Abstract
Therapeutic gene delivery mediated by retroviral vectors has the advantage of stable integration into the host genome. A major safety concern for gene delivery achieved by murine leukemia virus (MLV)-based retroviral vectors is the activation of adjacent cellular genes including oncogenes following integration into the host genome. Self-inactivating (SIN) vectors lacking viral enhancers/promoters in their 3′ long terminal repeat (LTR) have been proposed as a means of overcoming this safety concern. However the MLV-based SIN vectors currently used by laboratories to assess insertional mutagenesis, integration site selection, and the potency of transgene expression are not uniform in the composition of their 3′ LTRs. We constructed a series of SIN vectors representative of the currently employed vectors, but lacking an internal promoter. Green fluorescent protein (GFP) was used as a reporter gene. Target cells exposed to these vectors were evaluated for number of integrants and GFP expression at the messenger RNA (mRNA) level and protein level. We found that viral promoter activity in the 3′ LTR is not attenuated in many currently employed SIN vectors. These results suggest that the influence of strong residual promoter activity should be taken into consideration when interpreting experimental results obtained using SIN vectors in gene therapy research.
Introduction
Insertional mutagenesis has been observed when retroviral vectors derived from murine leukemia virus (MLV) were employed to correct X-linked severe combined immunodeficiency.1,2,3,4,5 In principle, the risk of oncogene activation can be eliminated using self-inactivating (SIN) retroviral vectors that lack viral promoter and enhancer activity in their 3′ long terminal repeat (LTR). A SIN vector with a high safety profile should have no residual promoter/enhancer activity in the LTRs of the integrated vector. The expression of a target gene in an ideal SIN vector relies solely on the internal promoter. However, some SIN vectors contain “TATA” and “CAAT” boxes representative of traditional promoter motifs. Previous studies have suggested that cryptic promoters/enhancers in addition to those present in the U3 region of the LTR may exist in some SIN vectors as well as in enhancer-deficient MLVs. Transcripts initiating from cryptic promoters have been described for SIN vectors and the presence of a cryptic enhancer within the integrated vector can mediate viral transcription.6 The presence of such an ancillary promoter in these MLV-derived SIN vectors and its ability to activate downstream gene expression needs to be considered when evaluating the results obtained with oncogene activation or genotoxicity mediated by SIN retroviral vectors in gene therapy studies. In the present study we sought to determine the existence, location, and strength of any residual promoter activity of SIN vectors containing various deletions in their U3 regions using sensitive flow cytometry assays and PCR analyses. We employed a MLV-based retroviral vector (RT43.2GFP) that exhibits high titer, efficient packaging and optimized splicing capacity.7 A gene encoding enhanced green fluorescent protein (GFP) served as the reporter gene. The U3 region of the 5′ LTR of MLV is replaced with cytomegalovirus immediate early promoter to eliminate the probability for LTR reconstitution in RT43.2GFP (Figure 1). We constructed various RT43.2GFP-based vectors containing different deletions in their U3 regions including deletions similar to those found in MLV-based SIN vectors employed in previous studies.8,9,10,11,12,13,14,15,16,17,18,19
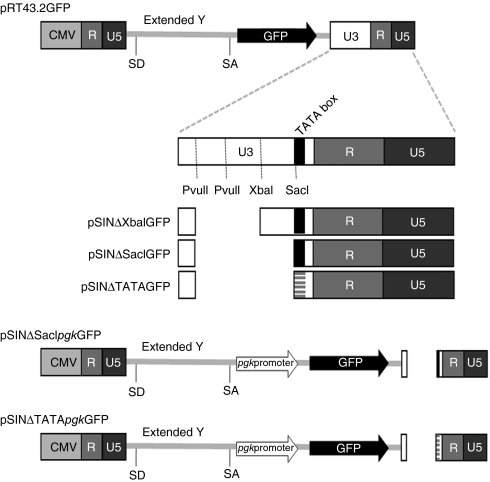
Figure 1.
Schematic representation of the MLV-based plasmids used in the studies. RT43.2GFP served as a template for the additional constructed plasmids. Diagram not to scale. The 3′ LTR of the murine leukemia virus (MLV) vectors derives from Moloney MLV. LTR, long terminal repeat.
Our assessment of MLV-based vectors containing various deletions in the U3 regions of their 3′ LTR showed that deletion of the enhancer regions is not sufficient to abolish viral promoter activity. Profound residual promoter activity was mapped to additional regions of the U3 other than the defined enhancer regions. We discovered that the deletion of an additional 129 bases of promoter sequences including the CAAT box was required for promoter attenuation. Additional disruption of the TATA box did not contribute to abrogation of viral promoter activity. The employment of these vectors containing the larger deletion in their 3′ LTR is required for effective inactivation of the viral promoter activity.
Results
SIN γ-retroviral vectors with deletion of the direct repeat enhancer region in the 3′ U3 retain the ability to efficiently promote GFP expression
To determine the minimal 3′ U3 region of a MLV vector that should be deleted to ensure the requisite attenuation of viral promoter activity to sufficiently diminish the risk of insertional gene activation by SIN vectors we assessed a number of MLV vectors with various deletions in this region. We used a vector named RT43.2GFP (Figure 1) with a MLV backbone as described by Finer et al. to construct different SIN MLV vectors.7 Quantitative GFP expression was used to assess the residual promoter activity in a SIN vector. Presently investigators employ either of two general types of MLV SIN vectors with different deletions in 3′ U3: SIN vectors in which the tandem repeats comprising the enhancer region are, for the most part deleted from the first PvuII site to the XbaI site, (Figure 1), referred to here as Type 1 SIN vectors,8,9,10,11,12,13,14,15,16,17,18 or SIN vectors of which most of the U3 region has been deleted from MLV vectors,20,21,22 or the equivalent deletion of 3′ U3 of spleen focus forming virus19 referred to here as Type 2 SIN vectors.
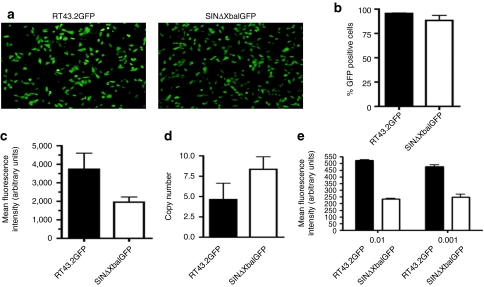
Three MLV-based SIN vectors were constructed that differed in their U3 sequence deletions. These constructs designated pSINΔXbaIGFP (a representative of Type 1 SIN vectors), pSINΔSacIGFP and pSINΔTATAGFP (representatives of Type 2 SIN vector) are schematically shown in Figure 1. All of these SIN constructs lacked any internal promoter upstream of the GFP gene. Mus dunni tail fibroblast (MDTF) cells were exposed to equivalent volume of vector supernatant containing either one of these genomes assembled with MLV gagpol and VSV-G as an envelope. GFP expression was monitored qualitatively using fluorescence microscopy (Figure 2a) and quantitatively using flow cytometric analysis (Figure 2b,c). Surprisingly, the percentage of MDTF cells expressing GFP efficiently after exposure to the SIN vector, SINΔXbaIGFP, was similar to that obtained using the wild type vector RT43.2GFP with >85% of the cells expressing GFP (P > 0.05, Figure 2b). A lower mean fluorescence intensity (MFI) of GFP, as an indicator of GFP protein expression level, was observed with MDTF cells transduced with SINΔXbaIGFP compared to MDTF cells transduced with RT43.2GFP vectors (Figure 2c). Cells exposed to SINΔXbaIGFP still exhibited about 50% MFI compared to cells exposed to RT43.2GFP indicating that deletion of the designated enhancer region of the MLV U3 does not extinguish the promoter activity in this SIN retroviral vector. To exclude the possibility that GFP expression from the integrated SINΔXbaIGFP vectors was attributable to substantially higher vector copy number compared RT43.2GFP, the number of integrants in MDTF cells exposed to RT43.2GFP and SINΔXbaIGFP vectors was analyzed using quantitative PCR. Genomic DNA was isolated from transduced MDTF cells, and primers specific for GFP were used to determine the number of vector integrants per cell. As shown in Figure 2d, no statistically significant difference was found in GFP copy number in the genomic DNA of MDTF cells transduced with SINΔXbaIGFP compared to RT43.2GFP vectors with less than 10 copies of integrants per cell detected in each of the two populations of transduced MDTF cells (P > 0.05).
Figure 2.
GFP expression in cells exposed to MLV-based SIN vectors and proviral copy number. (a) Representative fluorescence microscopy images of GFP expression of MDTF cells exposed to RT43.2GFP vectors (left panel) and SINΔXbaIGFP vectors (right panel). (b) Percent GFP positive MDTF cells exposed to RT43.2GFP or SINΔXbaIGFP vectors as measured by flow cytometry. (c) MFI of MDTF cells exposed to RT43.2GFP or SINΔXbaIGFP vectors as measured by flow cytometry. (d) Proviral integrant copy number. Genomic DNA was extracted from MDTF cells exposed to RT43.2GFP or SINΔXbaIGFP vectors. GFP specific primers were used to determine the copy number of vectors per cell with qPCR. (e) RT43.2GFP or SINΔXbaIGFP vectors were each diluted to 100 (shown as 0.01) and 1,000-fold (shown as 0.001) and then applied to MDTF cells. GFP expression was measured using MFI as an index. All results are depicted as mean ± SEM, n = 3. GFP, green fluorescent protein; MDTF, Mus dunni tail fibroblast; MFI, mean fluorescence intensity; MLV, murine leukemia virus; qPCR, quantitative PCR; SIN vectors, self-inactivating vectors.
To exclude the possibility that target cells were exposed to saturating concentrations of vectors, RT43.2GFP and SINΔXbaIGFP vectors were diluted 100- to 1,000-fold to ensure less than one copy of vector integrants per cell. An equivalent volume of diluted vector was applied to target cells and then the MFI of GFP expression was assessed using flow cytometry (Figure 2e). MFI mean values dropped approximately seven to eightfold for target cells exposed to both vectors after 100-fold dilution (3,735 compared to 523 for RT43.2GFP, 1,955 compared to 233 for SINΔXbaIGFP) (Figure 2c,e), however, no significant change in MFI was achieved upon further dilution of vectors to 1,000-fold. The MFI of GFP in MDTF cells transduced with either dilution of SINΔXbaIGFP vectors was about 50% of the MFI observed with cells exposed to the same dilutions of control RT43.2GFP (Figure 2e), which indicates that deletion of solely the enhancer region from the 3′ MLV U3 of MLV vectors only modestly attenuates the promoter activity.
The strong residual promoter activity observed in SIN vectors is not due to reversion to full length U3 nor to an internal cryptic promoter
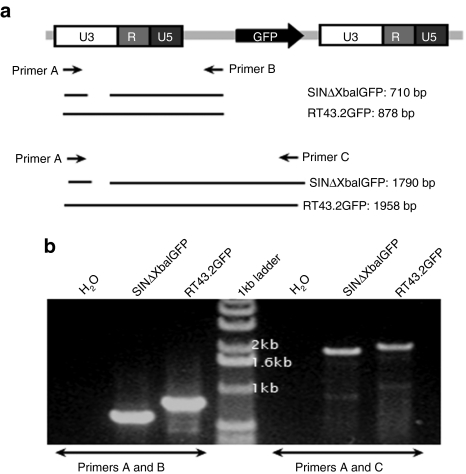
During reverse transcription of a SIN vector, deletions in the 3′ LTR should be incorporated in the 5′ LTR of the provirus. To determine whether the expression of GFP is the result of the failure to transfer 3′ U3 sequences with deletions to the 5′ LTR we employed PCR analyses of the genomic DNA of cells exposed to SINΔXbaIGFP vectors. We extracted high molecular weight genomic DNA from MDTF cells transduced with RT43.2GFP or SINΔXbaIGFP vectors and performed PCR using primers flanking the 5′ LTR (Figure 3a). Using two different primer sets, PCR products with sizes corresponding to deletions present in the U3 of the 3′ LTR of the SIN vectors were detected in the 5′ LTR of integrated vectors (Figure 3b). Sequencing of PCR products confirmed the presence of the deletion in the integrated 5′ LTR region. These results suggest that the deleted U3 was maintained in the reverse transcribed SINΔXbaIGFP proviral vector. Therefore, GFP expression that occurs in cells transduced by SINΔXbaIGFP vectors may be attributable to the retention of strong promoter activity in the 3′ U3 of SINΔXbaIGFP or to a cryptic promoter(s) within the vector genome as has been previously reported for lentiviral SIN vectors.6 To discriminate between these possibilities further deletion of a 129-nucleotide (nt) segment of U3 region was incorporated into SINΔSacIGFP and SINΔTATAGFP. The SINΔTATAGFP vector harbors a mutated TATA box in addition to the deleted 129 nts (Figure 1). GFP expression was assessed in transduced MDTF cells. We determined that GFP expression was abrogated in vectors harboring extended deletions in their U3 regions as exemplified by the loss of GFP expression in MDTF cells (Figures 4a and 2c).
Figure 3.
The 3′ U3 deletion of SINΔXbaIGFP is transferred to the 5′ LTR of integrated provirus. (a) Schematic depiction of RT43.2GFP provirus. Arrows indicate primers used to verify the SIN deletion in the 5′ LTR of the provirus, labeled primer A, B and C. Resulting PCR products with expected DNA sizes in base pairs (bp) are given below the schematic. Spaces in between the lines represent PCR product sequences deleted in RT43.2GFP to construct SINΔXbaIGFP. (b) Representative of ethidium bromide stained PCR products separated on an agarose gel from reactions performed with the indicated primer pairs on genomic DNA of MDTF cells exposed to RT43.2GFP or SINΔXbaIGFP. H2O indicates negative water control for PCR reaction. Center lane is 1 Kb DNA ladder. bp, base pair; LTR, long terminal repeat; MDTF, Mus dunni tail fibroblast; SIN vectors, self-inactivating vectors.
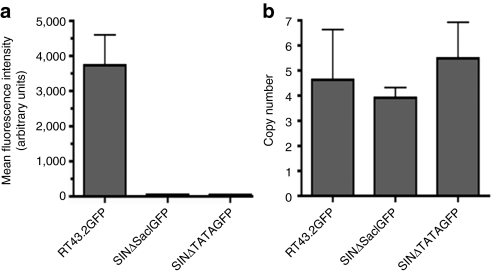
Figure 4.
GFP expression of MDTF cells exposed to SINΔSacIGFP or SINΔTATAGFP and proviral copy number. (a) MFI as measured by flow cytometry on MDTF cells exposed to RT43.2GFP, SINΔSacIGFP, and SINΔTATAGFP. (b) Copy number of vector integrants per cell determined with qPCR using genomic DNA extracted from MDTF cells exposed to various MLV-based vectors. Data is given as mean ± SEM, n = 3. GFP, green fluorescent protein; MDTF, Mus dunni tail fibroblast; MFI, mean fluorescence intensity; MLV, murine leukemia virus; qPCR, quantitative PCR; SIN vectors, self-inactivating vectors.
Reduced integrant copy number does not account for the attenuation of GFP expression in cells exposed to SINΔSacIGFP or SINΔTATAGFP vectors
MDTF cells were exposed to equivalent volume of SINΔSacIGFP or SINΔTATAGFP viral supernatants. GFP expression was not detected in MDTF cells exposed to either of these two vectors by fluorescence microscopy (data not shown). GFP expression represented by MFI was diminished over two orders of magnitude in cells exposed to SINΔSacIGFP or SINΔTATAGFP vectors as compared to RT43.2GFP or SINΔXbaIGFP as determined by flow cytometric analysis (Figures 4a and 2c).
Since the promoter activity in vectors with 3′ U3 region deletions up to the TATA box was decreased significantly compared to Type 1 SIN vectors containing solely deletion of the enhancer region, we reasoned it was possible that the reduction of GFP expression was due to a decreased number of integration events. Genomic DNA was therefore isolated from MDTF cells transduced with various vectors and quantitative PCR for GFP was performed. As shown in Figure 4b, no statistically significant differences were found in GFP copy number in the genomic DNA of MDTF cells exposed to RT43.2GFP, SINΔSacIGFP or SINΔTATAGFP vectors (P > 0.05). These results suggest that the efficient GFP expression associated with SINΔXbaIGFP vector is due to the residual promoter activity in the 3′ U3 between the direct repeat regions and the TATA box.
Decreased mRNA levels correspond to reduced of GFP expression in vectors with deletions in the 3′ U3 region
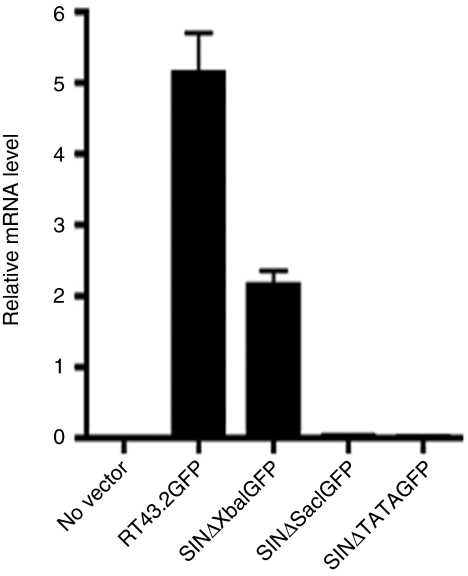
GFP expression decreased significantly in MDTF cells transduced with SINΔSacIGFP or SINΔTATAGFP compared to cells transduced with SINΔXbaIGFP without a concomitant decrease in number of integrants. To confirm that this decrease is due to diminished promoter activity, we tested GFP messenger RNA (mRNA) abundance by quantitative reverse transcripion-PCR on complementary DNA transcripts prepared from mRNA isolated from MDTF cells exposed to RT43.2GFP, SINΔXbaIGFP, SINΔSacIGFP or SINΔTATAGFP vectors. Using glyceraldehyde 3-phosphate dehydrogenase as an internal control, GFP mRNA levels in MDTF cells transduced by SINΔXbaIGFP (representing the Type 1 SIN vector) retained ~50% of RT43.2GFP vector transcription levels (Figure 5), in agreement with the retention of about 50% promoter activity observed using flow cytometry (Figure 2c,e). The GFP mRNA levels in MDTF cells exposed to SINΔSacIGFP or SINΔTATAGFP (as representatives of the Type 2 SIN vectors) were equivalent to control MDTF cells not exposed to vectors (P > 0.05, Figure 5). We did not observe significant differences of GFP mRNA levels in cells exposed to vectors with or without the TATA mutation (P > 0.05).
Figure 5.
Relative GFP mRNA levels in MDTF cells exposed to different MLV-based vectors. RNA was extracted from MDTF cells exposed to the indicated vectors. mRNA levels were determined by qRT-PCR. Relative GFP mRNA level was shown after normalization with GAPDH. Results are depicted as mean ± SEM, n = 3. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein; MDTF, Mus dunni tail fibroblast; MLV, murine leukemia virus; mRNA, messenger RNA; qRT-PCR, quantitative reverse transcription-PCR.
An internal promoter restores mRNA transcription levels in SINΔSacIGFP or SINΔTATAGFP
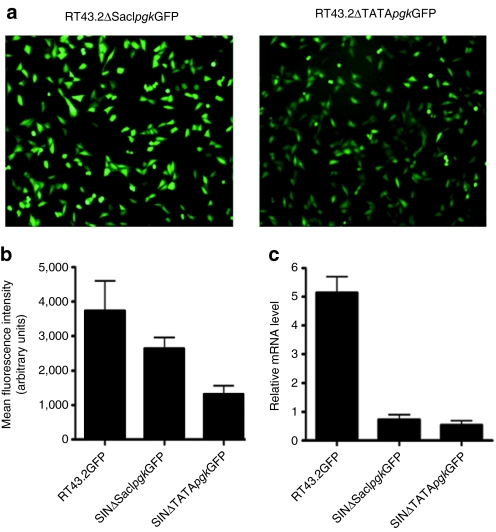
Since the promoter activity in the 3′ LTR of SINΔSacIGFP and SINΔTATAGFP was ostensibly abolished, we next assessed whether introducing an internal promoter, pgk, upstream of GFP in plasmids pSINΔSacIpgkGFP and pSINΔTATApgkGFP, respectively, can restore GFP expression. GFP expression was monitored with fluorescence microscopy (Figure 6a), flow cytometric analysis (Figure 6b), and quantitative reverse transcription-PCR (Figure 6c). Photo microscopic analysis and MFI evaluation of GFP protein expression clearly demonstrated the restoration of GFP expression in MDTF cells exposed to SINΔSacIpgkGFP or SINΔTATApgkGFP. However, the recovery of GFP expression after transduction with both internal promoter-containing SIN vectors did not reach to levels equivalent to that obtained in cells exposed to RT43.2GFP. Furthermore, the elimination of the TATA box by mutation appeared to have a deleterious effect on the GFP expression at both protein and RNA level in MDTF cells exposed to SINΔTATApgkGFP (Figure 6).
Figure 6.
An internal promoter, pgk, restored GFP expression in cells exposed to MLV-based vectors with larger deletions in the U3 region. (a) Representative fluorescence microscopic images of GFP expression in MDTF cells exposed to SINΔSacIpgkGFP vectors (left panel) or SINΔTATApgkGFP vectors (right panel). (b) MFI of MDTF cells exposed to RT43.2GFP, SINΔSacIpgkGFP or SINΔTATApgkGFP vectors as measured by flow cytometry. Results are depicted as mean ± SEM, n = 3. (c) Relative GFP mRNA level of MDTF cells exposed to RT43.2GFP, SINΔSacIpgkGFP or SINΔTATApgkGFP using qRT-PCR. Results are depicted as mean ± SEM, n = 3. GFP, green fluorescent protein; MDTF, Mus dunni tail fibroblast; MFI, mean fluorescence intensity; MLV, murine leukemia virus; mRNA, messenger RNA; qPT-PCR, quantitative reverse transcription-PCR; SIN vectors, self-inactivating vectors.
The promoter activity in the 3′ LTR of SIN lentiviral vectors is abolished
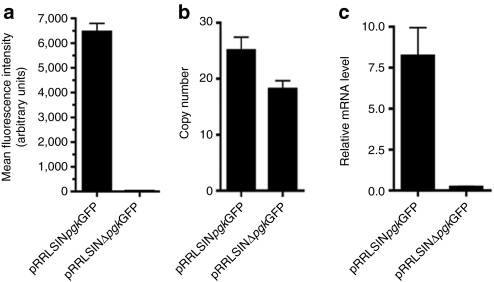
SIN vectors derived from HIV-1 vectors, originally described by Zufferey et al.23 contain deletions in the U3 of their 3′ LTR that are similar to the design of Type 2 γ-retroviral SIN vectors. This 400-nt deletion includes the enhancer and the TATA box, leaving just the 35 nts at the 5′ end of U3 to allow for integrase function and the 18 nts at the 3′ end of U3 required for polyadenylation.23 This RRLpgkGFP SIN vector,23 contains an internal pgk promoter to drive GFP expression. To determine whether the 400-nt deletion in the 3′ LTR region of the SIN lenviral vector abrogates promoter activity in a manner analogous to that observed with Type 2 γ-retroviral SIN vectors, the internal pgk promoter was deleted from RRLpgkGFP SIN vector to create RRLSINΔpgkGFP vector. Flow cytometric analysis of MDTF cells transduced with RRLSINΔpgkGFP demonstrated GFP expression was substantially attenuated compared to RRLSINpgkGFP vectors as shown in Figure 7a. The dramatic attenuation of GFP expression occurs despite multiple integrations of the RRLΔpgkGFP vector genomes (~20 integrants per cell) in MDTF cells (Figure 7b). No statistically significant difference was detected in the number of vector integrants per cell in MDTF exposed to RRLSINpgkGFP or RRLSINΔpgkGFP (P > 0.05, Figure 7b). Relative mRNA expression level of viral vector in target cells measured by quantitative reverse transcription-PCR also verified expression of GFP was substantially decreased at the mRNA level (Figure 7c). Therefore expression of GFP in cells containing integrated RRLSINpgkGFP vectors with a 400-nt deletion in the U3 within the 3′ LTR is dependent upon the internal pgk promoter, and thus these vectors were confirmed to be SIN.
Figure 7.
GFP expression and number of integrants in cells exposed to SIN lentiviral vectors. (a) MFI of MDTF cells exposed to RRLSINpgkGFP or RRLSINΔpgkGFP vectors as measured by flow cytometry. (b) Proviral integrants copy number. qPCR were used with GFP specific primers to determine the copy number of vectors per MDTF cell exposed to RRLSINpgkGFP or RRLSINΔpgkGFP vectors. (c) Relative GFP mRNA level of MDTF cells exposed to RT43.2GFP, RRLSINpgkGFP or RRLSINΔpgkGFP. All results are depicted as mean ± SEM, n = 3. GFP, green fluorescent protein; MDTF, Mus dunni tail fibroblast; MFI, mean fluorescence intensity; mRNA, messenger RNA; SIN vectors, self-inactivating vectors.
Discussion
To reduce the potential for clonal proliferation resulting from γ-retroviral vector-mediated therapeutic gene delivery, various SIN vectors have been developed. Early studies by Linney et al. showed the enhancer region with two copies of a 75-base pair (bp) element within the U3 region of the LTR of a MLV to be critical for promoting transcription of viral transcripts as well as indicator genes such as the gene encoding chloramphenicol acetyl transferase activity.24 This 207-bp U3 deletion consists of the entire enhancer segment containing the major determinants of MLV LTR promoter activity including most of the transcription factor binding sites.25 The deletion of this region of U3 was the basis for SIN vectors, such as those developed by Julius et al., to eliminate promoter activity that is responsible for activation of gene adjacent to proviral elements or vectors.14 Other SIN vectors such as those developed by Yu et al. delete a larger segment of the U3 encompassing the repetitive enhancer regions and adjacent sequences up to the TATA motif, a canonical sequence found within the core promoters of many viral and eukaryotic genes, resulting in a 299-bp deletion of U3 sequences.22 Subsequent reports determined that the SIN vectors developed by Yu et al. had reduced titers or were genetically unstable due to impeded retroviral transcription, transcript termination or polyadenylation.26 Furthermore Olson et al. found that the LTRs in these SIN vectors were frequently reconstituted via recombination or gene conversion.27
High performance SIN γ-retroviral vectors with improved viral titer and optimal transgene expression have since been developed and analyzed for their ability to safely and efficiently deliver genes. Many of the problems originally described as rate-limiting steps in the efficient delivery of genes by SIN vectors have been overcome using different strategies including but not limited to: replacing the use of NIH3T3-based packaging cell lines with the transient production of SIN vectors using plasmids optimally suited for high copy number expression in 293T cells;28 replacing 5′ LTR with a hybrid of cytomegalovirus enhancer and Moloney sarcoma virus promoter,7 and introducing woodchuck hepatitis post-transcriptional regulatory element into the 3′ untranslated region of the vector genome. Even though the design of SIN vectors has been significantly improved for efficient gene delivery, the safety of γ-retroviral SIN vectors remains a source of controversy. In part this controversy is attributable to variations in the regions of the U3 that are deleted among the different SIN vectors used in safety assessments.
SIN HIV-1 vectors have been developed with a 400-nt deletion in the 3′ LTR including the TATA box.23 These SIN lentiviral vectors are designed in a manner similar to Type 2 γ-retroviral vectors. Our results showed the promoter activity in the 3′ LTR of SIN lentiviral vector is extinguished to a level similar to that observed with Type 2 γ-retroviral vectors. SIN HIV-1 vectors have been used to assess insertional mutagenesis, integration site selection, and the potency of transgene expression as compared to MLV SIN vectors. However, these comparisons were based on the use of either of two types of MLV SIN vectors, Type 1 MLV SIN vectors lacking only enhancer sequences (analogous to the SINΔXbaIGFP vector used in our studies)8,9,10,11,12,13,14,15,16,17,18 or Type 2 SIN vectors lacking most of the U3 region of the 3′ LTR (analogous to the SINΔSacIGFP or SINΔTATAGFP vectors used here).19,20,21,22,29,30,31 Our results would suggest that the use of SIN γ-vectors lacking only enhancer sequences (Type 1) are not sufficiently attenuated with respect to viral promoter function and this finding may provide the rationale for the inconsistencies in the published reports of various deficiencies in MLV compared to HIV-1 SIN vectors with regard to adverse events and the overall efficiency of mediating therapeutic gene delivery.
Recent studies using Type 1 SIN MLV vectors showed them to be capable of immortalizing primary murine bone marrow cells by driving the expression of downstream oncogenes.8 The authors concluded that an enhancer element present in the internal pgk promoter was responsible for the dysregulation of oncogene expression caused by these vectors.8 A more likely alternative explanation for their findings is the residual promoter activity found in the 3′ LTR of Type 1 MLV SIN vectors. Earlier studies showed that Type 1 MLV SIN vectors, in contrast to lentiviral SIN vectors, exhibited a high efficiency of in vitro transgene expression due to 3′ readthrough of the SIN LTR.18 However, based on our findings, enhanced expression of 3′ genes could result from the retention of viral promoter in the 3′ LTR of these vectors. Type 1 MLV SIN vectors have also been compared to HIV SIN vector to demonstrate certain advantages of lentiviral over MLV SIN vectors in reducing the genotoxic potential of vectors using a tumor-prone mouse model to assess oncogene activation.17 Type 1 not Type 2 MLV SIN vectors were also found to trap cellular promoter more efficiently than HIV vectors. Enhanced promoter trapping signifies increased transcriptional interactions between the MLV SIN vector and its flanking genes at the integration site relative to HIV SIN vectors in different cell types.11 In contrast to these reports using MLV SIN vectors, gene marking results obtained using Type 2 MLV SIN vectors in murine hematopoietic stem cells demonstrated the absence of clonal dominance associated with in vivo tumor formation.30 Type 2 SIN vectors are currently being put forward for use in multi-institutional phase I/II trial proposed for severe combined immunodeficiency-X1 clinical trials,32 as appropriately effective clinical tools for the treatment of X-linked severe combined immunodeficiency,31 as well as treatment for a recessive dystrophic epidermolysis bullosa.21
Given these collective published findings on Type 1 and Type 2 MLV SIN vectors together with our results that Type 1 SIN vectors are not transcriptionally attenuated suggest that Type 2, not Type 1 MLV-based SIN vectors are best suited for gene therapy research and therapeutic gene delivery.
Materials and Methods
Plasmids. Plasmids pRT43.2GFP, pSINpgkΔXbaIGFP and pRRLSINpgkGFP were obtained from Tom Dull (Cell Genesys, San Francisco, CA) and have been described previously.7 pSINΔXbaIGFP was generated by removing 169-bp of the 3′ U3 region in pRT43.2GFP using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions with complementary primers 5′-GGTCAGGAACAGATGGAACA GAGCAGTTTCTAGAGAACCATCAG-3′ and 5′-CCAGTCCTTGTCTACCTT GTCTCGTCAAAGATCTCTTGGTAGTC-3′. The pSINΔSacIGFP and pSINΔSacIpgkGFP plasmids were similarly constructed using pLitmus 28i vector (New England Biolabs, Ipswich, MA) as a transfer vector. The 3′ U3 region of pSINΔXbaIGFP was subcloned into pLitmus after digesting pSINΔXbaIGFP and pLitmus 28i vector with AvrII and BamHI and ligating the appropriate fragments to generate pLitmus-SINΔXbaIU3. An additional 129-bp deletion to the SacI site was made in the U3 region of pLitmus-SINΔXbaIU3 using mutagenesis to make pLitmus-SINΔSacIU3. The complementary primers used were 5′-GAGAAGTTCAGATCAAGGTCAG GAACAGATGGAACAGAATAAAAGAGCC CACAACCCC-3′ and 5′-GGGGTTGTGGGCTCTTTTATTCTGTTCCAT CTGTTCCTGACCTTGATCTGAACTTCTC-3′. Plasmids pSINΔSacIGFP and pSINΔSacIpgkGFP were subsequently constructed by exchanging the 3′ U3 region with extended deletion to the SacI site from pLitmus-SINΔSacIU3 with the 3′ U3 of pSINΔXbaIGFP and pSINΔXbaIpgkGFP using AvrII and BamHI restriction sites. pSINΔTATAGFP and pSINΔTATApgkGFP were generated by mutating the TATA box in pSINΔSacIGFP and pSINΔSacIpgkGFP, respectively, using the following primers: 5′-GGAACAGATGGAACAGC CGCGGAAGAGCCCACAACC-3′ and 5′-CCTTGTCTACCTTGTCGGCG CCTTCTCGGGTGTTGG-3′. pRRLSINΔpgkGFP was generated by removing 483 nts of the pgk promoter in pRRLSINpgkGFP by mutagenesis using complementary primers 5′- GGT TGC GCC TTT TCC AAG GCA GCC CTG GGT TTG CGC GCC ACC ATG GTG AGC AAG GGC GAG GAG C -3′ and 5′- GCT CCT CGC CCT TGC TCA CCA TGG TGG CGC GCA AAC CCA GGG CTG CCT TGG AAA AGG CGC AAC C -3′. All DNA plasmids were sequence confirmed.
Cells. MDTF,33 and 293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 100 U/ml penicillin and 100 g/ml of streptomycin (Invitrogen, Carlsbad, CA) with 5% CO2 at 37 °C.
Retroviral vector production and transduction. 293T cells were plated at a density of 2 × 106 per 10 cm plate 1 day before DNA transfection. pVSV-G env, pMLV gag/pol, and pRT43.2GFP plasmids with different deletions in the 3′ U3 region were transfected using a Profection kit (Promega, Madison, WI) according to the instructions of the manufacturer. Supernatants were collected 48 hours post-transfection, filtered and stored at −80 °C before use. MDTF cells were plated at a density of 2 × 104/well in a 24-well plate the day before transduction. Viral supernatant, 0.5 ml, from individual vectors with different deletions was used to infect MDTF cells. After overnight exposure to vector supernatants, MDTF cells were washed with Dulbecco's modified Eagle medium and maintained at 37 °C for at least 2 weeks before being collected for various analyses.
Lentiviral vector production and transduction. pVSV-G env, plenti gag/pol/rev, and pRRLSINpgkGFP or pRRLSINΔpgkGFP plasmids were transfected into 293T cells using a Profection kit. Viral supernatants were collected and stored similarly as retroviral vectors. Lentiviral supernatants were concentrated 20× before transduction of MDTF to determine the expression level of GFP with multiple proviral integrants.
Flow cytometry. Transduced MDTF cells were collected and washed with phosphate buffered saline. These cells were then resuspended in Hank's buffered saline supplemented with 0.1% bovine serum albumin (w/v) and 0.1% sodium azide (w/v). Flow cytometry was performed using a FACSCalibur (BD Biosciences, San Jose, CA) and data was analyzed using CellQuest software (BD Biosciences).
Genomic DNA purification. Genomic DNA was isolated from 106 transduced MDTF cells using Wizard Genomic DNA Purification kit (Promega) according to the manufacturer's instruction. Genomic DNA was rehydrated in 50 µl dH2O and stored at 4 °C.
Extraction of RNA and reverse transcription. The total RNA from MDTF cell transduced with individual vector was collected using TRIzol (Invitrogen). Briefly cells were trypsinized and washed once with phosphate buffered saline. After cells were resuspended in 500 ml phosphate buffered saline, 160 ml TRIzol was added. Cells were gently mixed and 100 ml chloroform was added per sample. After incubation on ice for 3 minutes, samples were centrifuged for 15 minutes at 4 °C at maximum speed. Supernatants were transferred to RNAse-free microfuge tubes and an equal volume of 70% ethanol was added to RNA solution. Samples were gently mixed and further purified using an RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA concentrations were measured with a NanoVue (GE Healthcare, Waukesha, WI). Two milligrams of RNA was used for reverse transcription using SuperScript VILO cDNA Synthesis kit (Invitrogen) according to the manufacturer's instructions.
Quantitative reverse transcription-PCR. Quantitative reverse transcription-PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and a BioRad iCycler iQ detection system. Copy number of different vectors in genomic DNA was determined using the following GFP primers, sense: 5′-GGTCTTGTAGTTGCCGTGT-3′ and antisense: 5′- CCTGAAGTTCATCTGCACCA-3′. A standard curve was created using diluted GFP amplicons. A DNA mass of 3.25 pg per haploid mouse genome (http://www.argosbiotech.de/700/omics/genomics/mo/gp_musmusc.htm) was used to determine the absolute copy number per cell. To determine the mRNA level of retroviral or lentiviral vector after transduction, the same GFP primers mentioned above were used. Glyceraldehyde 3-phosphate dehydrogenase was used as an internal control using primer pairs: sense: 5′-CGACCCCTTCATTGACCTCAACTACATGG-3′ and antisense: 5′-CCCTTTTGGCCCCACCCTTCAGGTGAGCC-3′. Relative GFP mRNA level was obtained by normalizing to glyceraldehyde 3-phosphate dehydrogenase after quantitative reverse transcription-PCR.
Statistical analysis. Percentages of GFP positive cells and MFIs were calculated by BD CellQuest (BD Biosciences) software. Data from three separate experiments were analyzed by a Kruskal–Wallis test with a 95% confidence interval. Multi-comparison analysis was performed with Dunn's post-test.
Acknowledgments
We are greatly indebted to James Nagle and Deborah Kauffman at National Institute of Neurological Disorders and Stroke sequencing facility (National Institutes of Health) for sequencing of plasmid and genomic DNA. We are grateful to Arlette Kouwenhoven for her expert technical assistance and helpful comments. We thank Laura Li for experimental help. National Institute of Mental Health intramural funding supported this work. The authors declared no conflict of interest.
REFERENCES
- Hacein-Bey-Abina S, de Saint Basile G., and, Cavazzana-Calvo M. Gene therapy of X-linked severe combined immunodeficiency. Methods Mol Biol. 2003;215:247–259. doi: 10.1385/1-59259-345-3:247. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E.et al. (2008Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1 J Clin Invest 1183132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E.et al. (2003A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency N Engl J Med 348255–256. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H.et al. (2008Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients J Clin Invest 1183143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa H, Persons DA., and, Nienhuis AW. Mobilization and mechanism of transcription of integrated self-inactivating lentiviral vectors. J Virol. 2005;79:8410–8421. doi: 10.1128/JVI.79.13.8410-8421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer MH, Dull TJ, Qin L, Farson D., and, Roberts MR. kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- Bosticardo M, Ghosh A, Du Y, Jenkins NA, Copeland NG., and, Candotti F. Self-inactivating retroviral vector-mediated gene transfer induces oncogene activation and immortalization of primary murine bone marrow cells. Mol Ther. 2009;17:1910–1918. doi: 10.1038/mt.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce JW, Ahlquist P., and, Young JA. The host cell sulfonation pathway contributes to retroviral infection at a step coincident with provirus establishment. PLoS Pathog. 2008;4:e1000207. doi: 10.1371/journal.ppat.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani B, Montini E, Maruggi G, Ambrosi A, Mirolo M, Selleri S.et al. (2009Integration of retroviral vectors induces minor changes in the transcriptional activity of T cells from ADA-SCID patients treated with gene therapy Blood 1143546–3556. [DOI] [PubMed] [Google Scholar]
- De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E.et al. (2005Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells Blood 1052307–2315. [DOI] [PubMed] [Google Scholar]
- Hahm SH, Yi Y, Lee DK, Noh MJ, Yun L, Hwang S.et al. (2004Construction of retroviral vectors with enhanced efficiency of transgene expression J Virol Methods 121127–136. [DOI] [PubMed] [Google Scholar]
- Jaalouk DE, Crosato M, Brodt P., and, Galipeau J. Inhibition of histone deacetylation in 293GPG packaging cell line improves the production of self-inactivating MLV-derived retroviral vectors. Virol J. 2006;3:27. doi: 10.1186/1743-422X-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius MA, Yan Q, Zheng Z., and, Kitajewski J. Q vectors, bicistronic retroviral vectors for gene transfer. BioTechniques. 2000;28:702–708. doi: 10.2144/00284st06. [DOI] [PubMed] [Google Scholar]
- Modlich U, Bohne J, Schmidt M, von Kalle C, Knöss S, Schambach A.et al. (2006Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity Blood 1082545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M.et al. (2009The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy J Clin Invest 119964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C.et al. (2006Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration Nat Biotechnol 24687–696. [DOI] [PubMed] [Google Scholar]
- Zaiss AK, Son S., and, Chang LJ. RNA 3' readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J Virol. 2002;76:7209–7219. doi: 10.1128/JVI.76.14.7209-7219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraunus J, Schaumann DH, Meyer J, Modlich U, Fehse B, Brandenburg G.et al. (2004Self-inactivating retroviral vectors with improved RNA processing Gene Ther 111568–1578. [DOI] [PubMed] [Google Scholar]
- Marty L, Roux P, Royer M., and, Piechaczyk M. MoMuLV-derived self-inactivating retroviral vectors possessing multiple cloning sites and expressing the resistance to either G418 or hygromycin B. Biochimie. 1990;72:885–887. doi: 10.1016/0300-9084(90)90007-4. [DOI] [PubMed] [Google Scholar]
- Titeux M, Pendaries V, Zanta-Boussif MA, Décha A, Pironon N, Tonasso L.et al. (2010SIN retroviral vectors expressing COL7A1 under human promoters for ex vivo gene therapy of recessive dystrophic epidermolysis bullosa Mol Ther 181509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SF, von Rüden T, Kantoff PW, Garber C, Seiberg M, Rüther U.et al. (1986Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells Proc Natl Acad Sci USA 833194–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L.et al. (1998Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery J Virol 729873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E, Davis B, Overhauser J, Chao E., and, Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. Nature. 1984;308:470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Speck NA., and, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P, Nelson S., and, Dornburg R. Improved self-inactivating retroviral vectors derived from spleen necrosis virus. J Virol. 1994;68:7060–7066. doi: 10.1128/jvi.68.11.7060-7066.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P, Temin HM., and, Dornburg R. Unusually high frequency of reconstitution of long terminal repeats in U3-minus retrovirus vectors by DNA recombination or gene conversion. J Virol. 1992;66:1336–1343. doi: 10.1128/jvi.66.3.1336-1343.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA., and, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg R., and, Temin HM. Retroviral vector system for the study of cDNA gene formation. Mol Cell Biol. 1988;8:2328–2334. doi: 10.1128/mcb.8.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils K, Lange C, Schambach A, Brugman MH, Nowak R, Lioznov M.et al. (2009Stem cell marking with promotor-deprived self-inactivating retroviral vectors does not lead to induced clonal imbalance Mol Ther 17131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill SI, Schambach A, Howe SJ, Ulaganathan M, Grassman E, Williams D.et al. (2008Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency Mol Ther 16590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog RW. Gene therapy for SCID-X1: round 2. Mol Ther. 2010;18:1891. doi: 10.1038/mt.2010.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander MR., and, Chattopadhyay SK. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]