse strains (Fig. S3).
To evaluate the role of the EAT-2-adapters in CD244-mediated NK cell functions, EAT-2A/B−/−, EAT-2A−/− and EAT-2B−/− mice were injected i.p with CFSE-labeled RMAS that express CD48, the high affinity ligand for CD244. As controls RMAS/CD48− cells were used. After 18 hours, the number of RMAS/CD48+ tumor cells was significantly higher in the peritoneal cavity of EAT-2A/B−/− and EAT-2A−/− mice than in B6 mice (Fig. 1A and Fig. S4). However, the number of RMAS/CD48+ tumor cells recovered from the peritoneal cavity of EAT-2B−/− mice was comparable to that in wt mice (Fig. S4). As expected, without triggering of CD244 by its ligand CD48 the absence of EAT-2A/B in the NK cells had no effect (Fig. 1A).
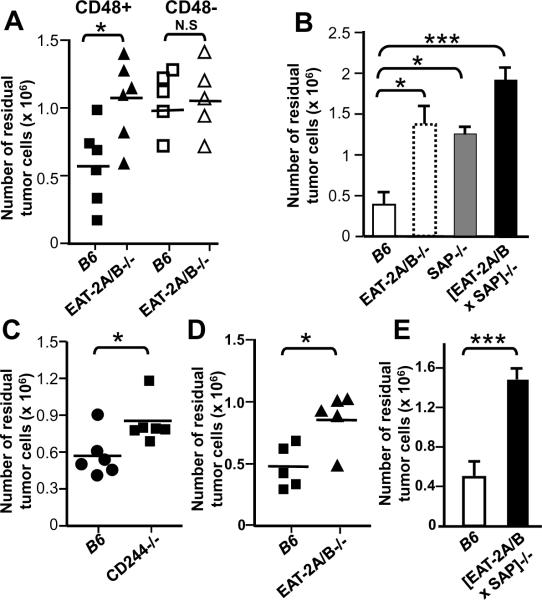
FIGURE 1. SAP family adaptors are required for in vivo clearance of CD48+ or CD84+ targets.
CFSE-labeled RMAS / CD48+, RMAS/CD48− cells [3×106] (A, B and C) or P815/CD84+ cells [3×106] (E and F) were injected in the peritoneum of wt, EAT-2A/B−/−, SAP−/−, EAT-2A/B × SAP−/− or CD244−/− B6 mice. After 18 hours, the tumor cells were recovered from the peritoneum and the number of tumor cells was calculated based on the percentage of CFSE+ cells by flow cytometry. The data are representative of three independent experiments. * p<0.05, ***p<0.001.
EAT-2A/B−/− and SAP−/− mice were equally impaired in their ability to remove the RMAS/CD48+ cells (Fig. 1B). The striking decrease of CD244-dependent cytotoxicity in EAT-2A/B−/− and SAP−/− mice raised the question whether NK cell functions would be more severely impaired by the loss of all the SAP-related adaptors. Indeed, in mice that lack all three adapters, i.e. EAT-2A/B−/− × SAP−/−, clearance of RMAS/CD48+ tumor cells mice was less than in either EAT-2A/B−/− or SAP−/− mice (Fig. 1B). These studies strongly suggest that in B6 mice, both EAT-2A/B and SAP are positive regulators of CD244-dependent in vivo NK cell killing and that these specific adapters may act synergistically.
In agreement with the notion that both the CD244-EAT-2A/B and CD244-SAP pathways represent an activating receptor-adapter system in B6 mice, is our observation that CD244−/− mice are also impaired in the in vivo clearance of RMAS / CD48+ tumor cells (Fig. 1C). When NK cells had been removed by treatment with anti-NK1.1 the EAT-2A/B mutation did not have any effect on the killing of RMAS/CD48+ cells (Fig. S5). Thus, like in humans, in B6 mice CD244 and its SAP-related adaptors predominately appear to be part of the activating system for NK cell cytotoxicity.
To determine whether NK killing by another SLAMF receptor, which binds EAT-2 (20) is also positively regulated by EAT-2A/B, in vivo killing of CD84 expressing P815 tumor cells was evaluated in EAT-2A/B−/− and SAP−/− mice. Again, in the absence of EAT-2A/B, the number of P815/CD84+ target cells was markedly increased as compared with wt B6 littermate controls (Fig.1D). Similarly, the number of P815/CD84+ target cells recovered in EAT-2A/B × SAP−/− mice was dramatically higher than in wt littermates (Fig. 1E). Collectively, these data strongly suggest that, like SAP, EAT-2A/B positively regulate CD244− and CD84–dependent NK cell functions in vivo. Furthermore, the data suggest that the contribution of EAT-2A is greater than that of EAT-2B.
EAT-2A/B are positive regulators of in vitro CD244 and CD84 dependent NK cell killing
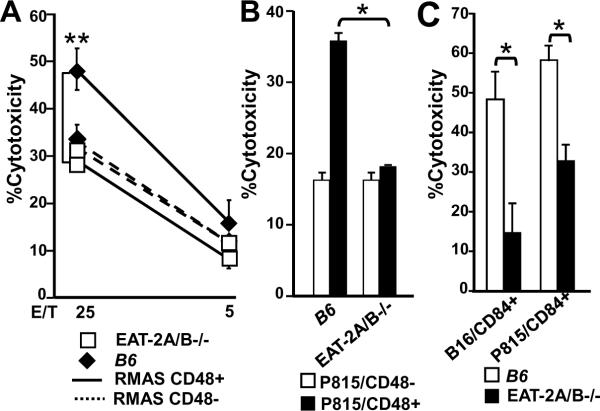
To evaluate whether the in vivo observations correlated with in vitro killing, RMAS, P815 and B16 target cells with or without CD48 or CD84 were used in either an in vitro [51Cr]-release assay or in an LDH-cytotoxicity assay. Compared to wt NK cells, EAT-2A/B−/− NK cells were impaired in their ability to in vitro lyse RMAS/CD48+, but not RMAS / CD48− target cells (Fig. 2A). Whereas EAT-2A/B−/− and EAT-2A−/− NK cells (Fig.2B and Fig. S6) had lost their ability to kill P815/CD48+ targets, the EAT-2B−/− mutation had no impact. Similarly, P815/CD84+ or B16/CD84+ targets were killed less efficiently by NK cells that lacked the EAT-2A/B genes (Fig. 2C). By contrast, NK cells derived from EAT-2A-, EAT-2B-, or EAT-2A/B-mutant mice efficiently killed the target cells YB2/0, YAC-1 and CHO (Fig. S7A, S7B, S7C). Thus, whereas EAT-2A positively regulates in vitro NK cell killing that is mediated by CD244 and CD84, the effect of the EAT-2B mutation is marginal.
FIGURE 2. Ligation of CD244 or CD84 by CD48 or CD84 expressing targets enhances NK cytotoxicity in B6, but not EAT-2A/B−/− mice.
NK cells isolated from the spleens of wt or EAT-2A/B−/− B6 were cultured in an IL-2 containing medium for seven days. Cytolytic activity was determined by the [51Cr]- or LDH-release assay. NK cell cytotoxicity against RMAS/CD48+ or RMAS/CD48− cells (LDH release assay) (A), P815/CD48+ or P815/CD48− cells (51Cr release assay) (B) or B16/CD84+ or P815/CD84+ cells (LDH release assay) (C). Pooled data from three independent experiments were shown. *p<0.05, **p<0.01. Error bars represent SD.
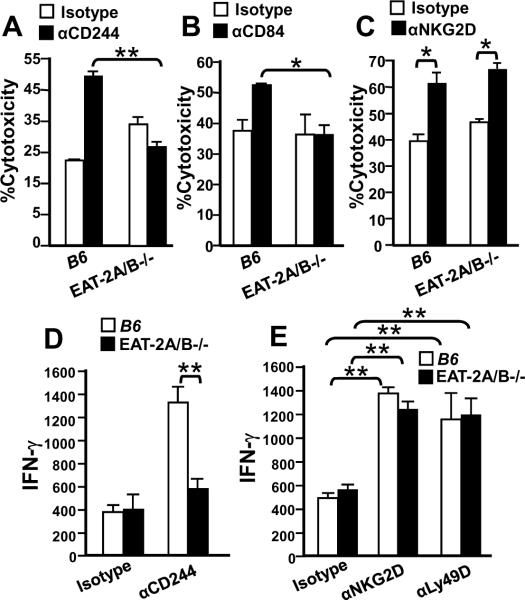
EAT-2A/B−/−, EAT-2A−/− and EAT-2B−/− NK cells are also defective in αCD244-dependent killing of [51Cr] -labeled P815 cells (Fig. 3A and Fig. S8A). Similarly, αCD84 coated P815 cells were more efficiently killed in a redirected killing assay by wt than by EAT-2A/B−/− NK cells (Fig. 3B). A defect in IFN-γ production by EAT-2A/B−/− NK cells was also observed by triggering of CD244 with αCD244 mAb (Fig. 3D)
FIGURE 3. Defective αCD244 or αCD84 redirected killing and IFN-γ production by EAT-2A/B−/− NK cells.
NK cells from the spleens of wt or EAT-2A/B−/− mice were were cultured with IL-2-containing medium for seven days and analyzed in a redirected killing assay against the FcγR+ P815 target cells either in the absence or presence of anti-CD244 (A) anti-CD84 (B) or anti-NKG2D (C) mAbs. The lytic activity of wt or EAT-2A/B−/− NK cells was tested against P815 target cells by measuring 51Cr (A and C) or LDH (B) released into the cell supernatants. Wt or EAT-2-A/B−/− NK cells purified from the spleens were cultured in the presence of IL-2. At day 7, the NK cells were stimulated with anti-CD244 (5μg/ml) (D), anti-NKG2D (5μg/ml) or anti-Ly49D (5μg/ml) (E) mAbs for 24 hours. Culture supernatants were harvested and IFN-γ production was quantified by ELISA. Pooled data from three independent experiments were shown. *p<0.05, **p<0.01. Error bars represent SD.
To exclude the possibility that the defective redirected cytotoxicity and IFN-γ production by EAT-2A/B−/− NK cells was caused by a global dysfunction of NK cells, monoclonal antibodies directed against the activating NK cell receptors NKG2D and Ly49D were used. EAT-2A/B−/−, EAT-2A−/− or EAT-2B−/− NK cells lysed [51Cr]-labeled P815 cells coated with αLy49D or αNKG2D equally efficiently as wt B6 NK cells (Fig. 3C and Fig. S8B, S8C). Thus, consistent with the absence of ITSM motifs in the cytoplasmic portions of these NK receptors the absence of EAT-2A and -2B did not affect their functions. This was confirmed by the finding that IFN-γ production induced by αLy49D or αNKG2D was comparable in EAT-2A−/−, EAT-2B−/− or EAT-2A/B−/− and B6 NK cells (Fig. 3E and Fig. S9).
Taken together, the outcomes of these experiments demonstrate that the lytic functions and IFN-γ production of EAT-2A−/− and EAT-2A/B−/− NK cells are defective in a CD244− and CD84− mediated manner. Our data, therefore, contrast with a previous report, in which 129 background EAT-2A−/− and EAT-2B−/− NK cells were found to have enhanced ability to kill xenogeneic target cells and also increase IFN-γ production upon triggering not only by CD244, but also by other NK cell activating receptors NKG2D and Ly49D (9). These differences may be due to different genetic background between 129 and B6 mice and to the absence of selection markers in the targeted allele. Whether and how the mouse genetic background affects EAT-2A and EAT-2B regulation of NK cell function will require further investigation.
Absence of EAT-2A and EAT-2B affects CD244-mediated phosphorylation of Vav
The precise mechanisms by which EAT-2A and EAT-2B are involved in the CD244-mediated signaling pathways are not well understood. Upon engagement of CD244 by αCD244 or CD48 expressing target cells the receptor is recruited to lipid rafts, where the tyrosines of the ITSM motifs are phosphorylated, leading to recruitment and activation of several downstream signaling molecules. In addition to SAP and EAT-2 these include Vav-1, SHIP, PI3K, Csk, PLCγ, SHP-1, SHP-2, and LAT (6, 15).
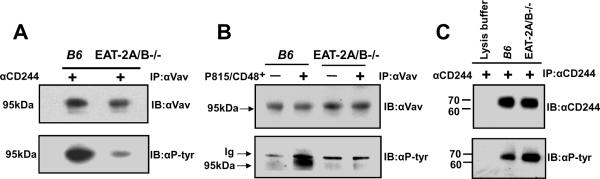
To assess whether the absence of EAT-2A/B would affect a downstream signaling molecule that could be responsible for the impaired CD244-mediated cytotoxicity, we focused on the guanine nucleotide exchange factor Vav-1 (16). To this end, wt and EAT-2A/B−/− cultured NK cells were stimulated with αCD244 mAb and NK cell lysates were used for immunoprecipitation with anti-Vav-1. Tyrosine phosphorylation of Vav-1 was significantly reduced in EAT-2A/B−/− NK cells compared to wt NK cells (Fig. 4A). Decreased phosphorylation of Vav-1 was also observed with EAT-2A/B−/− NK cells triggered by CD48-expressing targets (Fig. 4B). As the GEF activity of Vav proteins is activated by tyrosine phosphorylation and the phosphorylated Vav proteins are able to positively regulate NK cell-mediated killing, the reduced phosphorylation of Vav most likely contributes to impaired lysis in the EAT-2A/B−/− NK cells.
FIGURE 4. Phosphorylation of Vav-1 by αCD244 or by ligation with CD48 expressing targets is reduced in EAT-2A/B−/− NK cells.
A. IL-expanded splenic NK cells from wt or EAT-2A/B−/− mice were stimulated with αCD244 mAb for 15 min. Cell lysates were used for immunoprecipitation (IP) with αVav-1 mAb and were probed with anti-p-Tyr mAb (4G10) and reprobed with αVav-1. B, NK cells were stimulated with CD48-expressing P815 cells for 30 min. and analyzed, as in Fig. 4A. C, Lysates of αCD244 activated NK cells form wt or EAT-2A/B−/− mice were analyzed by probing αCD244 immunoprecipitates with anti-p-Tyr mAb (4G10). CD244 was quantitated by reprobing the membrane with αCD244.
This defect of Vav-phosphorylation is not dependent of phosphorylation of the receptor itself, because tyrosine phosphorylation of CD244 was not affected in EAT-2A/B−/− NK cells (Fig. 4C). As we know that phosphorylation of CD244 precedes SAP and EAT-2A/B binding to its cytoplasmic tail (17), and because SLAMF receptors can be phosphorylated in the absence of the adapters (6), the role of CD244 phosphorylation in the pathway towards Vav-1 is not immediately clear. In general, NK cell activation requires synergizing receptors, e.g. NKG2D and CD244, which is regulated at the level of Vav-1 by a hierarchy of mechanisms. Phosphorylation of phospholipase PLC-γ2, Ca2+ mobilization, and degranulation are involved. It is likely that c-Cbl plays an inhibitory role (18). Thus, as the pathway from CD244 to Vav will, like in humans, undoubtedly involve a number of factors that are recruited to the NK cell synapse by CD244, this requires a more detailed study.
Taken together, the outcomes of our studies demonstrate that EAT-2A/B and EAT-2A, like SAP, positively regulate CD244-mediated NK cell functions in B6 mice, which is different from the model that these adapters have dual roles in 129 NK cells. The diversity of EAT-2A and EAT-2B functions in NK cells may result from extensive polymorphism of SLAM family members between two mouse strains, influence of the presence of selection markers in gene targeted loci, a strain-dependent gender effect (19) or different environment conditions of animal facilities. Moreover, our studies also suggest EAT-2A and EAT-2B are involved in phosphorylation of downstream effector molecule Vav-1, which plays a critical role in natural cytotoxicity. The notion that EAT-2 is a positive regulatory molecule was first discovered using human cells (20) is consistent with SLAMF receptors functioning as positive regulators on human NK cells (6).
Supplementary Material
Acknowledgements
We thank Dr. Klaus Rajewsky for providing Bruce 4 ES cells and advice and Dr. Vinay Kumar for the RMAS/CD48+/− and P815/CD48+/− target cells.
The work was supported by National Institutes of Health Grant xxxx, (to C.T).
Abbreviations used in this paper
- SLAM
Signaling Lymphocyte Activation Molecule
- SAP
SLAM-Associated Protein
- EAT-2
EWS/FLI1 Activated Transcript 2
- ERT
EAT-2 Related Transcript
- SHP2
SH2 domain-containing tyrosine phosphatase 2
- NK
Natural Killer
Footnotes
Disclosures The authors have no financial conflicts of interest.
References
- 1.Orange JS, Ballas ZK. Natural killer cells in human health and disease. Clin Immunol. 2006;118:1–10. doi: 10.1016/j.clim.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Long EO. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 4.Colucci F, Di Santo JP, Leibson PJ. Natural killer cell activation in mice and men: different triggers for similar weapons? Nat. Immunol. 2002;3:807–813. doi: 10.1038/ni0902-807. [DOI] [PubMed] [Google Scholar]
- 5.Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol. 2010;32:157–171. doi: 10.1007/s00281-009-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C. The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 7.Dong Z, Cruz-Munoz ME, Zhong MC, Chen R, Latour S, Veillette A. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat Immunol. 2009;10:973–980. doi: 10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 8.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat Immunol. 2009;10:297–305. doi: 10.1038/ni.1693. [DOI] [PubMed] [Google Scholar]
- 9.Roncagalli R, Taylor JE, Zhang S, Shi X, Chen R, Cruz-Munoz ME, Yin L, Latour S, Veillette A. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nat Immunol. 2005;6:1002–1010. doi: 10.1038/ni1242. [DOI] [PubMed] [Google Scholar]
- 10.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Morel L L, et al. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu YY, George T, Dorfman JR, Roland J, Kumar V, Bennett M. The role of Ly49A and 5E6 (Ly49C) molecules in hybrid resistance mediated by murine natural killer cells against normal T cell blasts. Immunity. 1996;4:67–76. doi: 10.1016/s1074-7613(00)80299-x. [DOI] [PubMed] [Google Scholar]
- 13.Schatzle JD, Sheu S, Stepp SE, Mathew PA, Bennett M, Kumar V. Characterization of inhibitory and stimulatory forms of the murine natural killer cell receptor CD244. Proc Natl Acad Sci U S A. 1999;96:3870–3875. doi: 10.1073/pnas.96.7.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KM, McNerney ME, Stepp SE, Mathew PA, Schatzle JD, Bennett M, Kumar V. CD244 acts as a non-major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. J Exp Med. 2004;199:1245–1254. doi: 10.1084/jem.20031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNerney ME, Lee KM, Kumar V. 2B4 (CD244) is a non-MHC binding receptor with multiple functions on natural killer cells and CD8+ T cells. Mol Immunol. 2005;42:489–494. doi: 10.1016/j.molimm.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Cella M, Fujikawa K, Tassi I, Kim S, Latinis K, Nishi S, Yokoyama W, Colonna M, Swat W. Differential requirements for Vav proteins in DAP10- and ITAM-mediated NK cell cytotoxicity. J Exp Med. 2004;200:817–823. doi: 10.1084/jem.20031847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayós J, Nguyen KB, Wu C, Stepp SE, Howie D, Schatzle JD, Kumar V, Biron CA, Terhorst C. Potential pathways for regulation of NK and T cell responses: differential X-linked lymphoproliferative syndrome gene product SAP interactions with SLAM and 2B4. Int Immunol. 2000;12:1749–1757. doi: 10.1093/intimm/12.12.1749. [DOI] [PubMed] [Google Scholar]
- 18.Kim HS, Das A, Gross CC, Bryceson YT, Long EO. Synergistic signals for natural cytotoxicity are required to overcome inhibition by c-Cbl ubiquitin ligase. Immunity. 2010;32:175–186. doi: 10.1016/j.immuni.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya SV, Stepp SE, McNerney ME, Lee JK, Bennett M, Lee KM, Stewart CL, Kumar V, Mathew PA. Targeted disruption of the 2B4 gene in mice reveals an in vivo role of 2B4 (CD244) in the rejection of B16 melanoma cells. J Immunol. 2005;174:800–807. doi: 10.4049/jimmunol.174.2.800. [DOI] [PubMed] [Google Scholar]
- 20.Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J. 2001;20:5840–5852. doi: 10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.