Abstract
Enterococcus faecalis V583 was grown in a glucose-limited chemostat at three different growth rates (0.05, 0.15, and 0.4 h−1). The fermentation pattern changed with growth rate, from a mostly homolactic profile at a high growth rate to a fermentation dominated by formate, acetate, and ethanol production at a low growth rate. A number of amino acids were consumed at the lower growth rates but not by fast-growing cells. The change in metabolic profile was caused mainly by decreased flux through lactate dehydrogenase. The transcription of ldh-1, encoding the principal lactate dehydrogenase, showed very strong growth rate dependence and differed by three orders of magnitude between the highest and the lowest growth rates. Despite the increase in ldh-1 transcript, the content of the Ldh-1 protein was the same under all conditions. Using microarrays and quantitative PCR, the levels of 227 gene transcripts were found to be affected by the growth rate, and 56 differentially expressed proteins were found by proteomic analyses. Few genes or proteins showed a growth rate-dependent increase or decrease in expression across the whole range of conditions, and many showed a maximum or minimum at the middle growth rate (i.e., 0.15 h−1). For many gene products, a discrepancy between transcriptomic and proteomic data were seen, indicating posttranscriptional regulation of expression.
INTRODUCTION
A number of regulation mechanisms exist to control the various processes in a cell and to maintain homeostasis. During balanced growth the cells maintain a constant composition, implying the regulation of cellular processes tightly coordinated with growth rate. Such regulation is essential at all growth rates, but the cell's composition, size, and metabolism may respond to changes in growth rate. The effects of growth rate on cellular processes have been best studied in yeast (3, 25), where hundreds of genes are expressed in a growth rate-dependent fashion (17). In bacteria, growth rate-dependent control has been shown to involve regulation at the levels of transcription and translation, and an increase in growth rate is associated with an increase in cell size and number of ribosomes (11, 16, 24). In Lactococcus lactis, increases in growth rate have been reported to correlate with the altered transcription of 30% of the genes (6). Moreover, a shift to less efficient energy metabolism is frequently observed at higher growth rates (20). For lactic acid bacteria, a reduction in growth rate is specifically associated with a change from homolactic to mixed acid fermentation (9, 12, 19, 31). The metabolic and genetic control of glycolysis has been thoroughly studied (7, 8), but the mechanisms underlying the shift in metabolism are not completely understood (7, 23). Previously, we used a mutant unable to produce lactic acid to show that a change in metabolism was associated with large effects on the Enterococcus faecalis transcriptome and proteome profiles (18). In this work, we have studied the effects of growth rate on the transcriptome, metabolome, and proteome of E. faecalis grown under energy limitation in chemostat culture to obtain a better understanding of the mechanisms regulating the cell's metabolic processes.
MATERIALS AND METHODS
Growth conditions and analytical procedures.
Enterococcus faecalis V583 (26) was grown anaerobically at 37°C in the chemically defined medium named CDM-LAB. This contained (per liter) 1 g K2HPO4, 5 g KH2PO4, 0.6 g ammonium citrate, 1 g sodium acetate, 0.25 g tyrosine, 0.24 g alanine, 0.125 g arginine, 0.42 g aspartic acid, 0.13 g cysteine, 0.5 g glutamic acid, 0.15 g histidine, 0.21 g isoleucine, 0.475 g leucine, 0.44 g lysine, 0.275 phenylalanine, 0.675 g proline, 0.34 g serine, 0.225 g threonine, 0.05 g tryptophan, 0.325 g valine, 0.175 g glycine, 0.125 g methionine, 0.1 g asparagine, 0.2 g glutamine, 10 g glucose, 0.5 g l-ascorbic acid, 35 mg adenine sulfate, 27 mg guanine, 22 mg uracil, 50 mg cysteine, 50 mg xanthine, 2.5 mg d-biotin, 1 mg vitamin B12, 1 mg riboflavin, 5 mg pyridoxamine-HCl, 10 μg p-aminobenzoic acid, 1 mg pantothenate, 5 mg inosine, 1 mg nicotinic acid, 5 mg orotic acid, 2 mg pyridoxine, 1 mg thiamine, 2.5 mg lipoic acid, 5 mg thymidine, 200 mg MgCl2, 50 mg CaCl2, 16 mg MnCl2, 3 mg FeCl3, 5 mg FeCl2, 5 mg ZnSO4, 2.5 mg CoSO4, 2.5 mg CuSO4, and 2.5 mg (NH4)6Mo7O24 (9, 13).
Chemostat cultures were grown in a Biostatbplus fermentor (Sartorius Stedim Biotech) with a working volume of 750 ml at dilution rates (D) of 0.05, 0.15, and 0.4 h−1. The pH was kept constant at pH 6.4 by the automatic addition of 4 M NaOH. Cultivation was carried out under anaerobic condition (60 ml/min N2) with a stirring speed of 250 rpm. The cultures were considered to be in steady state when there was no detectable glucose in the culture supernatants and the optical density, cell dry weight, and product concentrations of the cultures were constant in samples taken on two consecutive days. Samples used for metabolite, transcriptomic, and proteomic analyses were taken from cultures grown for six generations after the sample confirming steady state had been taken. All experiments were performed in triplicate.
Culture samples of 20 to 50 ml were centrifuged at 4°C at 6,000 × g for 10 min, and pellets were either flash frozen in liquid nitrogen or treated according to the protocols for measuring the dry weight as previously described (1). Supernatants were frozen at −20°C until metabolite analysis.
Metabolite analyses.
Bacterial dry weight was measured as previously described (1). Glucose, pyruvate, lactate, formate, acetate, and ethanol were determined by high-pressure liquid chromatography (HPLC; LKB) with a Rezex organic acid analysis column (Phenomenex) at a temperature of 45°C with 7.2 mM H2SO4 as the eluent, using an RI 1530 refractive index detector (Jasco) and AZUR chromatography software for data integration (9). Lactate and glucose were measured by using Megazyme enzymatic kits (Wicklow-Ireland).
Amino acid analysis was performed using the Waters AccQ.Tag chemistry package. The analyses were performed on an Agilent 1200 series HPLC equipped with a Hitachi fluorimetric detector, operating at an excitation wavelength of 250 nm and detection of emission at 395 nm. The samples were separated on a 3.9- by 150-mm AccQ.Tag column at a temperature of 37°C. The sample volume was 5 μl. Amino acid derivatives were obtained and separated according to the standard procedure of the Waters Company. The mobile phase was composed of (i) aqueous buffer containing 1 to 10 parts Waters AccQ.Tag eluent A solution and (ii) a 60% acetonitrile-water solution. Conditions of the gradient elution are described in the AccQ.Tag chemistry package. The elution flow rate was 1 ml/min.
RNA isolation, cDNA synthesis, and transcriptional analyses.
RNA was isolated from flash-frozen cell pellets stored at −80°C as previously described (18). cDNA synthesis, labeling, and hybridization were performed according to Opsata et al. (22), and transcriptome analyses were done by microarrays as described by Solheim et al. (29), with some modifications. In brief, on each array a dilution rate of 0.15 (D0.15) was used as the reference, and the two treatments at D0.05 and D0.4 were analyzed as direct comparisons to D0.15. We also made an indirect comparison between the two treatments by considering the contrast between log2(D0.4/D0.15) and log2(D0.05/D0.15). In all cases an empirical Bayes smoothing of gene-wise variances was conducted according to Smyth et al. (28). For each gene, the P value was adjusted to control the false discovery rate (FDR); hence, all P values displayed are FDR adjusted.
A selection of transcripts was quantified by real-time quantitative PCR (RT-qPCR) analyses as previously described (18). The expression level of 23S RNA was used as an internal control.
Two-dimensional gel electrophoresis and proteomic analyses.
Proteins were isolated from flash-frozen pellets stored at −80°C. Protein isolation, two-dimensional gel analyses, quantification, and protein identification were performed as previously described (18).
Microarray data accession number.
The microarray data have been deposited in the ArrayExpression database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-725.
RESULTS AND DISCUSSION
Growth of E. faecalis V583 at three different growth rates: metabolite analysis.
To study the effects of growth rate on the physiology of E. faecalis V583, we grew the cells in continuous, glucose-limited cultures (chemostats), allowing the precise regulation of growth rate by the pump speed. The cells were grown at fast (0.4 h−1), slow (0.05 h−1), and intermediate (0.15 h−1) growth rates under anaerobic conditions. Metabolite analyses from the experiments are shown in Table 1. Glucose was undetectable in all of the culture supernatants, demonstrating that glucose was the growth-limiting factor at all growth rates. Furthermore, the pattern of metabolites formed changed with the growth rate. At the lowest dilution rate, most of the glucose was converted to formate, ethanol, and acetate. The contribution of this pathway decreased with increasing growth rate, and homolactic fermentation became dominating.
Table 1.
Metabolite production of E. faecalis V583 grown under different conditions
| Condition | Dilution rate (h−1) | Dry wt (g/liter) | Concn (mM) ± SD of: |
ATP yield | ||||
|---|---|---|---|---|---|---|---|---|
| Glucoseb | Ethanol | Acetate | Lactate | Formate | ||||
| Chemostat | 0.05 | 1.45 | 0 | 32.8 ± 2.8 | 36.3 ± 2.1 | 27.7 ± 2.2 | 67.4 ± 2.1 | 2.5 ± 0.1 |
| Chemostat | 0.15 | 1.55 | 0 | 26.2 ± 5.1 | 20.3 ± 8.0 | 64.2 ± 1.1 | 46.3 ± 7.3 | 2.3 ± 0.4 |
| Chemostat | 0.4 | 1.78 | 0 | 6.5 ± 0.9 | 8.2 ± 2.1 | 97.3 ± 1.4 | 16.3 ± 0.5 | 2.1 ± 0.1 |
| Batch culturea | 1.1 | 25.5 | 3.2 | 0 | 60.1 | 6.5 | 1.9 | |
Batch culture data are from reference 12.
Glucose left in supernatant. The growth medium contained 55.5 mM glucose and 16 mM acetate.
Homolactic fermentation gives 2 U of ATP per molecule of glucose, while mixed acid fermentation producing ethanol, formate, and acetate results in a net production of 3 U of ATP per glucose molecule. Despite a gradual increase in ATP produced per molecule of glucose consumed, as seen in the change from a homofermentative to mixed acid fermentation (Table 1), growth yield declined when we lowered the growth rate (Table 1). The apparent increase in ATP yield could not compensate for the relatively higher maintenance demands incurred by lowering the growth rate. Although a shift toward mixed acid fermentation is common in lactic acid bacteria (9, 12, 19, 31), differences are seen in the response to growth rate reduction, reflecting differences in the regulation of metabolism. Some strains of L. lactis make a more abrupt shift toward the mixed acid pathway, and unlike E. faecalis V583, they can even display an increase in growth yield in response to growth rate reduction (31).
In batch culture with no glucose limitation, the homolactic fermentation was even more dominating than that in cultures growing at rate of 0.4 h−1 (Table 1), and consequently ATP yield was lower. As shown in Table 2, the main contributor to the change in metabolic profile in these experiments was the lactate flux, which showed a strong increase with increasing growth rate. Thus, the specific in vivo lactate dehydrogenase (LDH) activity (lactate flux) in cells growing at the highest growth rate were 20 times higher than that in cells growing at the lowest growth rate. The metabolite flux through pyruvate formate lyase (PFL) was much less sensitive to changes in growth rate and varied less than 2-fold. The specific in vivo activity of PFL (flux) was highest in cells grown at a rate of 0.15 h−1 and was almost the same at rates of 0.4 and 0.05 h−1. The lower Km of PFL than that of LDH for pyruvate (10) could explain a higher lactate/formate flux ratio by the increased growth rate, but the formate flux maximum at the middle growth rate indicates the regulation of enzyme activity (see below).
Table 2.
Carbon flux in Enterococcus faecalis V583 at different growth rates
| Metabolite | Flux (mmol/g dry wt h−1) ± SD by growth rate |
||
|---|---|---|---|
| 0.05 h−1 | 0.15 h−1 | 0.4 h−1 | |
| Glucose | 2.0 ± 0.1 | 5.6 ± 0.1 | 12.5 ± 0.1 |
| Ethanol | 1.3 ± 0.1 | 2.5 ± 0.5 | 1.5 ± 0.2 |
| Acetate | 1.2 ± 0.2 | 2.1 ± 0.1 | 1.8 ± 0.5 |
| Lactate | 1.0 ± 0.1 | 6.4 ± 0.1 | 21.6 ± 0.4 |
| Formate | 2.4 ± 0.1 | 4.5 ± 0.7 | 3.6 ± 0.1 |
The changes in catabolism were not restricted to carbohydrate metabolism. As shown in Table 3, a number amino of acids were broken down at different rates depending on the growth rate. Under all conditions tested, more than 90 to 95% of serine and arginine amino acids were consumed. Genes for the degradation of these two amino acids are regulated by carbon catabolite repression (CCR) in E. faecalis (2, 15, 22), and the results show, as expected, that the cells are relieved of CCR when grown under glucose-limiting conditions. Arginine degradation provides the cells with one molecule of ATP per arginine molecule consumed, and the amino acid is thus an additional energy source. Serine enters the pyruvate pool after enzymatic conversion by l-serine dehydratase. In L. lactis grown in glucose-limited chemostats, about 10% of the carbon flow through pyruvate could be derived from serine (21). Serine breakdown also could provide additional ATP, as has been shown for Staphylococcus epidermidis (27) and suggested for L. lactis (30).
Table 3.
Amino acid flux and rate ratio in Enterococcus faecalis V583 grown at different growth rates
| Dilution rate | Flux (mmol/g dry wt h−1) by growth rate |
Flux/growth rate ratio by growth rate |
||||
|---|---|---|---|---|---|---|
| 0.05 h−1 | 0.15 h−1 | 0.4 h−1 | 0.05 h−1 | 0.15 h−1 | 0.4 h−1 | |
| Asparagine | 0.031 ± 0.002 | 0.093 ± 0.033 | 0.028 ± 0.035 | 0.62 ± 0.05 | 0.62 ± 0.31 | 0.07 ± 0.01 |
| Arginine | 0.013 ± 0.002 | 0.037 ± 0.003 | 0.081 ± 0.012 | 0.26 ± 0.05 | 0.25 ± 0.02 | 0.20 ± 0.04 |
| Alanine | 0.003 ± 0.001 | 0.006 ± 0.009 | 0.002 ± 0.001 | 0.06 ± 0.02 | 0.04 ± 0.08 | 0.00 ± 0.01 |
| Glutamine | 0.041 ± 0.004 | 0.112 ± 0.044 | 0.004 ± 0.025 | 0.82 ± 0.11 | 0.75 ± 0.41 | 0.01 ± 0.08 |
| Glycine | 0.074 ± 0.005 | 0.184 ± 0.022 | 0.114 ± 0.039 | 1.48 ± 0.14 | 1.27 ± 0.20 | 0.28 ± 0.13 |
| Histidine | 0.021 ± 0.001 | 0.055 ± 0.011 | 0.018 ± 0.025 | 0.42 ± 0.02 | 0.37 ± 0.10 | 0.04 ± 0.08 |
| Isoleucine | 0.022 ± 0.001 | 0.065 ± 0.010 | 0.054 ± 0.019 | 0.44 ± 0.02 | 0.44 ± 0.10 | 0.13 ± 0.06 |
| Leucine | 0.047 ± 0.002 | 0.141 ± 0.024 | 0.127 ± 0.052 | 0.94 ± 0.05 | 0.94 ± 0.22 | 0.31 ± 0.18 |
| Lysine | 0.015 ± 0.003 | 0.042 ± 0.022 | 0.000 ± 0.021 | 0.3 ± 0.08 | 0.28 ± 0.20 | 0 ± 0.07 |
| Proline | 0.058 ± 0.036 | 0.133 ± 0.075 | 0.000 ± 0.03 | 1.16 ± 1.01 | 0.89 ± 0.71 | 0 ± 0.10 |
| Phenylalanine | 0.025 ± 0.002 | 0.073 ± 0.011 | 0.057 ± 0.011 | 0.5 ± 0.05 | 0.49 ± 0.10 | 0.14 ± 0.03 |
| Serine | 0.090 ± 0.001 | 0.248 ± 0.010 | 0.499 ± 0.135 | 1.8 ± 0.02 | 1.66 ± 0.10 | 1.24 ± 0.47 |
| Threonine | 0.005 ± 0.001 | 0.021 ± 0.014 | 0.001 ± 0.016 | 0.1 ± 0.02 | 0.14 ± 0.13 | 0.00 ± 0.05 |
| Tyrosine | 0.026 ± 0.008 | 0.055 ± 0.001 | 0.096 ± 0.056 | 0.52 ± 0.02 | 0.37 ± 0.01 | 0.24 ± 0.19 |
| Valine | 0.042 ± 0.003 | 0.132 ± 0.029 | 0.017 ± 0.005 | 0.84 ± 0.08 | 0.88 ± 0.27 | 0.04 ± 0.01 |
Apart from arginine and serine, tyrosine was the only amino acid degraded to a large extent at the high growth rate. At lower growth rates, the breakdown of other amino acids made a significant contribution to carbon flow, and their degradation rates were nearly proportional to the growth rate (Table 3). Amino acids other than arginine and serine can act as energy sources for E. faecalis (32). The differences between the middle and the high growth rates reflect a transition from one lifestyle to another. The two lifestyles are characterized by a great difference in the catabolism of amino acids, which possibly are governed by differences in energy supply.
Transcriptome profiling of cells growing at different rates.
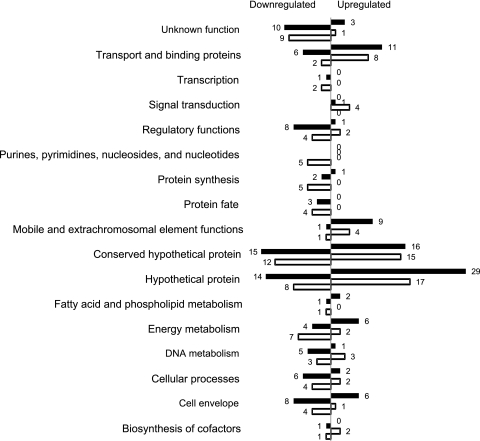
To assess the contribution of hierarchical regulation on the flux distribution, we undertook expression analysis at both transcriptome and proteome levels. Values from cells grown 0.15 h−1 were used as the reference in the transcriptome analyses. A tabulated review of the transcriptional results is presented in Table S1 in the supplemental material. As shown in Fig. 1, the expression of genes of all functional categories were affected by the changes in growth rate.
Fig 1.
Genes differently expressed by changes in growth rate (shaded bars, 0.05 to 0.15 h−1; open bars, 0.4 to 0.15 h−1). The genes are organized by functional category.
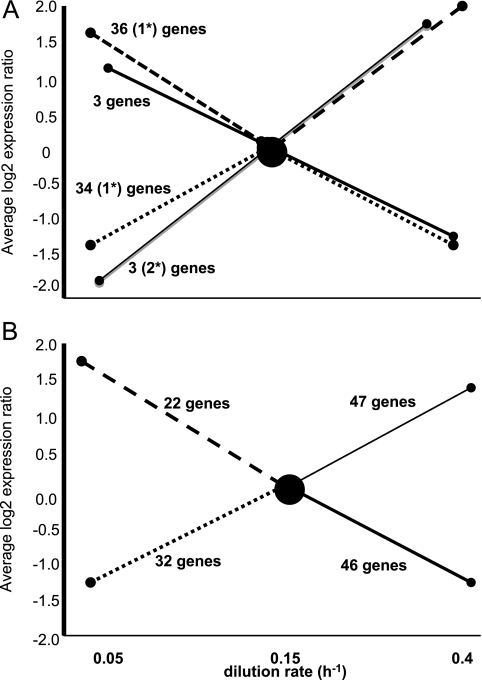
We found 223 gene transcripts to be altered more than 2-fold by changing the growth rate from 0.15 h−1 (P < 0.05). At the growth rate of 0.4 h−1, 88 genes were found to be upregulated and 84 were downregulated. At the lowest growth rate, 62 genes were upregulated and 82 were downregulated. Of the 77 genes represented by significant differences for both growth rate changes, most showed a maximum or minimum at the middle growth rate (Fig. 2).
Fig 2.
Average expression of genes differentially expressed at three growth rates. (A) Genes repressed by statistically significant data at all growth rates. The asterisk indicates that the gene(s) was analyzed by qRT-PCR. (B) Genes represented by statistically significant differences between two growth rates only. Shown are gene transcripts show increasing (dotted lines) or decreasing (dashed lines) abundance at the middle growth rate and increasing (gray lines) or decreasing (black lines) with growth rate.
We complemented the microarray analyses by the transcription analyses of a selection of genes by RT-qPCR, partly to confirm microarray data but also to investigate the transcription of genes for which the microarray data were of inadequate quality. This analysis included the transcription of genes encoding pyruvate formate-lyase activating enzyme (pflA; open reading frame [ORF] EF1612), a major facilitator family transporter (EF0082), glyceraldehyde-3-phosphate dehydrogenase (EF1964), bifunctional acetaldehyde-coenzyme A (CoA)/alcohol dehydrogenase (EF0900), and the principal lactate dehydrogenase (ldh-1; EF0255). As shown in Table 4, good agreement was found between the results obtained with microarray and RT-qPCR analyses for EF0900.
Table 4.
Effect of growth rate on transcription analyzed by qRT-PCR
| ORF | Gene | Gene product | Log2 growth rate (h−1) ratio |
|
|---|---|---|---|---|
| 0.05/0.15 | 0.4/0.15 | |||
| EF0255 | ldh-1 | Lactate dehydrogenase | −3.88 ± 0.14 | 6.88 ± 0.32 |
| EF1612 | pflA | Pyruvate formate lyase-activating enzyme | −0.89 ± 0.03 | 0.14 ± 0.09 |
| EF0082 | Major facilitator family transporter | −2.80 ± 0.13 | −0.16 ± 0.06 | |
| EF1964 | gap-2 | Glyceraldehyde-3-phosphate dehydrogenase | 0.17 ± 0.04 | 2.01 ± 0.10 |
| EF0900 | adhE | Bifunctional acetaldehyde-CoA/alcohol dehydrogenase | −1.06 ± 0.19 | −1.5 ± 0.13 |
Only 10 genes showed statistically significant differences in transcription between the lowest and the highest growth rates. These genes encode a putative membrane protein (EF0025), a major facilitator family transporter (EF0082), l-lactate dehydrogenase ldh-1 (EF0255), putative bacteriocin immunity protein PlnM (EF0439), conserved hypothetical protein (EF1541), pyruvate formate-lyase activating enzyme (EF1612), glyceraldehyde-3-phosphate dehydrogenase (EF1964), Cro/CI family transcriptional regulator (EF2291), hypothetical protein (EF2518), and resolvase family site-specific recombinase (EF2283).
The most remarkable result obtained by the transcription analysis was the dramatic effect seen with ldh-1 encoding the principal lactate dehydrogenase in E. faecalis (13). As shown in Table 4, the ldh-1 transcript increased with growth rate, and fast-growing cells contained about 1,700 times more ldh-1 transcript than cells growing at the lowest rate. No other transcript showed a comparable response. To our knowledge, a difference in ldh-1 transcription of this magnitude has not been reported previously.
Previous work has shown that ldh-1 transcription can be regulated by the global regulators redox-sensing regulator (Rex) and the catabolite control protein A (CcpA) (18, 22). However, other genes found to be controlled by these regulators did not show a strong response to changes in growth rate. Additional mechanisms, specifically regulating the level of ldh-1 transcripts over a wide dynamic range, appear to be involved in the growth rate response. Whether this involves the regulation of transcription, mRNA breakdown, or both is not known.
Effects of growth rate on the proteome and comparison to transcriptional effects.
The proteomes of cells grown at the three growth rates were analyzed by two-dimensional (2D) gel electrophoresis. By using 2D gel software, we were able to detect more than 400 spots on silver-stained gels. The analyses identified 56 differentially expressed proteins (P < 0. 05) (Table 5). The proteins most affected by the changes in growth rate were the general stress protein (EF1744) and the tRNA modification GTPase TrmE, encoded by EF3312. Both showed their lowest expression at a growth rate of 0.15 h−1. Altogether, 24 proteins showed their lowest abundance at this growth rate. Even 30S ribosomal protein S3 (EF0212) and 50S ribosomal protein L6 (EF0221) showed this behavior. Six proteins showed reduced abundance, and 13 showed an increase with growth rate over the range tested. In general, the effects on the protein level were smaller than the transcriptional effect. Of the 56 differentially expressed proteins, the quantity of 21 varied more than 2-fold between the middle and the high or low growth rates (Table 5). Enzymes involved in energy metabolism were the dominant category (16 proteins).
Table 5.
Proteins differentially expressed by change in growth rates
| Protein and category | Name or function | Log2 growth rate (h−1) ratio |
|
|---|---|---|---|
| 0.05/0.15 | 0.4/0.15 | ||
| Amino acid biosynthesis | |||
| EF2550 | Serine hydroxymethyltransferase | −1.04 | −0.79 |
| Biosynthesis of cofactors, prosthetic group, carriers | |||
| EF1860 | 3-Methyl-2-oxobutanoate hydroxymethyltransferase | 0.52 | 0.75 |
| EF0901 | Isopentenyl diphosphate delta isomerase, putative | 0.63 | 1.1 |
| Cell envelope | |||
| EF2193 | dTDP-4-dehydrorhamnose 3,5-epimerase | −0.30 | 0.79 |
| EF2194 | Glucose-1-phosphate thymidyltransferase | −0.26 | 0.59 |
| EF3183 | Cell wall surface anchor family protein | 1.00 | 0.44 |
| Cellular processes | |||
| EF0080 | Gls24 protein | −0.15 | −0.5 |
| EF1744 | General stress protein | 1.30 | 1.46 |
| EF1991 | Cold shock protein CspC | −0.25 | 0.42 |
| EF3312 | tRNA modification GTPase TrmE | 1.01 | 1.63 |
| DNA metabolism | |||
| EF0002 | DNA polymerase III, beta subunit | 0.65 | 0.32 |
| EF0883 | Primosomal DnaI | −0.60 | 1.15 |
| Energy metabolism | |||
| EF0020 | Phosphotransferase system, mannose-specific IIAB components | −0.15 | −1.3 |
| EF0195 | Phosphoglycerate mutase 1 | −0.33 | 0.52 |
| EF0949 | Phosphotransacetylase | 0.30 | 0.54 |
| EF1125 | Putative l-ascorbate 6-phosphate lactonase | 0.02 | 0.61 |
| EF1131 | l-Ribulose-5-phosphate 4-epimerase | 0.39 | 0.75 |
| EF1167 | Fructose-bisphosphate aldolase | 0.77 | 0.24 |
| EF1353 | Pyruvate dehydrogenase complex E1 component, alpha subunit | 0.30 | 0.94 |
| EF1354 | Pyruvate dehydrogenase complex E1 component, beta subunit | −0.12 | 0.65 |
| EF1356 | Dihydrolipoamide dehydrogenase | −0.53 | −1.64 |
| EF1416 | Glucose-6-phosphatase isomerase | −0.02 | −0.89 |
| EF1526 | Glyceraldehyde-3-phosphate dehydrogenase | 0.08 | −0.95 |
| EF1612 | Pyruvate formate-lyase activating enzyme | 0.24 | −1.18 |
| EF1962 | Triosephosphate isomerase | 0.04 | 0.73 |
| EF1963 | Phosphoglycetare kinase | −0.37 | −0.2 |
| EF1964 | Glyceraldehyde-3-phosphate dehydrogenase | 0.35 | −1.07 |
| EF0283 | 3-Oxoacyl-(acyl carrier protein) synthetase II | −0.12 | −0.91 |
| Fatty acid and phospholipid metabolism | |||
| EF2875 | Acetyl-CoA carboxylase subunit alpha | 0.00 | 0.89 |
| EF2881 | 3-Ketoacyl-(acyl carrier protein) reductase | −0.36 | 1.10 |
| EF2882 | Acyl carrier protein S-malonyltransferase | 0.00 | 0.54 |
| Hypothetical proteins | |||
| EF1227 | Hypothetical protein | −0.99 | 0.26 |
| EF1241 | Hypothetical protein | −0.14 | 0.11 |
| EF1560 | Hypothetical protein | 0.47 | 0.91 |
| Protein fate | |||
| EF2200 | Methionine aminopeptidase | −0.48 | 0.70 |
| F2898 | Peptidyl-prolyl cis trans isomerase | 0.75 | 0.74 |
| EF3066 | Peptide deformylase | 0.81 | 1.48 |
| Protein synthesis | |||
| EF0212 | 30S ribosomal protein S3 | 0.97 | 1.39 |
| EF0221 | 50S ribosomal protein L6 | 0.60 | 0.72 |
| EF0701 | Peptide chain release factor 3 | 0.36 | 1.22 |
| Purines, pyrimidines, nucleosides, and nucleotides | |||
| EF0228 | Adenylate kinase | 0.61 | 1.59 |
| EF1713 | Orotidine 5′-phosphate decarboxylase | 0.57 | 0.91 |
| EF1721 | Bifunctional pyrimidine regulatory protein PyrR uracil phosphoribosyltransferase | −1.16 | 0.61 |
| EF2549 | Uracil phosphoribosyltransferase | 0.04 | 0.60 |
| EF3293 | Inositol-5-monophosphate dehydrogenase | 0.08 | −0.79 |
| Regulatory functions, signal transduction, transcription, and transport and binding proteinsa | |||
| EF1741 | Catabolite control protein A | 0.25 | 0.38 |
| EF3289 | DNA-binding response regulator | 0.82 | 1.31 |
| EF1050 | DNA-binding response regulator | 0.38 | 1.04 |
| EF0233 | DNA direct RNA polymerase subunit alpha | 0.50 | −0.26 |
| EF0865 | Glycine betaine/carnitine/choline transporter, ATP-binding protein | −0.24 | −0.54 |
| Unknown function | |||
| EF0076 | Oxidoreductase short chain dehydrogenase/reductase family | 0.31 | −1.64 |
| EF0877 | Aldo-/keto-reductase family oxidoreductase | −0.64 | 0.48 |
| EF1138 | Aldo-/keto-reductase family oxidoreductase | −0.45 | 0.23 |
| EF2591 | Glyoxalase family protein | 0.54 | 1.07 |
| EF2656 | Flavoprotein family protein | 0.97 | 1.16 |
| EF2927 | Haloacid dehydrogenase superfamily hydrolase | 0.92 | 2.04 |
EF1741 and EF3289 have regulatory functions; EF1050 is involved in signal transduction; EF0233 is involved in signal transcription; and EF0865 is involved in transport and binding.
Surprisingly, the content of the major lactate dehydrogenase, Ldh-1 (EF0255), was the same at the three growth rates (results not shown) despite the huge differences in transcript levels. Thus, the synthesis of Ldh-1 appears to be controlled by a posttranscriptional mechanism. The amount of bifunctional acetaldehyde-CoA/alcohol dehydrogenase (EF0900) also was constant (data not shown), although the transcript levels varied. Other discrepancies between data from transcriptional and proteomic analyses were seen for enzymes such as glyceraldehyde 3-phosphate dehydrogenase (EF1964) and PflA, the pyruvate formate lyase-activating enzyme (EF1612). The downregulation of PflA expression is in line with the reduction of formate flux when the growth rate was increased from 0.15 and 0.4 h−1 (Table 2). Evidence for posttranscriptional regulation of enzymes in energy metabolism has been observed previously in L. lactis (5), Escherichia coli (14), and in our study of the Ldh mutant of E. faecalis (18).
In conclusion, the expression of a number of genes and proteins varied with the growth rate. Many showed a maximum or minimum of expression at the intermediate growth rate, coinciding with a shift in energy metabolism. The cells shifted toward a more energy-efficient catabolism upon transition from the highest to the intermediate growth rate, possibly triggered by lowered energy status in the cells. The best-studied mechanism for transcription regulation in response to energy status in Firmicutes is CCR (4), but as already mentioned, even the fast-growing cultures in our experiments were relieved of CCR due to glucose limitation. Thus, the differences in metabolism, transcriptome, and proteome seen in our experiments reflect the involvement of other regulatory mechanisms. The regulation of expression beyond the stage of transcription appears to be common, most strikingly seen with ldh-1 expression. High LDH activity is the hallmark of a lactic acid bacterium and is a prerequisite for its competitiveness in natural habitats. Its activity is regulated by several mechanisms, including the biochemical regulation of enzyme activity and genetic regulation. In this work, we have demonstrated the presence of a new transcriptional and posttranscriptional mechanism with the potential to exert strong control on LDH activity. Under the conditions used in these experiments, these mechanisms counteract each other to maintain a constant enzyme level, but high transcript levels of ldh-1 might be an important part of the strategy of a lactic acid bacterium to succeed in a (rapidly) changing environment.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the SysMO-LAB project, which is financed by the Norwegian Research Council and the Netherlands Organization for Scientific Research (NWO).
We acknowledge Maria Jönsson, Morten Skaugen, Linda H. Godager, and Mari C. Brekke for technical assistance.
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alexeeva S, de Kort B, Sawers G, Hellingwerf KJ, de Mattos MJ. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barcelona-Andrés B, Marina A, Rubio V. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boer VM, de Winde JH, Pronk JT, Piper MDW. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265–3274 [DOI] [PubMed] [Google Scholar]
- 4. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 5. Dressaire C, et al. 2009. Transcriptome and proteome exploration to model translation efficiency and protein stability in Lactococcus lactis. PLoS Comput. Biol. 5:e1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dressaire C, et al. 2008. Growth rate regulated genes and their wide involvement in the Lactococcus lactis stress responses. BMC Genomics 9:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Even S, Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1999. Pyruvate metabolism in Lactococcus lactis is dependent upon glyceraldehyde-3-phosphate dehydrogenase activity. Metab. Eng. 1:198–205 [DOI] [PubMed] [Google Scholar]
- 8. Even S, Lindley ND, Cocaign-Bousquet M. 2003. Transcriptional, translational and metabolic regulation of glycolysis in Lactococcus lactis subsp. cremoris MG 1363 grown in continuous acidic cultures. Microbiology 149:1935–1944 [DOI] [PubMed] [Google Scholar]
- 9. Fiedler T, et al. 2011. Characterization of three lactic acid bacteria and their isogenic ldh deletion mutants shows optimization for YATP (cell mass produced per mole of ATP) at their physiological pHs. Appl. Environ. Microbiol. 77:612–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645–677 [DOI] [PubMed] [Google Scholar]
- 12. Jensen NB, Melchiorsen CR, Jokumsen KV, Villadsen J. 2001. Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Appl. Environ. Microbiol. 67:2677–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jönsson M, Saleihan Z, Nes IF, Holo H. 2009. Construction and characterization of three lactate dehydrogenase-negative Enterococcus faecalis V583 mutants. Appl. Environ. Microbiol. 75:4901–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kramer G, et al. 2010. Proteome-wide alterations in Escherichia coli translation rates upon anaerobiosis. Mol. Cell Proteomics 9:2508–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leboeuf C, Leblanc L, Auffray Y, Hartke A. 2000. Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by CcpA. J. Bacteriol. 182:5799–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lemke JJ, et al. 2011. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U. S. A. 108:5712–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levy S, Barkai N. 2009. Coordination of gene expression with growth rate: a feedback or a feed-forward strategy? FEBS Lett. 583:3974–3978 [DOI] [PubMed] [Google Scholar]
- 18. Mehmeti I., et al. 2011. Transcriptome, proteome, and metabolite analyses of a lactate dehydrogenase-negative mutant of Enterococcus faecalis V583. Appl. Environ. Microbiol. 77:2406–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melchiorsen RC, Jokumsen KV, Villadsen J, Israelsen H, Arnau J. 2002. The level of pyruvate-formate lyase controls the shift from homolactic to mixed-acid product formation in Lactococcus lactis. Appl. Microbiol. Biotechnol. 58:338–344 [DOI] [PubMed] [Google Scholar]
- 20. Molenaar D, van Berlo R, de Ridder D, Teusink B. 2009. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novák L, Loubiere P. 2000. The metabolic network of Lactococcus lactis: distribution of 14C-labeled substrates between catabolic and anabolic pathways. J. Bacteriol. 182:1136–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Opsata M, Nes IF, Holo H. 2010. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Papagianni M, Avramidis N, Filiousis G. 2007. Glycolysis and the regulation of glucose transport in Lactococcus lactis spp. lactis in batch and fed-batch culture. Microb. Cell Fact. 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Picard F, Dressaire C, Girbal L, Cocaign-Bousquet M. 2009. Examination of post-transcriptional regulations in prokaryotes by integrative biology. C. R. Biol. 332:958–973 [DOI] [PubMed] [Google Scholar]
- 25. Regenberg B, et al. 2006. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 7:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sahm DF, et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sivakanesan R, Dawes EA. 1980. Anaerobic glucose and serine metabolism in Staphylococcus epidermidis. J. Gen. Microbiol. 118:143–157 [DOI] [PubMed] [Google Scholar]
- 28. Smyth GK, Michaud J, Scott HS. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075 [DOI] [PubMed] [Google Scholar]
- 29. Solheim M, Aakra A, Vebo H, Snipen L, Nes IF. 2007. Transcriptional responses of Enterococcus faecalis V583 to bovine bile and sodium dodecyl sulfate. Appl. Environ. Microbiol. 73:5767–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuart MR, Chou LS, Weimer BC. 1999. Influence of carbohydrate starvation and arginine on culturability and amino acid utilization of Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 65:665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas TD, Ellwood DC, Longyear VMC. 1979. Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J. Bacteriol. 138:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward DE, et al. 2000. Branched-chain alpha-keto acid catabolism via the gene products of the bkd operon in Enterococcus faecalis: a new, secreted metabolite serving as a temporary redox sink. J. Bacteriol. 182:3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.