Summary
microRNAs are endogenous non-coding small RNAs with important roles in many biological pathways; their generation and activity are under precise regulation [1–3]. Emerging evidence suggests that miRNA pathways are precisely modulated with controls at the level of transcription [4–8], processing [9–11] and stability [12,13], with miRNA deregulation linked with diseases [14] and neurodegenerative disorders [15]. In the Drosophila miRNA biogenesis pathway, long primary miRNA transcripts undergo sequential cleavage [16–18] to release the embedded miRNAs. Mature miRNAs are then loaded into Argonaute 1 (Ago1) within the RNA-induced silencing complex (RISC) [19,20]. Intriguingly, we found that Drosophila miR-34 displays multiple isoforms that differ at the 3'end, suggesting a novel biogenesis mechanism involving 3'end processing. To define the cellular factors responsible, we performed an RNAi screen and identified a putative 3'→5' exoribonuclease CG9247/nibbler essential for the generation of the smaller isoforms of miR-34. Nibbler (Nbr) interacts with Ago1 and processes miR-34 within RISC. Deep sequencing analysis revealed a larger set of multi-isoform miRNAs that are controlled by nibbler. These findings suggest that Nbr-mediated 3' end processing represents a critical step in miRNA maturation that impacts miRNA diversity.
Results and Discussion
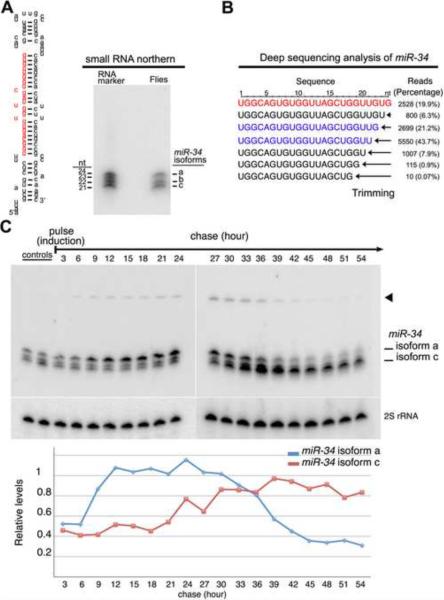
While miRNAs are typically annotated and observed as a single species, we found that miR-34 showed a pattern of three major isoforms of 24, 22 and 21 nt in Northern blots from adult Drosophila (Fig. 1A). Deep sequencing analysis [21] also showed that miR-34 is present in multiple forms that all bear the same 5' terminus but differ at their 3' ends, presenting a nested series (Fig. 1B). To assess the relationship among these, we designed a pulse-chase experiment to follow miR-34 biogenesis. Heat shock driven primary miR-34 was tightly induced for 30 min, then monitored over time in adult flies. The longest isoform, isoform a (24nt), was predominant initially, while the accumulation of the shorter isoforms was delayed, but then increased over time (Fig. 1C). Moreover, as the 21 nt isoform accumulated, the 24 nt form was lost in a seemingly reciprocal manner, suggesting that the 24-mer may be converted into the 21-mer.
Figure 1. Fly miR-34 Shows Multiple Isoforms Whose Generation Appears Dependent on 3' End Trimming.
(A) miR-34 has multiple forms in adult flies. Left, miR-34 precursor, mature 24-nt sequence in red. Right, Northern for miR-34. Isoforms of 24, 22 and 21nt, labeled a, b, c, respectively.
(B) miR-34 isoforms from a deep sequencing fly S2 cell dataset [21]. In red, 24nt isoform a. In blue, isoforms b and c. These are 99.1% of the miR-34 reads.
(C) Northern blot analysis of miR-34 isoform accumulation in vivo. Transient induction of pri-miR-34 by hs-GAL4 in adult flies leads to initial accumulation of isoform a, which is lost over time while the shorter isoforms accumulate. Arrowhead, pre-miR-34.
(D) Quantification of miR-34 isoforms from pulse-chase in C. Values normalized to 2S rRNA.
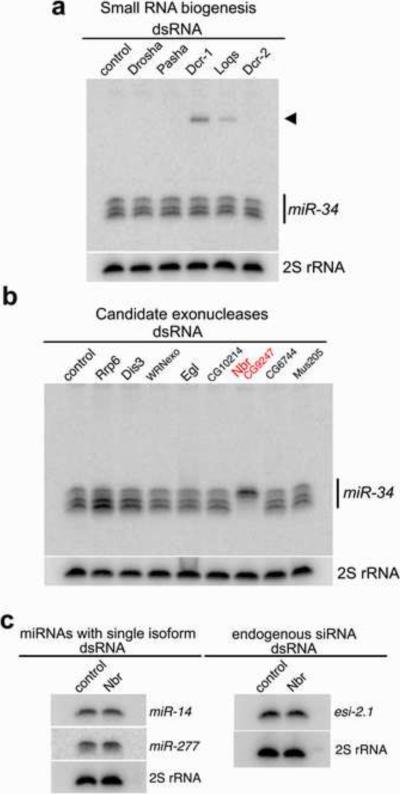
To define the mechanism, we treated cells with dsRNA targeting specific genes within the small RNA biogenesis pathways, and assessed the miR-34 pattern by Northern blot. Imprecise cleavage of the precursor transcript could result in the production of the multiple forms. However, reduction of either Drosha or Dcr-1, or their binding partners Pasha and Loquacious, or Dicer-2 (Dcr-2), responsible for siRNA generation, did not alter the pattern (Fig. 2A). Therefore, we reasoned that the smaller isoforms may instead be generated by an exonuclease that sequentially processes the longest isoform into the nested series observed. To test this hypothesis, we performed an RNAi screen against the predicted 3'→5'exonucleases in Drosophila, including components of the RNA exosome (Table S1). This identified one gene, CG9247 (which we named nibbler/nbr), with a striking effect: depletion of nbr led to a dramatic accumulation of the miR-34 large isoform with a concomitant loss of the shorter isoforms (Fig. 2B; Fig. S1). In contrast, loss of nbr did not appear to alter the sizes or levels of miRNAs that normally show a single isoform by Northern, such as miR-14 and miR-277 (Fig. 2C). We also examined whether nbr knockdown had an effect on endogenous siRNAs, but saw no impact on esi-2.1 (Fig. 2C). These data suggested that the novel putative exoribonuclease Nbr is required to generate the shorter isoforms of the multi-isoform miRNA miR-34, but is not required for general small RNA biogenesis.
Figure 2. nbr is Required to Generate the Isoforms of miR-34.
(A) Depletion of known factors in the small RNA biogenesis pathways has no effect on miR-34.
(B) Depletion of candidate exoribonucleases shows that loss of CG9247/Nbr (red) leads to an accumulation of the 24nt isoform, with dramatic reduction of the shorter isoforms.
(C) Cells depleted of Nbr are not altered in single isoform miRNAs or endogenous siRNA esi-2.1.
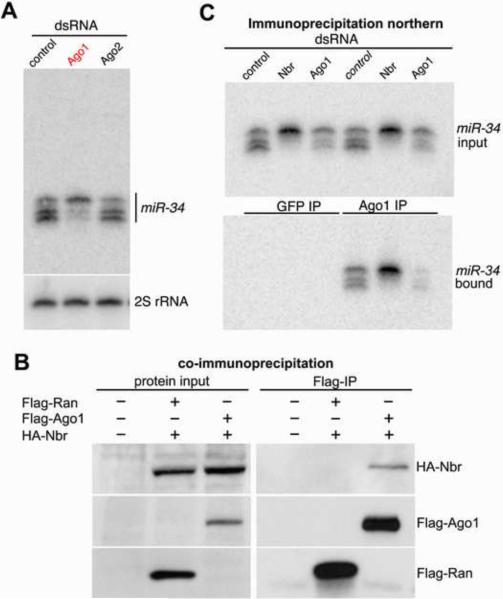
The Nbr exoribonuclease domain shows closest sequence homology to human EXD3, falling within the E. coli RNAse D protein family; this includes the Werner exoribonuclease and C. elegans Mut-7 involved in transposon silencing (Fig. S1;[22]). Nbr, however, showed no predicted RNA binding domain, suggesting that it may function with a partner with RNA binding capacity, to bring Nbr activity to RNA substrates. To define these, we then performed a second RNAi screen genes known to bind RNA or associate with small RNA silencing pathways, including the two somatic RISC-associated Argonautes (Table S1). Strikingly, loss of Ago1 phenocopied nbr depletion: accumulation of the 24 nt isoform occurred, with reduction of the shorter isoforms (Fig. 3A). Controls indicated that knockdown of Ago1 had no effect on nbr expression, and nbr knockdown had no effect on Ago1 expression. These data suggested that Ago1 is also required for trimming, and that Nbr and Ago1 may act in a complex. Co-immunoprecipitation (IP) studies indicated that HA-tagged Nbr associates with Flag-tagged Ago1, but not with a control protein (Flag-Ran) (Fig. 3B). RNase treatment indicated the association was not RNA-dependent (Fig. S2). Proteomic studies have identified both Ago1 and Nbr as small RNA associated proteins [23], underscoring the specificity of the interaction. Since Nbr associates with Ago1, we hypothesized that miR-34 3'end processing may occur in the context of RISC. Indeed, IP of Ago1 revealed that all miR-34 isoforms were bound (Fig. 3C). Furthermore, when Nbr was depleted, the longest miR-34 isoform remained bound to Ago1 (Fig. 3C). Altogether, these data suggest that the 24 nt miR-34 isoform is first generated by Dcr-1 then loaded into RISC. Next, Nbr, in association with Ago1, processes the long 24 nt isoform into shorter isoforms that remain loaded in RISC.
Figure 3. Nbr Interacts with Ago1-RISC.
(A) Small RNA Northern blot analysis of mir-34 isoforms. Depletion of Ago1 phenocopies Nbr knockdown.
(B) Ago1 and Nbr interact by co-IP. Cells were untreated or transfected with HA-Nbr and Flag-Ago1 or Flag-Ran (control). Following IP, interacting proteins were probed by immunoblot. Input, 10% of Flag-IP.
(C) All miR-34 isoforms co-IP with Ago1. Cells were treated with dsRNA to control (LacZ), Nbr or Ago1, and IPs were performed anti-GFP (control) or Ago1 antibodies. Input and IP-ed RNA were analyzed by Northern blotting for miR-34.
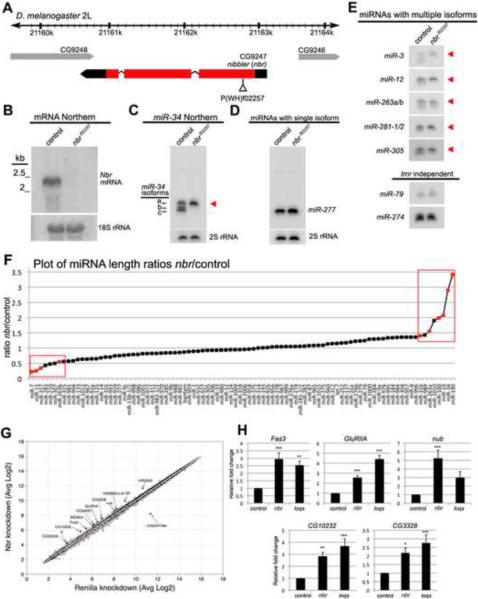
To assess the in vivo role of Nbr, we analyzed the expression and function of nbr in flies. Northerns revealed that nbr is expressed during development and in the adult, with peaks during the late larval/early pupal stage, and in adults (not shown). Analysis of nbr mRNA levels in animals with a transposon insertion in the coding region (nbrf02257) showed that homozygous mutants (nbr−/−) lacked nbr expression (Fig. 4A, B). nbr−/− flies were semi-lethal, and sterile, indicating that nbr function is critical. Given the homology to Mut-7, we examined levels of transposons, but found no evidence linking nbr to transposon silencing (not shown). Assessment of miR-34 expression in nbr−/− flies phenocopied cells treated with dsRNA: the shorter isoforms were abolished, while the 24-nt form accumulated (Fig. 4C). As in cells, there was no striking effect on single-form miRNAs like miR-277 (Fig. 4D). Furthermore, miR-34* levels and isoform distribution appeared unaffected (Fig. S1). These data indicated that nbr modulates the isoform abundance of miR-34.
Figure 4. nbr is Required in vivo to Process Select miRNAs and Silence Target mRNAs.
(A) Genomic map of the nbr locus. Coding region in red, with transposon insertion highlighted.
(B) Northern blot for nbr. The nbrf02257 mutant shows complete mRNA loss.
(C) Shorter isoforms of miR-34 are abolished in the nbr mutant. Arrow, isoform a.
(D) Northern blot of single-isoform miR-277, which is not altered in nbr−/−.
(E) Comparison of multiple-isoform miRNAs from control and nbrf02257 flies. Some miRNAs require nbr (red arrowheads), while others are nbr-independent.
(F) The ratio of the most frequent form of the miRNA in wild type, compared to the sum of all other forms, was generated for nbr and control. The ratios were compared (nbr ratio/control ratio), and plotted. The ratio was excessively large or low when isoform biogenesis is defective. Red boxes, miRNAs with extreme ratios that were further analyzed. Red symbols are confirmed Nbr-targets (Fig. S4).
(G) Scatterplot of microarray data from cells treated with dsRNA against Nbr or Renilla control. Highlighted, all of the genes >1.5 fold changed in either direction.
(H) Realtime PCR for mRNAs from nbr and loqs mutant flies. (4–6 experiments; *p<0.05, **p<0.01, ***p<0.001).
To assess the broader impact of Nbr function, we screened 65 miRNAs by Northern blot of RNA from cultured cells and flies. We identified 9 additional miRNAs with multiple isoforms: mir-2, miR-3, miR-12, miR-79, miR-263a/b, miR-274, miR-279, miR-281-1/2 and miR-305. The expression patterns of five of these were altered in nbr−/− mutants, exhibiting accumulation of the longest isoforms with concomitant loss of the shorter isoforms (Fig. 4E; miR-2 family not studied further due to cross hybridization between members). Analysis of small RNA profiling data from cells [21] confirmed that two of these (miR-263a, miR-305) had significant levels of multiple forms that differed at the 3'end (Table S2); miR-3, miR-12, miR-281, and miR-274 were too low for analysis. Three multiple-isoform miRNAs (miR-79, miR-274, and miR-279) did not show an altered pattern in nbr−/− flies (Fig. 4E). The deep sequencing dataset revealed that miR-279 displays a series of isoforms that do differ at the 3'end; since miR-279 processing is nbr-independent, nbr may be one member of a larger set of genes or mechanisms responsible for 3'end diversity. miR-79 isoforms differed at the 5'end, suggesting that mechanisms also exist for 5' end diversity of miRNAs.
We further investigated the extent to which trimming is involved in miRNA processing by deep sequencing the small RNAs from flies, comparing nbr mutants to controls. There was no major impact on the size distribution of small RNAs as a whole or miRNAs in particular (Fig. S3A–B). To more carefully assess isoforms, reads were mapped to the miRNA stem-loop sequences and analyzed for length. For each miRNA, we calculated a ratio of the most frequent length in wild type to the sum of all other lengths, and compared this ratio between nbr and control. The distribution of the length ratios highlighted a cohort of miRNAs with extreme differences between nbr−/− and control. At the two ends of the plot were miRNAs where the most common length isoform of the miRNA was present at a much higher or much lower level in nbr−/− than in wild type, reflecting an altered pattern of isoform distribution or relative abundance for these miRNAs in the absence of nbr. These included miRNAs we had defined as trimmed and modulated by nbr (miR-34, miR-263a, miR-263b), with additional candidates (Fig. 4F, red boxes). Northern blots were performed on the top and bottom 8 miRNAs that we had not tested; we confirmed 7 new nbr-dependent miRNAs (miR-7, miR-10, miR-11, miR-31b, miR-100, miR-190, miR-317; Fig. S3, Table S3, S4). Northern revealed some miRNAs that were trimmed that were not detected as so by deep sequencing and the reverse: for any given miRNA, the extent of trimming had to be greater than ~10% in isoform level to detect a consistent change by Northern blot, while deep sequence analysis suggested that not all isoforms were cloned with equal efficiency.
Trimming exerts a profound and diverse impact on miRNA sequence profiles: nbr promotes the diversity of some miRNAs (miR-34, miR-7, miR-317), and alters the relative abundance of the most prominent isoform of others (miR-190 and miR-10; Fig. S3C-F, Table S3). To identify potential Nbr-dependent miRNA targets, we performed transcriptional profiling of cells. This would allow identification of mRNA targets whose stability was altered by miRNA trimming, but not targets primarily controlled by translational repression [24]. This identified 12 genes whose levels were affected by nbr depletion by >1.5-fold (Fig. 4G, Table S5); of these, one was reduced (nbr), the others were upregulated. Assessing the levels of 8 of these by realtime PCR confirmed increased expression of 6/8 mRNAs (75%) in nbr-depleted cells (Table S5). Next, we assessed expression of 9 of these genes in nbr−/− flies, compared to wild type and loquacious mutant flies. loqsf00791 mutants are viable and show deficiency in miRNA maturation and function, thus allow assessment of miRNA function in adults [25]. We reasoned that genes regulated by miRNAs that are impacted by nbr-processing would also show dependence on loqs. We validated 5/9 genes (55%) as upregulated in both nbr−/− and loqsf00791 (two additional genes were upregulated, although did not reach statistical significance in nbr/−) (Fig. 4H, Table S5). Sequence analysis of these mRNA targets revealed that 4/7 genes (57%) have potential sites for the miRNAs that showed nbr-dependent processing (Table S5). It is unclear, however, if existing algorithms for miRNA targeting efficiently predict binding sites for miRNAs with 3'end diversity; targets for trimmed miRNAs may use non-canonical recognition motifs that are more dependent on 3'end pairing than seed complementarity.
These data provide evidence for a novel step in miRNA biogenesis: miRNA 3' end terminal trimming mediated by the 3'→5' exoribonuclease Nbr. Notably, small RNA deep sequencing has unveiled a rich pattern of miRNA sequence isoforms, although miRNAs have routinely been annotated as a single mature form. Our findings suggest that miRNA processing by Nbr alters the repertoire of at least a subset of miRNAs in cells and whole animals, contributing to the diversity of the small RNA profile and potentially impacting post-transcriptional gene regulation in Drosophila. Mechanistically, our data indicate that, upon nbr knockdown, miR-34 is still associated with RISC, thus trimming is not a pre-requisite to miR-34 loading, and likely occurs after loading.
The impact and biological consequences of trimming may be complex. Nbr may impact strand selection within RISC because strand selection is influenced by the extent of 3' overhang and degree of pairing for any miRNA-miRNA* duplex [26,27]. Nbr may impact miRNA stability, as previous studies have demonstrated that tailing and trimming of mature Drosophila miRNAs influences their turnover [28]. Trimming may also impact mRNA silencing by favoring alterative miRNA sites within mRNA targets. Although canonical miRNA-target specificity is thought to be driven largely by complementarity within the seed, non-canonical interactions can depend more heavily on 3' compensatory sites [29,30]. Therefore, differences in the length of the 3' end of miRNAs may influence both target selection, as well as silencing efficiency of targets that require extensive 3'end pairing. Future analysis of trimmed miRNAs and their range of targets will reveal rules governing miRNA-mRNA pairing specificity that may be impacted by 3'end heterogeneity. Given that some mammalian miRNAs also display multiple isoforms [31,32], miRNA 3'end processing may be conserved. Our studies focused on the role of nbr in miRNA pathway function; whether nbr plays a role in additional small RNA pathways remains an open question, although we did not observe effects on transposons suggesting it does not globally impact esiRNA or piRNA pathways. The modification of mature miRNAs and their precursors is an emerging facet of miRNA-mediated gene regulation [33]. Nbr may represent a central player in a larger spectrum of factors that shape miRNA repertoire and function, through the generation of multi-isoform miRNAs.
Supplementary Material
Highlights
A new fly gene, nibbler/CG9247 (nbr), is identified in miR-34 3'end processing.
Nbr forms a protein complex with Ago1 and both are required for trimming.
Nbr-mediated 3'end processing modulates a large number of miRNAs.
Nbr modulation of the miRNA profile impacts gene expression.
Acknowledgements
We thank the Bloomington and Harvard Stock Centers for fly lines. Work supported by the NINDS (R01-NS043578), Ellison Medical Foundation (to NMB), and the NIAID (R01-AI074951, U54-AI057168) (to SC). We thank Dr. John Tobias for help with microarray analysis, and Dr. Dan Beiting for scatterplot analysis. SC and NMB are co-senior authors. NMB is an investigator with the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information Supplemental Information includes three figures, five tables, and Supplemental Experimental Procedures, and can be found with this article online.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. doi:10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. doi:10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 4.Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- 5.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. doi:10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 6.Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. doi:10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, He X, Lim LP, de Stanchina E, Xuan Z, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. doi:10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. doi:10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. doi:10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. doi:10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes & Development. 2010;24:1086–1092. doi: 10.1101/gad.1919710. doi:10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. doi:10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. doi:10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 14.Chang T-C, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. doi:10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 15.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol. Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. doi:10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. doi:10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. doi:10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 18.Lee YS, Nakahara K, Pham JW, Kim K, He Z, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 19.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. doi:10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 20.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Development. 2004;18:1655–1666. doi: 10.1101/gad.1210204. doi:10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, et al. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. doi:10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 23.Gerbasi VR, Golden DE, Hurtado SB, Sontheimer EJ. Proteomics identification of Drosophila small interfering RNA-associated factors. Mol. Cell Proteomics. 2010;9:1866–1872. doi: 10.1074/mcp.M900614-MCP200. doi:10.1074/mcp.M900614-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. doi:10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 25.Jiang F, Ye X, Liu X, Fincher L, McKearin D, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & Development. 2005;19:1674–1679. doi: 10.1101/gad.1334005. doi:10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 28.Ameres SL, Horwich MD, Hung J-H, Xu J, Ghildiyal M, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. doi:10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. doi:10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. doi:10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheloufi S, Santos Dos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. doi:10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. doi:10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. doi:10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.