Abstract
Fungal laccases have been widely used in industry. The expression of laccase often is repressible by the primary carbon source glucose in many fungi. The underlying basis is largely unclear. We demonstrate here that a gene, TSP2-1, was required for laccase repression by glucose in the basidiomycete Cryptococcus neoformans. TSP2-1 encodes a Tsp2-type tetraspanin. The disruption of TSP2-1 resulted in constant melanin formation and the expression of the laccase gene LAC1. This derepression phenotype was restorable by 10 mM exogenous cyclic AMP (cAMP). A capsule defect in the mutant tsp2-1Δ also was restored by cAMP. The results indicate an interaction of Tsp2-1 with the cAMP-dependent protein kinase A (PKA) pathway that has been shown to modulate laccase repression and capsule biosynthesis in this fungus. Other roles of TSP2-1, e.g., in maintaining cell membrane integrity and stress resistance, also were defined. This work reveals a Tsp2-1-dependent glucose repression in C. neoformans. The function of Tsp2-type tetraspanin Tsp2-1 is described for the first time.
INTRODUCTION
Laccase (EC 1.10.3.2), a blue copper-containing phenolic oxidase, is widely distributed in higher plants and fungi, and it also is found in bacteria and insects (7, 23, 30, 37). Laccase was first discovered by Yoshida in the lacquer tree (Rhus spp.) (48). The enzyme has been used in a broad array of industrial and environmental applications (7, 10, 23, 37), e.g., in pulp and paper manufacture, textile whitening, food processing (4, 37), and in the bioremediation of soil and water (2, 41). Laccases produced by the white-rot fungi play a critical part in the degradation of plant litters (29, 30). Recent uses of laccase include the generation of new organic molecules, biomaterials, and biosensors and the production of biofuel ethanol from plant lignocellulose (5, 23, 31, 36). The main source for industrial laccase production is higher fungi. However, reactor-scale production in industry is hampered by several difficulties (10, 23). For instance, levels of laccase produced by native producers or by heterogeneous expression still are too low for industrial production (10, 23). Knowledge of the expression regulation of laccase in fungal hosts is accumulating. Some fungal laccase genes are constitutively expressed, while others are inducible (9). The enzyme usually is repressed during fungal growth and is inhibited by carbon sources, such as glucose, and on some occasions it is inactivated by certain compounds (i.e., proteases) (10).
Cryptococcus neoformans produces abundant laccase and has served as a system to investigate laccase expression (42, 51). C. neoformans is a soilborne basidiomycete and is particularly enriched in bird droppings and tropical trees (18). Cryptococcal laccase, a cell wall-associated enzyme, was demonstrated to be encoded mainly by LAC1 (32, 49, 51). The disruption of LAC1 results in undetectable laccase activity in glucose starvation (32, 51). Another homolog, LAC2, is indicated to be expressed under oxidative stress (32, 51). The function of LAC2 is unknown. The transcription of LAC1 is induced by glucose starvation (32, 51). In C. neoformans, laccase is the sole enzyme for the formation of the pigment melanin via oxidizing phenolic substrates, e.g., norepinephrine (NE) (42, 51). Mutants of lac1 do not generate melanin and form albino colonies. This becomes advantageous for the study of laccase expression in C. neoformans for comparisons to many other fungi that carry multiple copies of laccase genes (6, 9, 21). The pigmentation can be applicable in laccase mutant screening via colony color alteration (51). It is known that several environmental factors determine laccase expression in C. neoformans. The trace element copper is an effective inducer of the enzyme, and high temperature inhibits its expression (19, 20). Glucose is a primary repressor of laccase (21, 51). The repression occurs at the transcription level, i.e., the transcription of LAC1 is inhibited in the presence of glucose (20, 51). When high concentrations of glucose are present, e.g., 2%, the transcription of LAC1 is downregulated (34, 51). The nutrient-sensing heterotrimeric G protein and the cyclic AMP (cAMP)-dependent protein kinase A (PKA) signaling pathway is responsible for laccase repression and capsule biosynthesis (1). However, many steps in this pathway leading to laccase repression and capsule modulation have been missing from this fungus (22).
We employed Agrobacterium tumefaciens-mediated T-DNA random insertional mutagenesis (ATMT) (40) to generate glucose derepression mutants for laccase in C. neoformans. By taking advantage of a color-based screening procedure, transformants of interest are expected to form dark colonies in the presence of 2% glucose due to melanin formation, whereas the wild-type colonies remain white. The genes in the candidates that presumably are disrupted by T-DNA can be cloned by methods like inverse PCR. With this approach, we obtained more than 30 glucose-depression mutant isolates (unpublished data). Here we describe one, LZM36, in which laccase is not inhibited by glucose. The mutated gene in LZM36 was identified as a tetraspanin-encoding gene, designated TSP2-1, which belongs to the fungal Tsp2 tetraspanin family (17, 24). We prove here that TSP2-1 plays a key role in laccase repression by glucose. Also, we showed that the gene TSP2-1 is critical for the fungus to respond to various environmental conditions.
MATERIALS AND METHODS
Strains, media, and culture conditions.
C. neoformans serotype D strain JEC21 (MATα) was the wild type in this study (33, 47). JEC21FOA, a uracil auxotrophic mutant of JEC21 (47), was used as a recipient for making the targeted disruption of TSP2-1. Strain LZM36 was a T-DNA insertional mutant created from JEC21FOA which produced melanin on 2% glucose-containing agar. C. neoformans was grown in YPD (2% glucose, 2% Bacto peptone, 1% yeast extract, pH 6.0). Asparagine (Asn) salt agar (0.1% asparagine, 0.3% KH2PO4, pH 5.1) supplemented with 2% glucose or other carbon sources was the medium for melanin production or LAC1 induction in the presence of the laccase substrate norepinephrine (NE) (100 mg/liter) (42, 47). Cells were incubated at 30°C unless indicated otherwise.
Identification of TSP2-1 and its targeted disruption.
Agrobacterium tumefaciens-mediated T-DNA random insertional mutagenesis (ATMT) in C. neoformans was carried out by following a previous protocol (40). Dark transformants on 2% glucose-containing Asn agar were picked. One mutant, LZM36, out of 32 dark transformants was chosen for further analysis (Fig. 1A). Genomic sequences flanking the T-DNA integration site in LZM36 was retrieved by inverse PCR. Briefly, 2.0 μg genomic DNA of the mutant was digested with XbaI and ligated with T4 DNA ligase overnight at 14°C. Approximately 1 μl of the ligation reaction mixture was taken as the template in inverse PCR with two pairs of primers located on the T-DNA left and right boundaries: p1-S/p1-A, 5′-ACTGGAACAACACTCAACCCTA-3′ and 5′-TGGGGTTTCTACAGGACG-3′; and p2-S/p2-A, 5′-TGGCGGGTAAACCTAAGAG-3′ and 5′-CGCACAATCCCACTATCC-3′. PCR products were subject to sequencing and were used for blast searches against GenBank databases.
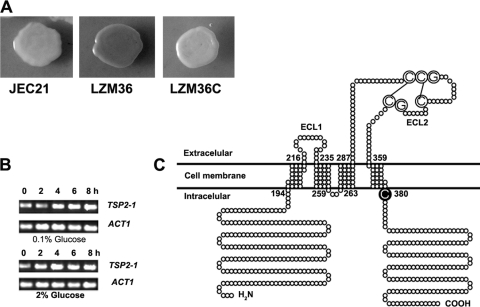
Fig 1.
Disruption of TSP2-1 in the mutant LZM36 is responsible for the laccase derepression phenotype. (A) LZM36 produced melanin in the presence of 2% glucose, the wild-type JEC21 showed an albino colony, and the complemented strain LZM36C restored the repression and produced little melanin. (B) TSP2-1 is a constitutively expressed gene, as shown by the RT-PCR amplification of TSP2-1 mRNA. Fresh cells of JEC21 were incubated in 0.1 or 2% glucose-containing Asn liquid medium at 30°C for 2, 4, 6, and 8 h as indicated. (C) Topological structure of Tsp2-1 showing four transmembrane domains (TM1 to TM4), three loops (ECL1, ICL, and ECL2), and a CCG motif on ECL2 (see Results for a description).
To create a deletion mutant of TSP2-1 via homologous recombination, two pairs of primers, i.e., Primer−969/Primer+54 (5′-TCTCTCGAGGGGATGGAAGAAGTGGAA and 5′-GGGATCGATCGAATTTGGATGCGAAG, respectively) and Primer+1752/Primer+2762 (5′-CCTGAATTCGTCATCAGGCGGACGAGT and 5′-TTAGGATCCCCTTTCTCCGCCACCATT, respectively), were designed to amplify 1-kb fragments of the 5′ and 3′ ends of TSP2-1 (Fig. 2A). The selection marker URA5 (1.9-kb) was ligated to the PCR products to create the disruption cassette pBS-TSP2-1-URA5. The disruption cassette (3.9 kb) then was amplified with primers Primer−969/Primer+2762 to make enough DNA to be transformed into JEC21FOA by electroporation. Transformants were screened by a PCR screening procedure with Primer−1859 (5′-TACGGATCCGCGTTGTGAGATTGCCTTGAG), located at 890 nucleotides upstream of the disruption cassette, and primer URA5 (5′-GCTGAATTCCGGTACTATGGTCGGTGCG), located in the open reading frame (ORF) of URA5 (Fig. 2A, bottom). A 2.8-kb PCR product was expected in the positive candidates that carried disrupted TSP2-1, while no product was expected from the wild type. The PCR product then was sequenced for verification. One positive candidate, strain tsp2-1Δ, was chosen for Southern blotting and reverse transcription-PCR (RT-PCR) verification with the primers Primer+943 (5′-GCCCCTGGCTACATTACCTACA) and Primer+1753 (5′-ACTTCCGCTGGCGATATGAG), generating a 0.81-kb product. The mutant was tested for melanin production on 2% glucose agar.
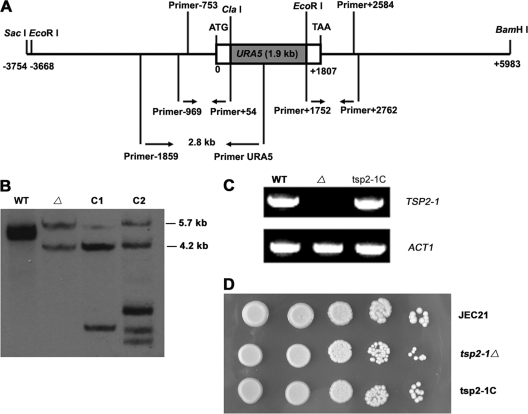
Fig 2.
Targeted disruption of TSP2-1. (A) Schematic diagram of the replacement of TSP2-1 by URA5 through homologous recombination. The locations of primers for making the disruption mutant and for PCR screening are indicated. Primer−969/Primer+54 were used to amplify the upstream fragment of TSP2-1, while Primer+1752/Primer+2762 were used to amplify the 3′ end of TSP2-1. PCR products were ligated to URA5 with ClaI and EcoRI restriction. Primer−1895/PrimerURA5 were used for mutant screening. (B) Southern analysis on the insertion site of URA5 in strain tsp2-1Δ (Δ). Genomic DNA was double digested with BamHI and EcoRI for wild-type JEC21 (lane 1), the targeted disruption mutant tsp2-1Δ (lane 2), and two complemented candidates (lanes 3 and 4; designated C1 and C2, respectively). The membrane was probed with a DIG-labeled 3.3-kb PCR fragment of the wild-type TSP2-1 gene (see Materials and Methods). As expected, the wild type had two bands, 4.9 and 4.7 kb. The mutant generated two bands of 5.7 and 4.2 kb in size. The complemented strains obtained extra TSP2-1 bands plus the two original bands, 5.7 and 4.2 kb, in tsp2-1Δ. C2 was used in the experiments depicted in panels C and D. (C) TSP2-1 mRNA was not detected in tsp2-1Δ. The reconstituted strain tsp2-1C restored the expression of TSP2-1. (D) TSP2-1 is dispensable for growth. Cells were spotted in 10-fold serial dilutions and grown on Asn agar with 2% glucose at 30°C.
The complemented strains were made by the cotransformation of a 3.3-kb PCR fragment of the wild-type TSP2-1 with primers Primer−753/Primer+2584 (ATAGGTACCTCGTTAGCTTGGGAGGCA/ATAGGATCCGCCCCTAAGGATTTGTTG) and a 2.3-kb HYG expression cassette into the mutant LZM36 and tsp2-1Δ strains on hygromycin-containing agar (200 μg/ml). Transformants were incubated on Asn salt agar containing 2% glucose and NE. The complemented strains exhibiting white colonies were subjected to Southern blotting and PCR confirmation together with the tsp2-1Δ mutant.
DNA preparation and Southern blotting.
C. neoformans cells were grown in 100 ml YPD to stationary phase at 30°C. DNA extraction was carried out as described previously (47, 50). Digested DNA with the appropriate restriction enzymes was subjected to separation in 0.8% agarose gels, and blotting was conducted on N+-Magaprobe nylon transfer membrane by following the manufacturer's instructions (GE Osmonics Inc., MN). The 3.3-kb PCR fragment of TSP2-1 DNA was used as the probe and was labeled with digoxigenin (DIG) High Primer DNA labeling and detection starter kit II (Roche China, Shanghai, China).
RNA preparation and RT-PCR.
To examine LAC1 transcription, yeast cells were grown in 2% glucose-containing Asn salt liquid at 30°C for 8 h. Total mRNA was prepared by following the protocol provided by the RNAiso plus kit (TaKaRa Biotech Co. Ltd., Dalian, China). The first chain of cDNA was synthesized by Moloney murine leukemia virus (M-MLV) reverse transcriptase with approximately 1 μg mRNA as the template and oligo(dT15) as the primer (TaKaRa Biotech Co. Ltd., Dalian, China). The level of LAC1 cDNA was determined by PCR amplification with primers LAC1-S (5′-ATGAAATCTCCATCGCACCC) and LAC1-A (5′-CGTCCTATCACCTGGAACTCG). A 0.57-kb LAC1 fragment was expected. The mRNA of ACT1, the actin-encoding gene, served as an internal control. A 0.52-kb ACT1 fragment was generated by the primers ACT-S (5′-ACCTCTTCACCACCTCTGC) and ACT-A (5′-GGTGGACGATTGAGGGAC).
Assays for melanin production and capsule biosynthesis.
Strains were examined for pigmentation on 2% glucose. Fresh cells were grown overnight in liquid YPD, washed three times with sterile double-distilled water (ddH2O), and resuspended in ddH2O to 2 × 108 cells/ml. Approximately 5 μl of the suspension was dropped onto Asn agar containing 2% glucose and NE at 30°C for 2 days for melanin formation. For capsule observation, fresh cells taken from YPD culture were transferred to maltose agar and incubated at 30°C for 5 days. Cells were subject to India ink staining microscopy (47). To examine the effects of different carbon sources on melanin production in the mutant strain tsp2-1Δ, fresh cells were spotted on Asn salt agar containing NE. Tested carbon sources included 2% sucrose, maltose, d-fructose, d-galactose, d-mannose, and lactose (in the place of dextrose) at 30°C for 16 or 6 h according to they type of pigmentation.
For the measurement of laccase enzymatic activity, fresh yeast cells were harvested from Asn salt plates (2% glucose) after incubation at 30°C for 16 h. The enzymatic activity was measured by the oxidation of NE (100 mg/liter) with 107 cells/ml in 0.05 M sodium phosphate (pH 6.5) at 37°C for 30 min. The OD at 475 nm was recorded. One enzymatic unit equals an A475 of 0.001. The assay was conducted in triplicate, and the standard deviations were calculated (42).
Stress tolerance assays and CFW staining microscopy.
Cells were cultured on YPD agar at 30°C for 2 days, washed by sterile water twice, and resuspended in sterile ddH2O again. The cell concentration was determined by a hemocytometer. Starting with 105 cells in 5 μl, cells were spotted on YPD agar in 10-fold serial dilutions to the last spot containing 10 cells. YPD plates were supplemented with 0.01% SDS, 1% Congo red, 1.0 M KCl, 2.0 M NaCl, 2.0 M sorbitol, 1 mM H2O2, or 1 mM NaNO2 at 30°C for 1 to 3 days. To examine chitin biosynthesis, aliquots of the cells were stained with calcofluor white (CFW; Sigma, St. Louis, MO) as described previously (47). Cells were examined with a Nikon Eclipse 80i fluorescence microscope using a 4,6-diamidino-2-phenylindole filter.
RESULTS
A Tsp2 tetraspanin gene was disrupted in LZM36.
LZM36 was a T-DNA insertional transformant that produced melanin in the presence of 2% glucose, while under this condition melanin production was inhibited in the wild-type strain JEC21 (Fig. 1A). Probing with T-DNA and Southern blotting verified that only one copy of T-DNA was inserted in the genome of LZM36 (data not shown). Inverse PCR revealed that T-DNA was integrated 54 bp downstream of the start codon of an ORF (AAW45945) that putatively encodes a fungal Tsp2 tetraspanin (17). Thus, we named the gene TSP2-1. The product of TSP2-1 consists of 533 amino acids (aa). TSP2-1 is the only tetraspanin gene in the genome of C. neoformans (17, 24). TSP2-1 is constitutively expressed regardless of glucose concentration in the medium (Fig. 1B).
A targeted disruption mutant, strain tsp2-1Δ, was created to confirm the role of TSP2-1 in the laccase derepression phenotype in LZM36. The ORF of TSP2-1 was replaced by URA5 via homologous recombination as a routine practice in this fungus (Fig. 2A; also see Materials and Methods). By screening with PCR, one TSP2-1 disruptant candidate, strain tsp2-1Δ, was obtained. Mutant strains LZM36 and tsp2-1Δ were complemented with a 3.3-kb PCR fragment of the wild-type TSP2-1 gene. LZM36, tsp2-1Δ, and their complemented strains (LZM36C and tsp2-1C, respectively) were verified by Southern blotting (only the tsp2-1Δ strain is shown in Fig. 2B; data not shown for LZM36). The reconstituted strain tsp2-1C (designated C2 in Fig. 2B) expressed TSP2-1 mRNA again (Fig. 2C). The disruption of TSP2-1 did not have an obvious effect on yeast growth at 30°C (Fig. 2D).
Consistently with the phenotype of LZM36, the targeted mutant strain tsp2-1Δ formed a dark colony on 2% glucose (Fig. 3A). The complemented strain tsp2-1C restored the repression of melanin production in a white colony (Fig. 3A). A laccase enzymatic activity assay confirmed the derepression of laccase in the mutant tsp2-1Δ (Fig. 3B). Laccase activity in 108 cells produced 291 ± 8 U in the tsp2-1Δ strain, whereas the values for JEC21 and the complemented strain were 109 ± 3 and 141 ± 41 U, respectively. These results clearly verify that the repression of laccase activity by 2% glucose is abolished by the disruption of TSP2-1.
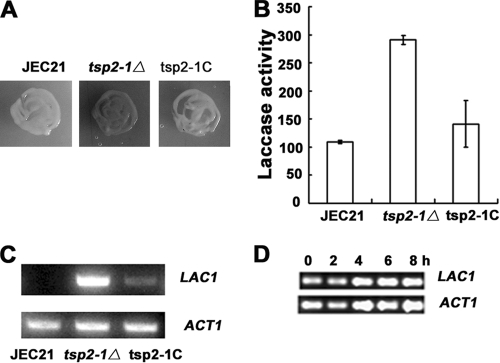
Fig 3.
TSP2-1-mediated repression of laccase on LAC1 transcription. (A) The targeted knockout mutant strain tsp2-1Δ exhibited a phenotype similar to that of LZM36, producing melanin in 2% glucose (30°C, 2 days). The wild-type JEC21 was used as a control. The reconstituted strain tsp2-1C restored the repression of laccase and formed an albino colony. (B) Laccase enzymatic activity increased dramatically in strain tsp2-1Δ in 2% glucose (30°C, 16 h) compared to that of the control JEC21 and the complement. Laccase activity in strain tsp2-1Δ was 291 ± 8 U (108 cells), whereas the wild-type JEC21 and the complemented strain tsp2-1C had 109 ± 3 and 141 ± 41 U (108 cells), respectively. (C and D) TSP2-1-mediated laccase inhibition at the transcription level by RT-PCR. (C) In the mutant tsp2-1Δ, LAC1 transcription was released from repression and expressed at a high level at 8 h. JEC21 and tsp2-1C served as controls. (D) Time course for LAC1 transcription reinforced the observations for panel C. LAC1 was constantly expressed from 0 to 8 h at a similar level. Total RNA was prepared from cells grown in 2% glucose Asn salt liquid at 30°C for 8 h (C) or in a time course at 0, 2, 4, 6, or 8 h (D). ACT1 mRNA served as the control. RT-PCR was carried out as described in Materials and Methods.
Laccase derepression in tsp2-1Δ mutant takes place at the transcriptional level.
To see at what stage the derepression of laccase takes place in the tsp2-1Δ mutant strain, RT-PCR was adopted to amplify the mRNA of LAC1 in the mutant (9). JEC21 and the complement served as controls in parallel. LAC1 was expressed in tsp2-1Δ, while no mRNA of LAC1 was detected in wild-type JEC21, and the complemented strain obviously restored the inhibition of LAC1 transcription (Fig. 3C). Further, LAC1 was constantly expressed from 0 to 8 h in the mutant strain tsp2-1Δ in a time course test (Fig. 3D). As a control, the mRNA of the actin-encoding gene ACT1 displayed a close level of transcription in each assay. These results manifest that the derepression of laccase in tsp2-1Δ occurs at the transcription level.
More sugar inhibition on laccase depends on TSP2-1.
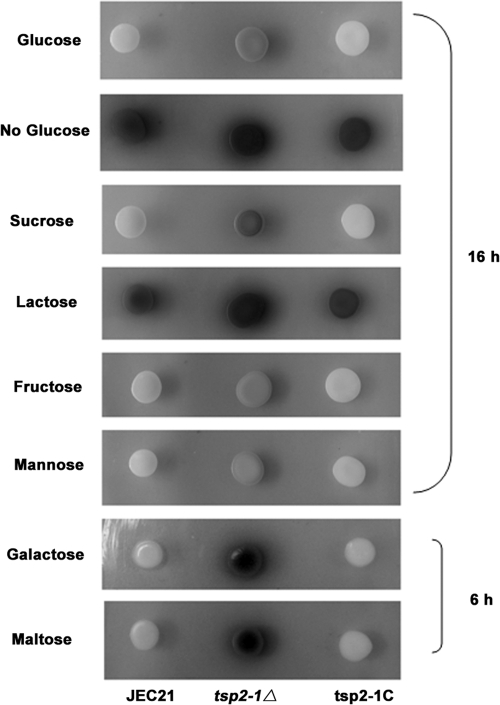
We further found that the inhibition of melanin production by some other carbon sources depended on Tsp2-1 as well. As shown in Fig. 4, five of the six tested sugars (lactose is the exception) exhibited a repression effect on laccase/melanin production in wild-type JEC21 (the spots on the left in each panel). Repression by these sugars required TSP2-1 function, as the mutant strain tsp2-1Δ produced significantly more melanin than JEC21 did (the spots in the middle of each panel). The complemented strain restored the repression of melanin formation (the spots on the right in each panel). As a control, laccase was highly expressed when there is no sugar in the medium (second panel from the top). This result suggests that sugar repression of laccase activity/melanin production shares the common Tsp2-associated step in C. neoformans.
Fig 4.
Laccase inhibition by other sugars also depends on TSP2-1. Sucrose, fructose, mannose, galactose, and maltose, but not lactose, exhibited a repression effect on melanin production in JEC21 (spots on the left in each panel). The disruption of TSP2-1 released the repression (central spots in each panel). The complemented strain was restored to the repression status (spots on the right in each panel). Yeast cells were dropped on Asn salt agar containing NE and 2% of each carbon source at 30°C for 16 or 6 h (for galactose and maltose only, due to fast pigmentation) for melanin production.
Exogenous cAMP restores glucose repression of melanin production in tsp2-1Δ strain.
The disruption of the G protein α subunit gene GPA1 causes a defect in melanin/laccase activity. This defect can be restored by the addition of cAMP (1, 12). We thus tested whether exogenous cAMP could restore melanin repression in mutant strain tsp2-1Δ. When cAMP was added to Asn salt agar, melanin production in tsp2-1Δ was inhibited. At 10 mM cAMP, melanin production in tsp2-1Δ was inhibited to a level similar to that of the wild type (Fig. 5A). This observation suggests that cAMP homeostasis is critical for laccase repression. The homeostasis likely is impaired in the mutant strain tsp2-1Δ.
Fig 5.
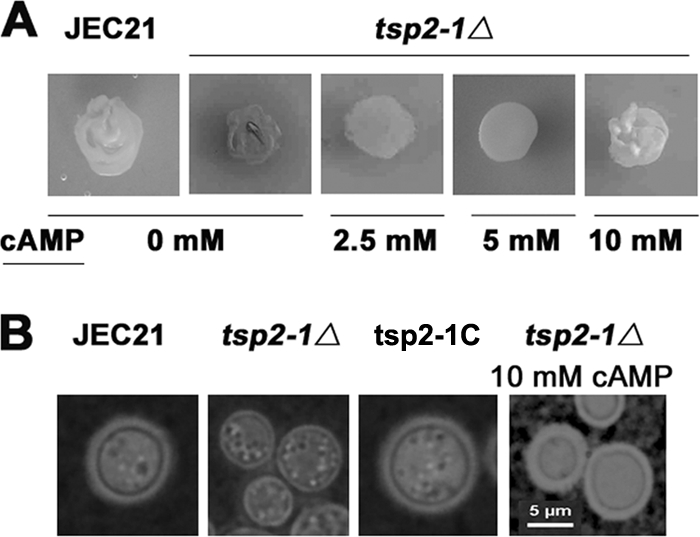
Restoration of melanin repression and capsule production by exogenous cAMP in tsp2-1Δ. (A) Addition of cAMP to the media restored the inhibition of melanin in the mutant tsp2-1Δ. Less pigmentation was achieved with increasing concentrations of cAMP added. Cells were incubated on 2% glucose Asn agar containing NE, with or without cAMP, at 30°C for 24 h (0, 2.5, 5, or 10 mM). (B) Likewise, 10 mM cAMP restored capsule production in tsp2-1Δ. The mutant strain tsp2-1Δ produced significantly less capsule (second image from the left). The complemented strain restored capsule production. Cells were incubated on 2% glucose Asn agar supplemented with 10 mM cAMP at 30°C for 5 days. Capsules were observed by India ink staining microscopy.
Capsule defect in tsp2-1Δ and its restoration by exogenous cAMP.
Since the Gα-cAMP-PKA pathway also controls capsule production in C. neoformans (1, 12), we examined the biosynthesis of capsule in the mutant strain tsp2-1Δ by India ink staining microscopy. As shown in Fig. 5B, capsule was significantly diminished in the tsp2-1Δ strain compared to that in the wild-type strain JEC21. A copy of wild-type TSP2-1 restored the production of capsule in the complemented strain (Fig. 5B and C). Again, 10 mM cAMP recovered capsule production in mutant strain tsp2-1Δ. This result supports the view that Tsp2-1 modulates capsule biosynthesis likely via the Gα-cAMP-PKA pathway.
Sensitivity of tsp2-1Δ to SDS, osmotic and oxidative/nitrosative stress, and heat.
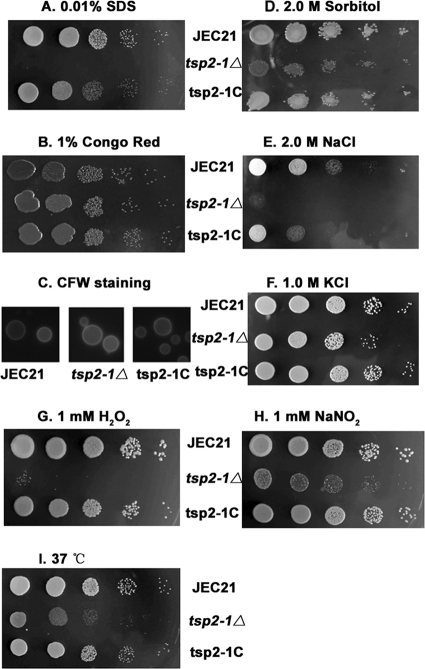
Considering that Tsp2-1 is a transmembrane protein, we examined the growth of tsp2-1Δ in the presence of several cell-disturbing agents. As shown in Fig. 6A, tsp2-1Δ displayed a severe sensitivity to 0.01% SDS. The complemented strain tsp2-1C had restored growth on SDS agar. This result suggests that defects were present in the cell membrane in tsp2-1Δ cells (14). All the strains had similar growth on 1% Congo red agar, indicating that the cell wall is intact in the mutant strain tsp2-1Δ (38). Cell morphology remained normal under examination by CFW microscopy (Fig. 6B and C). The mutant tsp2-1Δ exhibited an apparent sensitivity to 2 M sorbitol, 1 M KCl, and 2 M NaCl (Fig. 6D to F) and to 1 mM H2O2 and NaNO2 (Fig. 6G and H). We also found that TSP2-1 was required for growth at elevated temperatures, as tsp2-1Δ grew significantly more slowly than the wild type did at 37°C or higher temperatures (Fig. 6I). The wild-type strain JEC21 served as the control in these tests. The complemented strain restored the growth (at least partially restored in the case of 2 M NaCl) to the wild-type level under these stress conditions. The results demonstrate that TSP2-1 plays a crucial role in stress resistance in C. neoformans.
Fig 6.
Requirement of TSP2-1 for stress resistance. Yeast cells were serially diluted in 5 μl containing 105, 104, 103, 102, and 101 cells and separately spotted on 2% glucose agar supplement with testing agents at 30°C for 2 days. The mutant strain tsp2-1Δ was highly sensitive to SDS (0.01%) (A) but not to 1% Congo red (B). (C) CFW staining epifluorescence microscopy showed a normal cell wall chitin distribution pattern in the mutant. (D to F) tsp2-1Δ displayed hypersensitivity to osmotic stress from 2.0 M sorbitol, 2.0 M NaCl, or 1.0 M KCl. (G and H) tsp2-1Δ is sensitive to H2O2 (1 mM) and NaNO2 (1 mM). H2O2 was added to the medium at a temperature of 55 to 60°C after being autoclaved. (I) Disruption of TSP2-1 resulted in a growth defect at 37°C.
DISCUSSION
In this study, we identified a gene, TSP2-1, from the mutant LZM36 of C. neoformans which exhibited a phenotype of laccase derepression in 2% glucose (Fig. 1A and 3A and B). TSP2-1 was previously suggested, based on sequence homology, to be a Tsp2-type tetraspanin-encoding gene (17). The putative product of Tsp2-1 harbors characteristic features shared by members of the tetraspanin family (11, 17, 24). These include four transmembrane domains, a small extracellular loop (ECL1; 19 aa), a small intracellular loop (ICL; 4 aa), and a large extracellular loop (ECL2; 72 aa) with a typical cysteine-based motif [CCGY-x(12)-CY-x(5)-GCK] (Fig. 1C). One palmitoylation site is located at the end of TM4, proximal to the inner side of the membrane (a cysteine residue in Fig. 1C). Additionally, Tsp2-1 has long N-terminal (194 aa) and C-terminal cytoplasmic tails (153 aa) that were found only on Tsp2 tetraspanins (11, 17, 24). Tsp2 homologs are found in basidiomycetes by searching the genome project databases (17, 24). Tsp2-1 is the only tetraspanin in C. neoformans (17, 24). Although they share common features in three-dimensional structure, members from the different fungal tetraspanin families Pls1, Tsp2, and Tsp3 share little homology in peptide sequences (17, 24).
TSP2-1 is the first glucose-dependent repressor of laccase identified so far in C. neoformans. A significant finding for TSP2-1 is that the disruption of TSP2-1 activates LAC1 transcription (Fig. 3C and D). Considering that Tsp2-1 is a membrane protein, it is unlikely that it is involved in LAC1 transcription. We further found that exogenous 10 mM cAMP restored the inhibitory phenotype of laccase in the tsp2-1Δ mutant strain (Fig. 5A). Capsule defect in the tsp2-1Δ strain also was restored by cAMP in the mutant (Fig. 5B). It has been established in C. neoformans that the classic G protein-cAMP-PKA pathway controls capsule production and laccase induction in C. neoformans (1, 12, 22). Likewise, laccase and capsule defects caused by mutations in this pathway are restorable by cAMP (1, 12). In this respect, we speculate that Tsp2-1 interacts with the pathway at a step upstream of cAMP biosynthesis. It is intriguing to think that Tsp2-1 senses glucose on the plasma membrane and initiates a signal to the cAMP-PKA pathway. This speculation is supported by evidence from mammalian studies. Human tetraspanins provide molecular scaffolds for other proteins to form complexes and microdomains that are required for cellular signaling (16, 26, 39). A G protein-coupled receptor in human kidney cells, GPR56/TM7XN1, and the heterotrimeric G protein subunits, Gαq, Gα11, and Gβ, interplay specifically with tetraspanins CD9 and CD81 (27).
It should be noted that a canonic G protein-coupled receptor for glucose sensing (GPCR) is missing from C. neoformans (22). In Saccharomyces cerevisiae, a GPCR Gpr1, as a sugar receptor, senses glucose and sucrose to activate Gpa2, which further activates adenylyl cyclase for cAMP biosynthesis (25, 45, 46). In the genome of C. neoformans, seven GPCR-like proteins with structural similarity to Gpr1 have been reported (44). Unfortunately, none of them played a role in glucose sensing. The deletion of these GPCRs generates no obvious phenotype, except for Gpr4, which is an amino acid sensor. The finding of TSP2-1 in laccase repression may facilitate the finding of a glucose sensor in C. neoformans.
Other findings include roles of Tsp2-1 in stress resistance and in maintaining the integrity of the plasma membrane in C. neoformans. The mutant strain tsp2-1Δ displayed hypersensitivity to 0.01% SDS, suggesting a defect in the plasma membrane (14, 32). tsp2-1Δ was not sensitive to Congo red, which interacts specifically with β-d-glucan (37), implying that the cell wall integrity did not require TSP2-1 (14, 43). Consistently, no detectable changes were seen in chitin distribution in tsp2-1Δ by calcofluor white staining epifluorescence microscopy (Fig. 6C). The mutant tsp2-1Δ displayed growth retardation on plates supplemented with high concentrations of osmotic reagents, e.g., 2.0 M sorbitol, 2.0 M NaCl, and 1.0 M KCl (Fig. 6D, E, and F). Since the high-osmolarity glycerol (HOG) pathway controls the resistance to osmotic stress in C. neoformans (3), sensitivity to osmotic stress implies an interaction between Tsp2-1 and the HOG pathway.
Furthermore, tsp2-1Δ exhibited a hypersensitivity to 1 mM H2O2 or a nitrosative pressure and elevated temperature (37°C) (Fig. 6G, H, and I). Sensitivity to H2O2 indicates that catalase function was demolished in the mutant. Hypersensitivity to elevated temperature may be caused by various reasons. For example, mutations in the PKC pathway caused defects in the cell wall heat sensitivity of the yeast (14). A calcineurin gene, CNA1, is essential for heat resistance (35). The vacuolar H+-ATPase subunit VPH1 and the 5′-activated protein kinase subunit SNF1 also are required for growth at high temperatures (13, 47). The action of Tsp2-1 in heat resistance and oxidative/nitrosative stress remains to be illustrated.
In this work, we identified a tetraspanin-encoding gene, TSP2-1, from the basidiomycetous yeast C. neoformans, and we demonstrated that it is a glucose-dependent repressor of laccase. Tsp2-1 is also a key component for the integrity of the plasma membrane. The characterization of Tsp2-1 reveals a significant divergence in functions among fungal tetraspanins (8, 15, 24, 28). Questions are raised by this work, e.g., whether tetraspanin-dependent laccase repression is present in other fungi. The work may help to illustrate the molecular basis of laccase repression by glucose in C. neoformans.
ACKNOWLEDGMENTS
This work was supported in part by the Natural Science Foundation of China (NSFC grant 31170138) and a National Basic Research Program (973 Program 2007CB707801).
Footnotes
Published ahead of print 21 October 2011
REFERENCES
- 1. Alspaugh JA, et al. 2002. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asgher M, Bhatti HN, Ashraf M, Legge RL. 2008. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation 19:771–783 [DOI] [PubMed] [Google Scholar]
- 3. Bahn YS, Kojima K, Cox GM, Heitman J. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bajpai P, Anand A, Bajpai PK. 2006. Bleaching with lignin-oxidizing enzymes. Biotechnol. Annu. Rev. 12:349–378 [DOI] [PubMed] [Google Scholar]
- 5. Balan V, et al. 2008. Mushroom spent straw: a potential substrate for an ethanol-based biorefinery. J. Ind. Microbiol. Biotechnol. 35:293–301 [DOI] [PubMed] [Google Scholar]
- 6. Blackwood CB, Waldrop MP, Zak DR, Sinsabaugh RL. 2007. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environ. Microbiol. 9:1306–1316 [DOI] [PubMed] [Google Scholar]
- 7. Brijwani K, Rigdon A, Vadlani PV. 2010. Fungal laccases: production, function, and applications in food processing. Enzyme Res. 2010:149748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clergeot PH, et al. 2001. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. U. S. A. 98:6963–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins PJ, Dobson ADW. 1997. Regulation of laccase gene transcription in Trametes versicolor. Appl. Environ. Microbiol. 63:3444–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Couto SR, Toca-Herrera JL. 2007. Laccase production at reactor scale by filamentous fungi. Biotechnol. Adv. 25:558–569 [DOI] [PubMed] [Google Scholar]
- 11. DeSalle R, Mares R, Garcia-España A. 2010. Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol. Phylogenet. Evol. 56:486–491 [DOI] [PubMed] [Google Scholar]
- 12. D'Souza CA, et al. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erickson T, et al. 2001. Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Mol. Microbiol. 42:1121–1131 [DOI] [PubMed] [Google Scholar]
- 14. Gerik KJ, et al. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393–408 [DOI] [PubMed] [Google Scholar]
- 15. Gourgues M, Brunet-Simon A, Lebrun MH, Levis C. 2004. The tetraspanin BcPls1 is required for appressorium-mediated penetration of Botrytis cinerea into host plant leaves. Mol. Microbiol. 51:619–629 [DOI] [PubMed] [Google Scholar]
- 16. Hemler ME. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801–811 [DOI] [PubMed] [Google Scholar]
- 17. Huang S, et al. 2005. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86:674–684 [DOI] [PubMed] [Google Scholar]
- 18. Idnurm A, et al. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753–764 [DOI] [PubMed] [Google Scholar]
- 19. Jiang N, et al. 2011. Regulation of copper homeostasis by Cuf1 associates with its subcellular localization in the pathogenic yeast Cryptococcus neoformans H99. FEMS Yeast Res. doi:10.1111/j.1567-1364.2011.00733.x. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 20. Jiang N, et al. 2009. Negative roles of a novel nitrogen metabolite repression-related gene, TAR1, in laccase production and nitrate utilization by the basidiomycete Cryptococcus neoformans. Appl. Environ. Microbiol. 75:6777–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kojima Y, et al. 1990. Cloning, sequence analysis, and expression of lignolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J. Biol. Chem. 265:15224–15230 [PubMed] [Google Scholar]
- 22. Kozubowski L, Lee SC, Heitman J. 2009. Signaling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 11:370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kunamneni A, Plou FJ, Ballesteros A, Alcalde M. 2008. Laccases and their applications: a patent review. Recent Pat. Biotechnol. 2:10–24 [DOI] [PubMed] [Google Scholar]
- 24. Lambou K, et al. 2008. Fungi have three tetraspanin families with distinct functions. BMC Genomics 9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16:293–299 [DOI] [PubMed] [Google Scholar]
- 26. Levy S, Shoham T. 2005. Protein-protein interactions in the tetraspanin web. Physiology 20:218–224 [DOI] [PubMed] [Google Scholar]
- 27. Little KD, Hemler ME, Stipp CS. 2004. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol. Biol. Cell 15:2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malagnac F, et al. 2008. Convergent evolution of morphogenetic processes in fungi: role of tetraspanins and NADPH oxidases 2 in plant pathogens and saprobes. Commun. Integr. Biol. 1:180–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martínez AT, et al. 2005. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 8:195–204 [PubMed] [Google Scholar]
- 30. Mayer AM, Staples RC. 2002. Laccase: new functions for an old enzyme. Phytochemistry 60:551–565 [DOI] [PubMed] [Google Scholar]
- 31. Mikolasch A, Schauer F. 2009. Fungal laccases as tools for the synthesis of new hybrid molecules and biomaterials. Appl. Microbiol. Biotechnol. 82:605–624 [DOI] [PubMed] [Google Scholar]
- 32. Missall TA, Moran JM, Corbett JA, Lodge JK. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsal. Eukaryot. Cell 4:202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore TD, Edman JC. 1993. The alpha-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nurudeen TA, Ahearn DG. 1979. Regulation of melanin production by Cryptococcus neoformans. J. Clin. Microbiol. 10:724–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Odom A, et al. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Riva S. 2006. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 24:219–226 [DOI] [PubMed] [Google Scholar]
- 37. Rodríguez Couto S, Toca Herrera JL. 2006. Industrial and biotechnological applications of laccases: a review. Biotechnol. Adv. 24:500–513 [DOI] [PubMed] [Google Scholar]
- 38. Roncero C, Duran A. 1985. Effect of calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singethan K, Scherner-Schaulies J. 2008. Tetraspanins. Comm. Int. Biol. 1:11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walton FJ, Idnurm A, Heitman J. 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 57:1381–1396 [DOI] [PubMed] [Google Scholar]
- 41. Wesenberg D, Kyriakides I, Agathos SN. 2003. White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol. Adv. 22:161–187 [DOI] [PubMed] [Google Scholar]
- 42. Williamson PR. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wood PJ, Fulcher RG. 1983. Dye interactions. A basis for specific detection and histochemistry of polysaccharides. J. Histochem. Cytochem. 31:823–826 [DOI] [PubMed] [Google Scholar]
- 44. Xue C, Bahn YS, Cox GM, Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xue C, Hsueh YP, Chen L, Heitman J. 2008. The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans. Mol. Microbiol. 70:379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xue Y, Batlle M, Hirsch JP. 1998. GPR1 encodes a putative G protein-coupled receptor that associaTsp2-1 with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 17:1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang J, et al. 2010. Regulation of virulence factors, carbon utilization and virulence by SNF1 in Cryptococcus neoformans JEC21 and divergent actions of SNF1 between cryptococcal strains. Fungal Genet. Biol. 12:994–1000 [DOI] [PubMed] [Google Scholar]
- 48. Yoshida H. 1883. Chemistry of lacquer (urushi). J. Chem. Soc. 43:472–486 [Google Scholar]
- 49. Zhu X, Gibbons J, Garcia-Rovera J, Casadevall A, Williamson PR. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 69:5589–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu X, Williamson PR. 2003. A CLC-type chloride channel gene is required for laccase activity and virulence in Cryptococcus neoformans. Mol. Microbiol. 50:1271–1281 [DOI] [PubMed] [Google Scholar]
- 51. Zhu X, Williamson PR. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 5:1–10 [DOI] [PubMed] [Google Scholar]