Abstract
Experimental evolution via continuous culture is a powerful approach to the alteration of complex phenotypes, such as optimal/maximal growth temperatures. The benefit of this approach is that phenotypic selection is tied to growth rate, allowing the production of optimized strains. Herein, we demonstrate the use of a recently described long-term culture apparatus called the Evolugator for the generation of a thermophilic descendant from a mesophilic ancestor (Escherichia coli MG1655). In addition, we used whole-genome sequencing of sequentially isolated strains throughout the thermal adaptation process to characterize the evolutionary history of the resultant genotype, identifying 31 genetic alterations that may contribute to thermotolerance, although some of these mutations may be adaptive for off-target environmental parameters, such as rich medium. We undertook preliminary phenotypic analysis of mutations identified in the glpF and fabA genes. Deletion of glpF in a mesophilic wild-type background conferred significantly improved growth rates in the 43-to-48°C temperature range and altered optimal growth temperature from 37°C to 43°C. In addition, transforming our evolved thermotolerant strain (EVG1064) with a wild-type allele of glpF reduced fitness at high temperatures. On the other hand, the mutation in fabA predictably increased the degree of saturation in membrane lipids, which is a known adaptation to elevated temperature. However, transforming EVG1064 with a wild-type fabA allele had only modest effects on fitness at intermediate temperatures. The Evolugator is fully automated and demonstrates the potential to accelerate the selection for complex traits by experimental evolution and significantly decrease development time for new industrial strains.
INTRODUCTION
Many industrial processes that rely on microbial biocatalysts are limited by the absence of microbes that can efficiently catalyze the appropriate chemistry (i.e., phenotype) under the industrial conditions that are required for process optimization. A classic example of this problem is thermotolerance, where the efficiency of biocatalysis, and therefore the economic viability of a particular process, is often hindered by the inability of the necessary microbe to thrive at the temperatures encountered during process scale-up (47). Consequently, there is significant interest in producing microbes whose thermal growth parameters match those needed for particular industrial applications. In addition, there is also interest in studying the acquisition of thermotolerance from a more fundamental perspective. Based on the theory of the last universal common ancestor, thermophiles either evolved from mesophiles or vice versa (13). Thus, it is of considerable importance for evolutionary biology to study the mechanisms by which microbes can adapt to different thermal environments.
The most common approach to altering the thermal growth parameters of microbes is genetic engineering, and a panoply of molecular biological tools have been brought to bear on the problem (1, 8, 16, 22, 34, 38, 55, 62). The most success has been achieved using advanced high-throughput recombinant engineering techniques, because they either sample many random genetic variations (some of which may have originated in thermophiles) or affect highly pleiotropic genes and thus can access the genetic or biochemical diversity required to significantly alter complex traits, such as optimal growth temperature (Topt) or maximal growth temperature (Tmax). Moreover, these techniques do not require a priori knowledge about how to alter a particular trait, although they do require a certain degree of compatibility (codon usage, mRNA stability, protein folding, etc.) between the inserted functionality and the host that is not guaranteed. However, all genetic engineering methods share the same inherent problem: while they can alter a particular phenotype, they cannot simultaneously select for the most robust strain with that phenotype. Innovative screens, such as selection for colony size, have been developed that lessen the impact of this limitation, but the problem persists nevertheless (35, 61). More critically, engineered alterations in one phenotype frequently come at the expense of other critical phenotypes, such as growth rate (39). Thus, engineered strains are often phenotypically competent but growth attenuated. Since, from an economic perspective, yield in time can be as important as yield in space, the decreased growth rates of engineered strains can eliminate potential gains made through engineering, making them less interesting from a practical perspective. Finally, genetic engineering is not always a viable option in industry. For example, the means to genetically modify a microbe of interest may not already exist (33, 42), requiring significant time and resources for developing the necessary molecular biological tools prior to embarking on a strain development program. Finally, the microbe of interest may be intended for release into or have a high potential for escape into the wild, and for-profit entities—whose goal is to deploy a microbe and eventually earn a profit—must acknowledge the reality of proposing the use of genetically modified organisms (GMOs) for such purposes. Setting aside the debate surrounding the risks associated with introducing GMOs versus organisms modified by more natural means (e.g., breeding, natural selection), GMOs have been singled out in the legal sense, and the reality is that the use of GMOs may preclude deployment into certain lucrative markets for regulatory reasons. This must also be considered prior to planning a strain development program.
An alternative approach for the modification of complex phenotypes, such as Topt or Tmax, employs experimental evolution (29), in which cells are continuously maintained in an actively growing state under the desired environmental conditions such that adaptive variants emerge with new genotypes that allow robust growth under the culture conditions. This methodology is ideal for altering complex phenotypes, because no assumptions need to be made about which or how many mutations will ultimately be successful. It is also ideal for industrial strain development, since no complex molecular biological tools are necessary—just the ability to culture the cells in question. More importantly, the output strain is simultaneously optimized for growth under the conditions to which it has been adapted, which can be tailored to mimic industrial conditions. Finally, if natural mutation rate is the only source for genetic variation, the resulting strains are naturally occurring genetic variants of the parent strain and are, therefore, not considered GMOs.
Experimental evolution is vastly underutilized for industrial strain development (7), partly because the results are essentially irreproducible without genomic data. If the evolved strains are subsequently misplaced or change phenotype, then the information regarding the nature of the adaptation is lost. More importantly for industry, without sequence data it is more difficult to protect intellectual property. However, the advent of new high-throughput sequencing methods has changed this dynamic (3, 5, 25), and experimental evolution is increasingly being recognized as a potent method for altering the phenotypes of industrially important microbes (11, 17).
There is a considerable body of literature describing the elegant use of experimental evolution for thermal adaptation (9, 10, 29, 40, 45, 46, 54). However, almost all of this literature consists of adaptation either to transient thermal stress or to constant, but stressful, temperatures rather than to steadily increasing temperatures. In one recent example of the latter, a thermophile was rendered mesophilic via the replacement of an essential enzyme with a homolog from a mesophile (40). Thermophily was then reacquired through gradually raising the temperature in a serial batch process. However, while this study elegantly showed that serial batch culture could be used to improve the thermal properties of individual enzymes, no attempt was made to alter the overall thermal properties of the original organism (although alteration in individual enzymes could conceivably have a significant impact on global thermal properties). In a more salient example, Rudolph et al. recently reported the use of experimental evolution over the course of 2 years of traditional serial batch culture to adapt Escherichia coli K-12 MG1655 to steadily increasing temperatures (49), resulting in a thermotolerant strain that could grow at 48.5°C, well above the Tmax of the parent strain (58). However, while they reported proteomic characterization of the resulting strain, they did not perform any sequencing to characterize the adaptive genotype. Thus, many questions remain about how E. coli can be made to adapt to temperatures that are lethal to the wild-type (WT) strain.
Concurrent with the work of Rudolph et al., we used a recently described type of long-term culture bioreactor called the Evolugator to perform a nearly identical experiment. Thus, the publication by Rudolph et al. provided the ideal opportunity to compare results and methodologies. Our evolved thermotolerant strain of E. coli grows robustly at temperatures as high as 49.7°C in the Evolugator growth chamber. Moreover, the Topt for this strain has increased from 37°C to 46°C, making it a facultative thermophile by widely accepted definitions of the term (6, 30). More importantly, the results presented herein are accompanied by whole-genome sequencing to map the pathway of the evolutionary adaptation, and 31 potentially adaptive mutations were identified, at least two of which appear to affect thermal properties in isolation. More importantly, these results were achieved in only 8 months by using the Evolugator, less than half the time it took Rudolph et al. by using traditional serial batch culture. The Evolugator is fully automated, is resistant to issues of contamination, and counterselects against wall growth, making it ideal for long-term experimental evolution (11, 12). From a practical perspective, the ability to perform robust experimental evolution in the Evolugator could potentially reduce the time needed to develop strains with complex phenotypes. From a more fundamental perspective, our data suggest that the pathway to thermal adaptation in this study differed significantly from that found by Rudolph et al. and that multiple viable mechanisms for the acquisition of thermophily may exist.
MATERIALS AND METHODS
Strains and culture conditions.
The input strain MG1655 was obtained from the E. coli Genetic Stock Center (Yale, CT). The wild-type BW25113 and the glpF deletion mutant (ΔglpF mutant) were obtained from the Keio collection (4). fabA, glpF, and folE expression plasmids were obtained from the ASKA library (24) and transformed into our experimentally evolved thermotolerant strains (EVG1058 and EVG1064) using standard methods. LB and M9 minimal media were made according to Molecular Cloning: A Laboratory Manual (50). Plates containing minimal M9 agar were supplemented with dextrose, glycerol, or maltose as carbon sources at final concentrations of 0.4% (wt/vol). In order to grow on M9 minimal medium, evolved thermotolerant strain EVG1064 needed the following supplements based on a formulation from reference 64: 500 μM l-phenylalanine, 250 μM l-tyrosine, 200 μM l-tryptophan, 6 μM p-aminobenzoate, 6 μM p-hydroxybenzoate, 50 μM 2,3-dihydroxybenzoate, 10 μM pyridoxal, and 100 μM glycolaldehyde. Growth on various solid media was determined by restreaking single colonies from LB agar plates grown at 37°C onto plates that were preequilibrated at either 30°C, 37°C, 43°C, 46°C, or 48.5°C and incubated in a UVP SI-950 high-thermal accuracy incubator. Plates at 30°C and 37°C were incubated overnight, and plates incubated at higher temperatures were incubated for 24 to 36 h.
For liquid growth curves, overnight cultures were grown in LB at 37°C (MG1655) or 47°C (EVG1064), normalized for optical density, and reinoculated into medium that had been preequilibrated at various temperatures. Growth was monitored by measuring optical density at 600 nm (OD600). For gene complementation studies using plasmids from the ASKA collection, 24 μg/ml chloramphenicol was added to the LB medium to select for plasmid retention. Mean generation times were determined by plotting ln OD600 versus time and measuring the slope of the line during logarithmic phase. Generation time is equal to ln 2/slope. Growth curves were performed in triplicate in a shaking incubator set to 180 rpm. Mean generation times for each strain were calculated and plotted versus temperature. Error bars indicate ±1 standard deviation.
For thermal killing assays, overnight cultures were grown at either 37°C (MG1655) or 47°C (EVG1064). A total of 100 μl of each culture was pipetted into 6 PCR tubes. The tubes were placed in a Bio-Rad gradient iCycler thermocycler and incubated for 30 min using a temperature gradient with 6 steps from 48°C to 60°C. A total of 5 μl of 1×, 0.1×, and 0.01× dilutions were then spotted onto LB plates and incubated at 37°C to recover. Due to the possibility that EVG1064 suffers from antagonistic pleiotropy when grown at lower temperatures, the same experiment was repeated with the exception that EVG1064 was allowed to recover at 48.5°C instead of 37°C. The results were the same regardless of recovery temperature, and recovery at 48.5°C is shown in Fig. 2D. At 48°C, 36 to 48 h was required to see growth on plates.
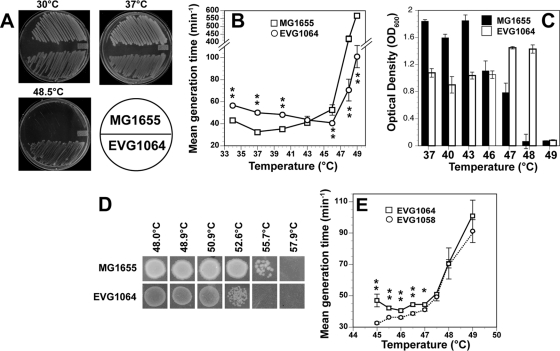
Fig 2.
Thermotolerance of wild type, EVG1064, and EVG1058. (A) Growth of MG1655 and EVG1064 E. coli strains on LB plates at 30°C, 37°C, and 48.5°C; (B) mean generation times of MG1655 and EVG1064 in liquid LB medium plotted as a function of temperature; (C) final cell densities, as measured by OD600, at 24 hours for MG1655 and EVG1064 grown in liquid LB medium at various temperatures; (D) resistance to 30-min exposures to elevated temperatures for MG1655 and EVG1064 as measured by growth on LB plates; (E) mean generation times of EVG1058 and EVG1064 in liquid LB medium plotted as a function of temperature. For all panels, error bars indicate ±1 standard deviation. For panels B and E, asterisks denote temperatures at which the growth rates of the two compared strains are significantly different using a 2-tailed, type 2 t test. **, P < 0.005; *, P < 0.05.
Experimental evolution.
The details of how the Evolugator works have been extensively described elsewhere (11, 12). Briefly, the traditional culture chamber has been replaced with a length of transparent flexible tubing. The tubing is filled with medium, autoclaved, loaded onto the Evolugator, and divided into sections by gates that clamp the tubing and prevent the flow of medium and cells from one section to another. The “growth chamber” is the section of tubing in the center of the machine (10-ml total volume, 7-ml liquid volume with a gas bubble) that is fed through two turbidimeters that, by virtue of the transparent nature of the tubing, can read optical density in the growth chamber in real time. Cells are inoculated into the growth chamber via injection through the tubing with a sterile syringe. Upon dilution, a belt moves the tubing and gates in unison half the length of the growth chamber, forcing the medium inside to move by peristaltic action. The new growth chamber consists of half of the tubing from the old growth chamber and half new tubing. By replacing the entire growth chamber wall every two dilutions, the Evolugator counterselects against wall growth. In addition, half of the saturated medium in the old growth chamber is replaced with fresh medium from upstream tubing. The other half of the tubing and saturated medium from the old growth chamber (5-ml total volume) is separated from the new growth chamber by a gate and is now in what is referred to as the “sampling chamber.” Because the sampling chamber is physically separated from the new growth chamber by a gate, a sample of the culture can be extracted from the sampling chamber by syringe without risking contamination of the new growth chamber. A gas bubble can be included in the growth chamber as well as upstream chambers so that, upon dilution, a fresh bubble of gas (in this case air) is brought into the growth chamber with every dilution. Therefore, the number of possible dilutions in a particular experiment is only limited by the length of tubing. Since the tubing is sterilized and sealed at the beginning, there is no contact with the outside until the tubing is used up and must be replaced. A computer controls the belt, and dilution is automatically initiated when the culture reaches the desired optical density or user initiated when the severity of selective pressure results in extremely poor growth of the culture. The entire growth chamber, belt and all, is encased in an environmentally controlled box in which temperature is monitored using a PT100 probe (IEC/Din class A) and regulated via a proportional-integral-derivative controller (West P6100). Agitation is achieved by rocking the entire environmentally controlled box, causing the bubble to move back and forth in the growth chamber. For this experiment, the Evolugator culture chamber was filled with LB medium and inoculated with a preculture of MG1655 grown in LB overnight. The temperature of the culture chamber was increased gradually from the permissive temperature of 44°C to 49.7°C. Automated dilutions—initiated as often as every 35 to 40 min—sufficed when adapting cells to nonlethal temperatures or when growth of the cultures was generally stable. User-initiated dilutions were often necessary when ramping up the temperature or when we encountered thermal barriers or temperatures at which small increases in temperature (sometimes as small as 0.1°C) resulted in profound reductions in growth rate or even negative growth in the chamber. To pass through these barriers, we often had to reduce temperature to allow the cultures to recover and then periodically challenge them with short bursts of heat shock.
Genetic analysis.
Plugs for pulsed-field gel electrophoresis (PFGE) were prepared exactly as published (43) and restriction fragment length polymorphism analysis (RFLP) was performed using XbaI. Electrophoresis was performed using a CHEF Mapper (Bio-Rad) apparatus, as follows: initial switch time value of 2.16 s, final switch time of 54.17 s at a gradient of 6 V/cm, and an included angle of 120°. Electrophoresis was performed for 19 h. Subsequent to electrophoresis, the gel was stained in ethidium bromide solution (40 μg/ml [wt/vol]) for 30 min and then destained three times for 15 min each in deionized water.
Genomes were sequenced using the Illumina sequencing platform. Briefly, genomic DNA preparations were made using the DNEasy kit (Qiagen). Genome libraries of each strain were generated using the genomic DNA sample prep kit as described by the manufacturer's directions. Sequencing was performed in a 36-cycle single-end run (Core Facility, Oregon State University). Sequence coverage for each strain was as follows: MG1655, 86×; EVG1031, 52×; EVG1041, 38×; EVG1058, 38×; and EVG1064, 48×. Single nucleotide polymorphisms (SNPs) were identified using both CLC genomics workbench version 3.6.5 (CLC Bio, MA) and Maq software (27). SNPs were independently verified by Sanger sequencing (University of Florida Core Sequencing Facility). Primers for Sanger sequencing are listed in Table 1. Genome sequences are available upon request.
Table 1.
Oligonucleotides used to amplify specific regions from the relevant strain's chromosome for the purpose of Sanger sequencing
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| rpoD_fwd | ATGGAGCAAAACCCGCAGTC |
| rpoD_rev | TTAATCGTCCAGGAAGCTAC |
| ylbE_fwd | ATGTTTACATCAGTGGCGCA |
| ylbE_rev | TCACTTCCCCTGCTCCAGTA |
| kdpD_fwd | ATGAATAACGAACCCTTACG |
| kdpD_rev | TCACATATCCTCATGAAATT |
| ybhN_fwd | ATGAGTAAATCACACCCGCG |
| ybhN_rev | TCACATCGCCGCTTCATTTT |
| rpsA_fwd | ATGACTGAATCTTTTGCTCA |
| rpsA_rev | TTACTCGCCTTTAGCTGCTT |
| pncB_fwd | ATGACACAATTCGCTTCTCC |
| pncB_rev | TTAACTGGCTTTTTTAATAT |
| fabA_fwd | ATGGTAGATAAACGCGAATC |
| fabA_rev | TCAGAAGGCAGACGTATCCT |
| yddB_fwd | ATGAAGCGAGTTCTTATTCC |
| yddB_rev | TTAAAATTTCATGCTGACAT |
| dgsA_fwd | GTGGTTGCTGAAAACCAGCC |
| dgsA_rev | TTAACCCTGCAACAGACGAA |
| pykF_fwd | ATGAAAAAGACCAAAATTGT |
| pykF_rev | TTACAGGACGTGAACAGATG |
| yejM_fwd | ATGGTAACTCATCGTCAGCG |
| yejM_rev | TCAGTTAGCGATAAAACGCT |
| tktB_fwd | ATGTCCCGAAAAGACCTTGC |
| tktB_rev | TCAGGCACCTTTCACTCCCA |
| mreD_fwd | GTGGCGAGCTATCGTAGCCA |
| mreD_rev | TTATTGCACTGCAAACTGCT |
| rpsJ_fwd | ATGCAGAACCAAAGAATCCG |
| rpsJ_rev | TTAACCCAGGCTGATCTGCA |
| perR_fwd | ATGAAGCTCTTAGCAAAAGC |
| perR_rev | TCAACGAATTTTACCCAGAT |
| malQ_fwd | ATGGAAAGCAAACGTCTGGA |
| malQ_rev | CTACTTCTTCTTCGCTGCAG |
| malT_fwd | ATGCTGATTCCGTCAAAACT |
| malT_rev | TTACACGCCGTACCCCATCA |
| yhhZ_fwd | ATGAGTAATATTGTTTACCT |
| yhhZ_rev | TCATTTTGTGTGGTCCATAA |
| malS_fwd | ATGAAACTCGCCGCCTGTTT |
| malS_rev | TTACTGTTGCCCTGCCCAGA |
| spoT_fwd | TTGTATCTGTTTGAAAGCCT |
| spoT_rev | TTAATTTCGGTTTCGGGTGA |
| wzzE_fwd | ATGACACAACCAATGCCTGG |
| wzzE_rev | CTATTTCGAGCAACGGCGGG |
| rffT_fwd | ATGACTGTACTGATTCACGT |
| rffT_rev | TCATGCGACCTCCCTGGCGG |
| glpF_fwd | ATGAGTTAAACATCAACCTT |
| glpF_rev | TTACAGCGAAGCTTTTTGTT |
| treB_fwd | ATGATGAGCAAAATAAACCA |
| treB_rev | TTAAACAATGTCCAGCGTGC |
| idi_fwd | ATGCAAACGGAACACGTCAT |
| idi_rev | TTATTTAAGCTGGGTAAATG |
| yidE_upstream_fwd | CCAATACCTAATCCTATGCC |
| yidE_upstream_rev | TCGTAAACGGTTTACTGCAT |
| ppiC_upstream_fwd | AGCTTGCCGAAATCGGCCCC |
| ppiC upstream_rev | CTTACAGAGGGTATCTTAAT |
| yegTfbaB_upstream_fwd | TCATGTCCGGGGAGATAAAG |
| yegTfbaB_upstream_rev | AAACCGCTTTTACTTAACCA |
| rydC_upstream_fwd | CGCATGATGCCGCGTAAACG |
| rydC_upstream_rev | TGTGAGATCCCCCCTTTCGA |
| yajD_upstream_fwd | TGGCATCTGCGTTGGCTCTG |
| yajD_upstream_rev | AACTCGCGGGAACAGCGACC |
| gltP_upstream_fwd | TATGGCAAAAAGTGATGGAT |
| gltP_upstream_rev | TCGCGGCTGTCGCTATGGTA |
| yqjF_upstream_fwd | ATCCTAATATGCTGGTCCGC |
| yqjF_upstream_rev | GTACCCGCGTAGCCAGTAAT |
Upon sequencing our strain of Escherichia coli K-12 MG1655, we identified an A-to-G polymorphism at position 547694 of the genome that differs from the published MG1655 genome sequence. This polymorphism results in a synonymous substitution at position 114 of the ylbE gene and is retained in all strains, including EVG1064.
Fatty acid analysis.
Fatty acid methyl ester analysis (FAME) was performed at the Bacterial Identification & Fatty Acid Analysis Lab in the Plant Pathology Department at the University of Florida. Briefly, strains were streaked onto agar plates and grown at 37°C or 48°C. Following 24 h of growth, the plates were provided to the laboratory, where transesterification and analysis by GC were performed using the Sherlock System developed by MIDI, Inc. Only fatty acids that comprise >1% of the total are included in Table 5; however, all saturated and unsaturated fatty acids were included in the calculation of the saturated/unsaturated ratio. Summed feature 3 in the chromatogram is assigned to the abundant fatty acid C16:1 Δ9c, although the exotic fatty acid C15:0 iso 2OH coelutes and may also contribute. Summed feature 2 in the chromatogram is assigned to C14:0 3OH, which is an abundant component of E. coli lipid A, although the exotic fatty acid C16:1 iso I coelutes and may contribute. Summed feature 4 in the chromatogram is assigned to C17:1 Iso I, although C17:1 anteiso B/i I coelutes. This experiment was performed in triplicate, and values are reported as averages ±1 standard deviation. The significance of the difference between selected averages was determined with 2-tailed, unmatched t tests.
Table 5.
Analysis of fatty acids from EVG1058 and EVG1064a
| Fatty acid | % total fatty acids ± SD |
|||||
|---|---|---|---|---|---|---|
| 37°C |
48°C |
|||||
| EVG1058 | EVG1064 | EVG1058 | EVG1064 | EVG1064 fabA strain | EVG1064 folE strain | |
| Saturated | ||||||
| C12:0 | 4.6 ± 0.3 | 4.4 ± 0.2 | 6.3 ± 0.6**‡ | 5.0 ± 0.4**‡ | 4.3 ± 0.0*§ | 3.8 ± 0.1*§ |
| C14:0 | 9.4 ± 1.6 | 9.8 ± 1.4 | 11.4 ± 1.0 | 12.2 ± 0.1 | 8.3 ± 0.4**§ | 9.2 ± 0.2**§ |
| C16:0 | 30.1 ± 1.3 | 30.8 ± 2.4 | 36.3 ± 2.8**‡ | 41.6 ± 1.3**‡ | 39.8 ± 2.4**§ | 44.6 ± 1.1**§ |
| C17:0 | 1.0 ± 0.9 | 1.0 ± 1.0 | 0.6 ± 0.3 | 0.4 ± 0.1 | ND | ND |
| C18:0 | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.8 ± 0.2 | 0.9 ± 0.1 | 1.2 ± 0.1 | 1.5 ± 0.2 |
| Unsaturated | ||||||
| C16:1 Δ9 | 13.8 ± 2.6 | 11.3 ± 5.2 | 16.1 ± 0.8**‡ | 13.4 ± 1.1**‡ | 8.6 ± 1.2 | 6.2 ± 1.2 |
| C16:1 Δ11 | 0.21 ± 0.03**† | 0.15 ± 0.02**† | ND | ND | 0.5 ± 0.0 | ND |
| C18:1 Δ11 | 11.5 ± 0.8 | 9.1 ± 1.8 | 6.7 ± 2.4**‡ | 2.8 ± 0.3**‡ | 6.9 ± 1.5**§ | 2.1 ± 0.5**§ |
| Other | ||||||
| 3-OH-C14:0 | 9.7 ± 0.4 | 9.3 ± 0.9 | 13.4 ± 2.5 | 11.3 ± 1.5 | 10.6 ± 0.3 | 9.8 ± 0.6 |
| C17:0 cyc | 15.6 ± 2.6 | 18.4 ± 4.8 | 6.8 ± 2.3 | 11.1 ± 1.2 | 14.4 ± 0.3*§ | 17.2 ± 0.5*§ |
| C17:1 Iso I | ND | ND | ND | ND | 2.7 ± 0.8 | 3.4 ± 1.1 |
| C19:0 cyc ω8c | 1.2 ± 0.5 | 1.5 ± 1.0 | 0.1 ± 0.2 | 0.2 ± 0.2 | 1.3 ± 0.2 | 1.0 ± 0.1 |
| Saturated/unsaturated | 1.8 ± 0.3 | 2.4 ± 0.8 | 2.5 ± 0.3**‡ | 3.7 ± 0.3**‡ | 3.4 ± 0.7**§ | 7.4 ± 1.6**§ |
| Saturated/unsaturated + cyc | 1.1 ± 0.0 | 1.2 ± 0.1 | 1.9 ± 0.2**‡ | 2.2 ± 0.02**‡ | 2.2 ± 0.2**§ | 2.6 ± 0.2**§ |
The same symbols in a row (†, ‡, §) indicate paired values that are significantly different when compared using t tests. Asterisks indicate P values: *, P < 0.005; **, P ≤ 0.05. Error bars indicate ±1 standard deviation. cyc, cyclopropane fatty acid; Δ, position of the double bond or cyclopropane ring relative to the carboxyl group; 3OH, β-hydroxyl group; Iso I, a terminally branched-chain fatty acid; ND, not detected.
RESULTS
Strain acquisition.
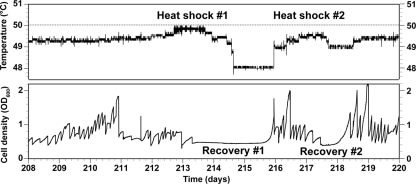
E. coli K-12 MG1655 was inoculated into the growth chamber containing LB, and the temperature was slowly increased from 44°C to 49.7°C over the course of 8 months of automated cycles of growth and dilution. It would be impractical to graphically depict the growth and chamber temperature versus time for the entire 8-month experiment. However, Fig. 1 shows a graph of the temperature and optical density (OD600) as a function of time for a representative 12-day period, between days 208 and 220 of the experiment. Occasionally, upon an increase in temperature, optical density either did not increase or decreased, indicating that variants with adaptive mutations had not yet arisen in the population. Under these circumstances, the temperature (and, consequently, selective pressure) was decreased to allow the culture to recover. Such an event is depicted in Fig. 1, where the temperature was increased, resulting in a decrease in the fitness of cells that was not reversed until the temperature was reduced. Such thermal barriers were also described by Rudolph et al (49). To pass through such barriers, cultures were often exposed to short bursts of heat shock to increase the likelihood that adaptive mutations might arise before continuing the increase in culture temperature. Indeed, two examples of the use of heat shock are shown in Fig. 1. The average temperatures of the culture chamber for the 8 days prior (days 200 to 208) and subsequent (days 220 to 228) to the time period in Fig. 1 were 49.1°C ± 0.6 and 49.5°C ± 0.1, respectively (P = 1.3E−285 for a 2-tailed, type 2 t test), indicating that the cells had adapted to a higher set temperature due to the events. Samples were periodically taken during the adaptation process and cryogenically stored (−80°C). Twice, an increase in temperature killed the culture, and the last sampled strain was reinoculated into the growth chamber at or below the Tmax of the sampled strain.
Fig 1.
Graphical depiction of adaptation process. The top panel shows the temperature of the Evolugator culture chamber as a function of time (days) for a short 12-day window of the experiment, which lasted a total of 234 days. The bottom panel shows the optical density of the culture in the growth chamber during the same time period. Two examples of heat shock are shown in this time period. During these heat shocks, the temperature is increased to stress the cells. There is a subsequent decrease in the optical density of the cultures due to heat shock. The temperature is then rapidly decreased, allowing the cultures to recover and optical density to increase again. Temperature is then increased to select for mutants that might have arisen that are more thermotolerant.
Four of the strains that were sequentially sampled from the Evolugator at consecutively higher temperatures were chosen for further characterization (named EVG1031, EVG1041, EVG1058, and EVG1064). The evolved strains were restreaked from collection at 37°C, and their thermal properties were analyzed outside the Evolugator culture chamber. Table 2 shows the growth of each strain on LB plates at various temperatures. MG1655 cannot grow on LB plates at temperatures at or exceeding 46°C. This is consistent with reports that MG1655 grows very poorly in liquid LB at 46°C (58). Strain EVG1031 was sampled on day 77 when consistent and robust growth was seen in the Evolugator culture chamber at 46.9°C. However, this strain did not prove to be more significantly thermotolerant than MG1655 on LB plates (Table 2). On day 122, when growth in the Evolugator culture chamber had reached 49.3°C, we sampled EVG1041, which could grow on LB plates at 46°C but was not viable at 48.5°C. EVG1058 was taken from the culture chamber on day 190 when consistent growth was seen at 49.6°C. This strain was capable of growing on LB plates at 48.5°C but not at 49°C. The same was true of the last strain to be sampled, EVG1064, which was taken from the machine on day 234 when the internal temperature had reached 49.7°C (Table 2, Fig. 2A). EVG1064 was capable of robust growth in liquid LB at 48°C, while MG1655 was not (Fig. 2B). It is notable that EVG1064 grows to a much lower cell density than MG1655 at 37°C (Fig. 2C). The significance of this will be discussed later. Mean generation times for MG1655 and EVG1064 were determined in batch LB culture at various temperatures to determine Topt (Fig. 2B and Fig. 2E). As expected based on previous data (21, 53), the Topt for the wild type is approximately 37°C. On the other hand, the Topt for EVG1064 has clearly increased to 46°C. The determination of Tmax in liquid culture is a more complicated endeavor. At issue is the fact that cells can grow, albeit at significantly lower growth rates, for brief periods of time, even at lethal temperatures. Thus, growth rates can be calculated even at temperatures where the cells do not achieve appreciable cell densities. This is how a growth rate for MG1655 was obtained at 49°C, which is a lethal temperature. Thus, it is difficult to rely on growth rates to determine Tmax. Instead, we relied on cell density at 24 h to determine Tmax. This allowed us to determine that MG1655 is incapable of growth at temperatures of >47°C, while EVG1064 is affected only above 48°C (Fig. 2C). Intriguingly, EVG1064 is killed by 30 min of exposure to ∼53°C, while the ancestral MG1655 can sustain 30 min at ∼56°C (Fig. 2D).
Table 2.
Growth of wild-type and thermotolerant strains at various temperatures on LB plates
| Strain | Growtha |
|||||
|---|---|---|---|---|---|---|
| 30°C | 37°C | 43°C | 46°C | 48.5°C | 49°C | |
| MG1655 | + | + | + | − | − | − |
| EVG1031 | + | + | + | − | − | − |
| EVG1041 | + | + | + | + | − | − |
| EVG1058 | + | + | + | + | + | − |
| EVG1064 | + | + | + | + | + | − |
+, growth; –, no growth.
Growth on plates or in liquid media at the highest temperatures failed to elucidate a significant difference between EVG1064 and EVG1058. However, a detailed comparison of growth rates revealed that the Topt for EVG1058 was below 45°C (Fig. 2E).
It must be noted that there are differences in the Tmax for EVG1064 between the Evolugator growth chamber (49.7°C), plates (48.5°C), and batch culture (48°C). Rudolph et al. also noted that their evolved E. coli cells were significantly less thermotolerant in batch liquid culture than on plates. Thus, these differences are likely due to variations in the environment, such as osmolarity, agitation, or heat exchange. However, we would also like to point out that there will always be variations in the ability to maintain accurate and constant temperatures among different incubators and bioreactor types. Even fluctuations of 0.1°C could significantly alter growth rates when one approaches Tmax. This is particularly problematic for traditional batch cultures in which the incubator must be routinely opened for sampling and growth rate determination, resulting in relatively large temperature drops when the unit is opened and relative large temperature spikes upon closing, when the heater attempts to offset the drop. That said, selection for thermotolerance in the Evolugator growth chamber produces strains that can grow at higher temperatures in batch culture outside the Evolugator, thus validating our approach to improving the thermal properties of industrially important microbes.
Genetic analysis.
To assess the possibility of genomic rearrangements associated with thermal adaptation, MG1655 and EVG1064 were analyzed for restriction fragment length polymorphisms (RFLP) using pulsed-field gel electrophoresis (PFGE). As far as can be determined by this method, there were no chromosomal recombination events during strain adaption (Fig. 3). The Illumina sequencing platform was used to identify potentially adaptive genetic alterations accumulated in EVG1064 and other intermediate strains. A comparison of MG1655 and EVG1064 revealed 31 single nucleotide substitutions that were confirmed by Sanger sequencing. A single additional mutation in proP was identified during the evolutionary process that was lost prior to the isolation of EVG1064, probably due to outcompetition. Whole-genome sequencing of intermediate strains allowed the correlation of experimental conditions (temperature) with the intermediate strains in which each substitution first appeared, revealing a detailed evolutionary map of each potential adaptation in EVG1064 (Table 3).
Fig 3.

Pulsed-field gel electrophoresis of XbaI-digested genomic DNA from MG1655 and EVG1064. Lanes: 1, lambda ladder; 2, MG1655; 3, EVG1064; 4, low-range ladder; 5, midrange ladder; 6, MG1655; 7, EVG1064; 8, lambda ladder. Notice the lack of difference between the fragmentation patters between MG1655 and EVG1064, which is indicative of the absence of large chromosomal rearrangements between the two strains.
Table 3.
Identification of SNPs and their evolutionary history
| Tmax (°C)a | Mutation in strainb |
Gene | Position of SNP in gene (gene length) | Genome position | Mutation | Aa changec | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MG1655 | EVG1031 | EVG1041 | EVG1058 | EVG1064 | ||||||
| – | X | X | X | X | X | ylbE | 114 (1,258) | 547694 | A→G | |
| 46.9 | X | X | X | X | kdpD | 1448 (2,685) | 722190 | A→G | Val 483 Ala | |
| X | X | X | X | dgsA | 607 (1,221) | 1665982 | C→T | Glu 203 Lys | ||
| X | X | X | X | rpoD | 293 (1,842) | 3211361 | T→A | Val 98 Glu | ||
| X | X | X | X | rpsJ | 95 (312) | 3451198 | G→C | Thr 32 Ser | ||
| X | X | X | X | yhhZ | 1057 (1,179) | 3580942 | A→G | Thr 353 Ala | ||
| X | X | X | X | spoT | 1091 (2,109) | 3821513 | A→G | Glu 364 Gly | ||
| X | X | X | X | Upstream of yidE | 3864388 | A→G | ||||
| X | X | X | X | treB | 338 (1,422) | 4463866 | A→T | Val 113 Glu | ||
| X | X | X | X | perR | 778 (894) | 268629 | C→T | Glu 260 Lys | ||
| 49.3 | X | X | X | malQ | 1769 (2,085) | 3546324 | A→G | Leu 590 Pro | ||
| X | X | X | wzzE | 595 (1,047) | 3967648 | G→C | Ala 199 Pro | |||
| X | X | X | rpsA | 812 (1,674) | 692029 | C→T | Ala 271 Val | |||
| X | X | X | pykF | 342 (1,413) | 1754063 | T→G | ||||
| 49.6 | X | proP | 850 (1,503) | 4229239 | A→C | Ser 284 Arg | ||||
| X | X | ybhN | 67 (957) | 821655 | C→T | Val 23 Met | ||||
| X | X | yddB | 2047 (2,373) | 1573597 | G→T | His 683 Asn | ||||
| X | X | pncB | 52 (1,203) | 989528 | A→G | Tyr 18 His | ||||
| X | X | mreD | 335 (489) | 3396563 | A→G | Leu 112 Pro | ||||
| 49.7 | X | malT | 1751 (2,706) | 3552857 | C→G | Ala 584 Gly | ||||
| X | malS | 1134 (2,031) | 3736653 | A→G | ||||||
| X | Upstream of ppiC | 3957957 | C→T | |||||||
| X | rffT | 180 (1,080) | 3975727 | G→A | ||||||
| X | glpF | 7 (846) | 4116107 | G→A | Gln 3 Stp | |||||
| X | Upstream of gltP | 4292389 | A→T | |||||||
| X | Upstream of yajD | 429789 | A→G | |||||||
| X | fabA | 108 (519) | 1015586 | C→T | Met 36 Ile | |||||
| X | Upstream of rydC | 1489550 | T→C | |||||||
| X | Upstream of yegT and fbaB | 2176752 | T→C | |||||||
| X | yejM | 1069 (1,761) | 2283466 | G→A | Ala 357 Thr | |||||
| X | tktB | 1499 (2,004) | 2579156 | T→G | Val 500 Gly | |||||
| X | idi | 395 (549) | 3031481 | A→T | Asn 132 Ile | |||||
| X | Upstream of yqjF | 3248421 | C→T | |||||||
Tmax indicates the temperature of the Evolugator incubation chamber when the strain was removed. —, original strain (not isolated from the machine).
Shaded X, the strain in which the mutation was first observed; X, strains that retain the mutation.
Stp, stop codon.
Growth in minimal medium.
Many of the adaptations that occurred during the evolutionary process may be neutral with respect to thermotolerance yet adaptive for some other experimental parameter, such as long-term growth in LB medium, which is rich in amino acids and other critical nutrients but deficient in carbohydrates (52). Table 4 demonstrates that while the WT can grow on solid M9 minimal medium with maltose as the sole carbon source, none of the evolved strains can. This indicates that the ability to utilize maltose was lost early in the evolutionary process. Similarly, EVG1064 cannot grow in minimal medium with maltose, dextrose, or glycerol as the carbon sources. However, at temperatures below 48°C, growth of EVG1064 on dextrose can be rescued by adding certain aromatic amino acids and vitamins (see Materials and Methods) to the medium, indicating the strain had become auxotrophic. Finally, neither EVG1058 nor EVG1064 can grow at all in minimal medium at 48.5°C—a temperature at which both can grow in LB. Growth could not be rescued by supplementation with aromatic amino acids or vitamins.
Table 4.
Growth of wild-type and thermotolerant mutant strains on M9 minimal plates
| Strain | Growtha |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30°C, without |
37°C |
43°C |
46°C |
48.5°C |
||||||||||||||||||
| Without |
With |
Without |
With |
Without |
With |
Without |
With |
|||||||||||||||
| D | M | D | G | M | D | G | D | G | M | D | G | D | G | M | D | G | D | G | M | D | G | |
| MG1655 | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − |
| EVG1031 | + | − | + | + | − | + | + | + | + | − | + | + | − | − | − | − | − | − | − | − | − | − |
| EVG1041 | + | − | + | + | − | + | + | + | + | − | + | + | + | + | − | + | + | − | − | − | − | − |
| EVG1058 | + | − | + | + | − | + | + | + | + | − | + | + | + | + | − | + | + | − | − | − | − | − |
| EVG1064 | − | − | − | − | − | + | − | − | − | − | + | − | − | − | − | + | − | − | − | − | − | − |
D, M, and G define the carbon sources used in the minimal medium. D, dextrose; G, glycerol; M, maltose. + and – refer to growth or no growth on plates. M9 medium is either unsupplemented (without) or supplemented (with) with the amino acids and cofactors defined in Materials and Methods.
glpF analysis.
We performed preliminary phenotypic analysis on glpF to determine if the mutations identified in this gene played any role in improved strain performance at higher temperature. The glpF mutation identified in EVG1064 introduces a stop codon at position 3 of the coding sequence, making it equivalent to a gene deletion. glpF encodes a glyceroporin responsible for passive transport of glycerol in and out of the cell and is essential for glycerol utilization (44). Not surprisingly, EVG1064 cannot grow on minimal medium (either unsupplemented or supplemented with aromatic amino acids and vitamins) with glycerol as the sole source of carbon (see Table 4), while EVG1058 can. This confirms the inactivation of the glpF gene in EVG1064.
To test if the deletion of glpF might contribute to the thermotolerance of E. coli, we determined doubling times as a function of temperature for a glpF deletion mutant (Keio collection) and its corresponding wild type (BW25113). The doubling time for the glpF mutant was significantly lower than the doubling time for the wild type at temperatures between 43°C and 48°C (Fig. 4A). In addition, the Topt for the glpF mutant increased from 37°C in BW25113 to ∼43°C in the glpF mutant. We put the wild-type glpF gene (ASKA collection) back into EVG1064 (glpF mutant) to determine the effect of complementation on doubling times at various temperatures. We also overexpressed the folE gene on the same expression plasmid in EVG1064 to control for antibiotic, plasmid, or gene expression effects. The data indicate a significant difference in growth between the glpF-expressing plasmid and the folE-expressing control at temperatures of ≥46°C (Fig. 4B).
Fig 4.
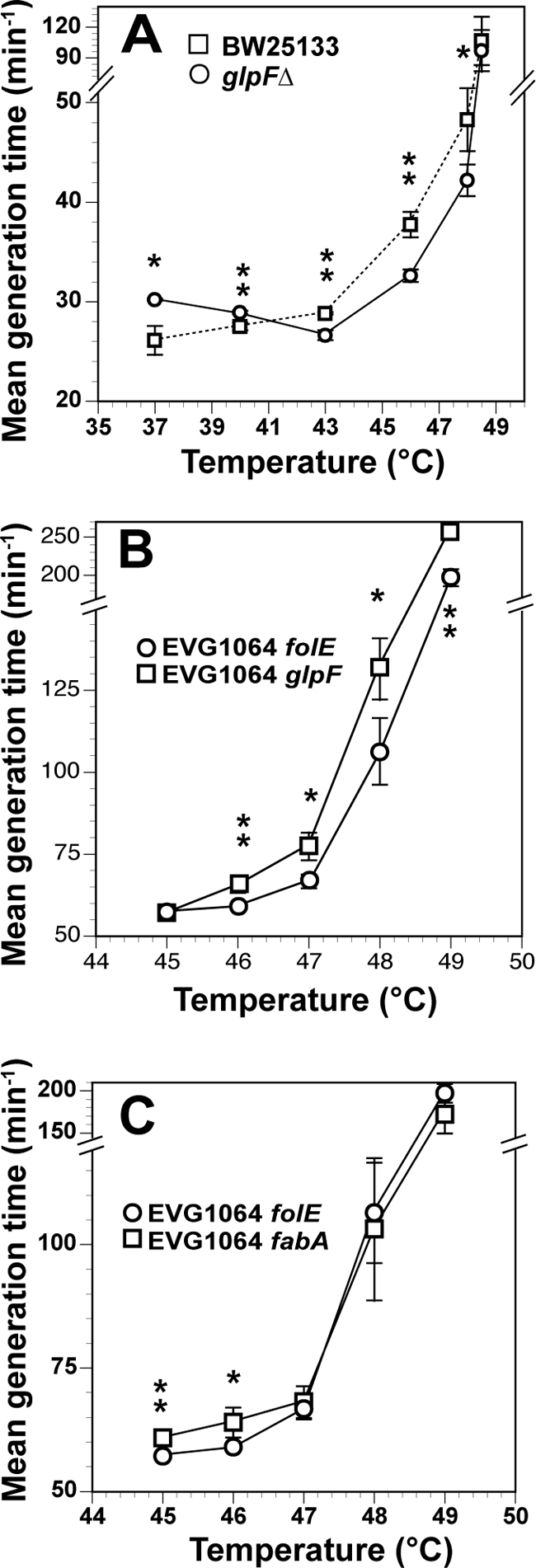
Phenotypic analysis of glpF and fabA. All growth curves were obtained in liquid LB medium. (A) Mean generation times of wild-type BW25113 and its corresponding glpF deletion mutant (ΔglpF mutant) plotted as a function of temperature; (B) mean generation times of EVG1064 carrying either the glpF or folE expression plasmids as a function of temperature; (C) mean generation times of EVG1064 carrying either the fabA or folE expression plasmids as a function of temperature. For all panels, error bars indicate ±1 standard deviation. Asterisks denote temperatures at which the growth rates of the two strains depicted are significantly different using a 2-tailed, type 2 t test. **, P < 0.005; *, P < 0.05. For panels B and C, 24 μg/ml chloramphenicol is added to the LB to select for plasmid retention.
fabA analysis.
EVG1064 also acquired a mutation at codon 36 in fabA, which encodes the fatty acid desaturase/isomerase responsible for incorporating double bonds into membrane fatty acids (31). DETECTER analysis (19) of the FabA protein family reveals that Met36 is conserved in homologs from over 300 bacterial genomes, strongly suggesting that the Met36Ile mutation in EGV1064 affects function. To determine if this mutation affected the ability of EVG1064 to make unsaturated fatty acids, we obtained fatty acid profiles from EVG1064 (fabA mutant) and EVG1058 (fabA wild type) at 37°C and 48°C. A semiquantitative comparison of fatty acids at 48°C revealed significantly higher ratios of saturated/unsaturated fatty acids in EVG1064 than in EVG1058 (Table 5). This difference is due largely to significantly more palmitate (C16:0) and significantly less cis-palmitoleate (C16:1 Δ9c) and cis-vaccenate (C18:1 Δ11c). Since cyclopropane fatty acids are derived from and are believed to perform a role similar to that of unsaturated fatty acids in regulating membrane fluidity (20), we also calculated the ratio of saturated to unsaturated and cyclopropane fatty acids and found that EVG1064 had a significantly higher ratio than EVG1058.
In order to complement the phenotype of increased saturation in EVG1064, we transformed this strain with the wild-type fabA- and folE-expressing plasmids. EVG1064 cells carrying the fabA plasmid rendered EVG1064 nearly equivalent to EVG1058 with respect to fabA, while those carrying the folE control are meant to control for the presence of antibiotic to ensure plasmid retention and to control for any effects on fatty acids that might arise from general protein overexpression. As is shown in Table 5, the presence of wild-type fabA in EVG1064 produces a strain with a lower ratio of saturated to unsaturated and saturated to unsaturated and cyclopropane fatty acids. However, it is important to note that restoration of the wild-type fabA allele does not produce a strain with a fatty acid profile identical to that of EVG1058. Rather, it appears to have a fatty acid profile that is intermediate between EVG1064 and EVG1058. Moreover, EVG1064 cells that carried a protein expression plasmid (either fabA or folE) accumulated larger quantities of branched and cyclopropane fatty acids at 48°C relative to untransformed cells. In particular, it appears as though a significant portion of palmitoleic acid (C16:1 Δ9) has been converted to C17:0 cyclopropane fatty acid. Since the alteration of branched and cyclopropane fatty acids has been shown to be an adaptation to the exposure of bacteria to phenolic compounds (48, 57), it is likely that this effect may be related to the addition of chloramphenicol to the LB and not due to thermal adaptation.
fabA is an essential gene, so we could not analyze the thermal properties of a ΔfabA mutant using the Keio collection. However, we did transform EVG1064 with a plasmid containing the wild-type fabA allele from the ASKA collection. Figure 4C shows that transforming EVG1064 with the wild-type fabA plasmid did result in a slight, but significant, reduction in the growth rate relative to the folE plasmid at 45°C and 46°C.
DISCUSSION
Herein, we present the evolutionary adaptation of E. coli K-12 MG1655 for improved growth at higher temperatures through experimental evolution using a new method of maintaining continuous cultures called the Evolugator. Not only did we improve Tmax from 46°C to 48°C, but Topt was raised from 37°C to over 46°C, indicating that the resulting temperature-adapted strain is now a facultative thermophile based on widely accepted definitions of the term (6, 30). Intriguingly, despite the elevated Tmax/Topt, the resulting strain is surprisingly less resistant than the wild type to transient thermal stress. EVG1064 also displays reduced growth rates relative to the wild type at temperatures below 43°C and reduced biomass yield at 37°C. These findings suggest significant specialization associated with the thermal adaptation.
This study represents a purely biotechnological application in which the desired goal is the production of a growth-optimized strain with alterations in a complex phenotype. The identification of individual mutations associated with the thermal adaptation was performed for intellectual property reasons and so that the results could be reproduced independently in the event the strains were lost or changed phenotype. This is especially important considering the fact that it is highly improbable that a repeat of this experiment would result in an identical evolutionary pathway to adaptation. Nevertheless, it is important to briefly discuss the nature of the identified mutations and how they may relate to the adaptive process.
The 7:1 ratio of nonsynonymous to synonymous mutations is indicative of a strong adaptive signal (28); however, in a background of manifold genetic differences, it is notoriously difficult—sometimes even impossible—to quantify the extent to which individual genetic variations contribute to changes in complex phenotypes. This is because the contributions of individual adaptive mutations to the thermotolerant phenotype are either additive or epistatic (32), and in both cases the effects of individual mutations are likely to be negligible. For example, while additive mutations individually contribute to elevated Tmax/Topt, mutations that result in large adaptive changes are rare (15). On the other hand, epistatic mutations only improve Tmax/Topt in conjunction with another mutation and may even be deleterious in isolation, in which case we would need to assess each mutation in combination with all of the other mutations. With 31 point mutations identified, the number of mutational permutations rapidly becomes intractable.
Compounding this problem is the fact that many of the mutations are likely to be neutral (or even deleterious) with respect to thermotolerance yet adaptive for some other experimental parameter. For example, the longer cells are cultured on a single unvarying carbon source, the more likely they are to lose the ability to utilize other carbon sources. Alternatively, the presence of certain biochemical building blocks in the medium (amino acids, vitamins, etc.) relaxes the selective pressure to retain genes involved in the synthesis of these building blocks, increasing the likelihood that adapted cells will become auxotrophs. LB medium is a rich medium with high concentrations of amino acids and vitamins and low concentrations of carbohydrates. Thus, the long-term culture of E. coli on LB might be expected to produce a strain that has lost the ability to utilize certain carbon sources and that has acquired auxotrophies for components of the medium. Indeed, adaptation for LB rather than temperature is more likely to be the case for the mutations acquired in EVG1031, which is not significantly more thermotolerant than the wild type when grown on LB plates (see Table 2). Not surprisingly, several of the genes (pykF, dgsA, spoT, and malT) mutated early in the adaptive process were previously identified as genes that were mutated during long-term adaptation to dextrose limitation (36, 41). In particular, DgsA is known to regulate maltose utilization (36). Accordingly, EVG1031 and all subsequent strains with the mutation in the dgsA gene have lost the ability to grow on M9 minimal medium with maltose as the sole carbon source (Table 4). Another intriguing mutation that is unique to EVG1064 and may be related to adaptation to rich medium occurred in the tktB (transketolase B) gene. An intact pentose phosphate pathway is essential for the biosynthesis of aromatic biosynthetic intermediates, and strains lacking transketolase activity are auxotrophic for specific aromatic amino acids and vitamins (64). Consistent with the tktB mutation resulting in loss of transketolase activity, EVG1064 cannot grow on minimal medium at any temperature (regardless of carbon source) unless these aromatic amino acids are added to the medium (Table 4). Interestingly, neither EVG1058 nor EVG1064 grows at 48.5°C in minimal medium, even with aromatic amino acid and vitamin supplementation, suggesting that these strains have an additional temperature-sensitive auxotrophy that may or may not be associated with adaptation to rich medium. Moreover, EVG1064 grows to a much lower cell density in LB than does the WT, suggesting that LB has become limiting for some nutrient with respect to EVG1064. In order to better identify which mutations are solely adaptive for rich medium, we might have adapted the strain to LB first before adapting for temperature. However, it must be pointed out that for such an experiment to be meaningful, there must be an endpoint for the adaptation to LB. While it is true that the magnitude of increases in fitness decreases over time (26), the adaptation process never truly ends, meaning that we could still be faced with trying to determine which mutations are adaptive for rich medium.
Despite the complexity of the adaptive process, some mutations are easy to theoretically connect to the thermotolerant phenotype. For example, published work already links some genes with alterations in thermal parameters, including yddB, rydC, yejM, and rpoD (2, 14, 51, 56). We decided to focus on two mutations for further phenotypic analysis: glpF and fabA.
The mutation in glpF results in a nonfunctional protein and impacts the ability of glycerol to pass through cellular membranes. While glycerol is a well-studied cryoprotectant, it also appears to play a role in resistance to heat shock (18, 23, 37). Indeed, the ΔglpF mutant is more thermotolerant than wild-type E. coli at higher temperatures and shows an increase in Topt from 37°C to 43°C (see Fig. 3). This result suggests that glpF deletion can have a large effect on thermal properties; however, it is important to note that the effect may also be due to the kanamycin resistance cassette used to delete the glpF gene, even though kanamycin was not added to the medium.
Complementation of EVG1064 with a wild-type glpF allele is complicated by several factors. First, EVG1058, which has a wild-type glpF allele, differs from EVG1064 at 13 other loci. Thus, one would not expect complementation of a single gene to render EVG1064 identical to EVG1058 with respect to thermal parameters. Second, EVG1058 does not show significantly decreased fitness at the highest temperatures relative to that of EVG1064, so measuring growth rates at high temperature would not distinguish EVG1058 from EVG1064. Surprisingly, however, Fig. 4B shows that restoring a wild-type glpF allele to EVG1064 does significantly reduce fitness at temperatures above 43°C relative to the same strain carrying a control plasmid (folE). These data suggest that glpF inactivation may play an important role in determining the thermal properties of EVG1064.
fabA encodes a fatty acid desaturase/isomerase that is essential for synthesizing cis-unsaturated fatty acids (31), and the M36I mutation is predicted to have an impact on the activity of this enzyme and in the degree of saturation of fatty acids. This is not surprising, since increasing the degree of saturation in membrane lipids is a well-known adaptation to exposure to high temperatures that has even been demonstrated in E. coli (63). Therefore, it is not surprising that the membranes of EVG1064 (fabA mutant) contain a greater ratio of saturated fatty acids than those of EVG1058 (fabA wild type) and that this phenotype could be partially complemented by putting the WT fabA allele back into EVG1064 (Table 5). However, EVG1064 cells expressing the WT fabA allele still have a higher ratio of saturated to unsaturated and saturated to unsaturated and cyclopropane fatty acids than EVG1058. Thus, it is possible that, without inducer, the fabA expression plasmid from the ASKA collection does not sufficiently express functional FabA to fully complement the phenotype.
Analysis of thermal properties suggest that the wild-type fabA allele did not significantly affect growth rates at the highest temperature; however, it did seem to have a small, but significant, negative effect on growth rates at 45°C and 46°C. In addition, data suggest that the Topt for EVG1064 might be slightly reduced when the wild-type fabA plasmid is present, and this is consistent with the reduced Topt for EVG1058 relative to EVG1064. However, it is important to note that EVG1064 carrying the folE plasmid also shows a reduced Topt, suggesting that the conditions required to carry the plasmid may significantly affect thermal parameters, making it impossible to directly compare the thermal properties of plasmid-bearing EVG1064 strains with those of EVG1064 and EVG1058. While these data suggest that complementation of EVG1064 with wild-type fabA has relatively little effect on growth at higher temperatures, this may be due to incomplete complementation. Thus, more research needs to be done to explore the effects of this mutation.
In conclusion, we coupled experimental evolution with whole-genome sequencing to simultaneously select and optimize for a complex phenotype as well as to characterize the resulting genotype. From an academic perspective, the evolutionary map of these mutations can eventually be used to dissect the relationship between each mutation and thermotolerance in order to better understand the molecular mechanisms that allow thermophily. That said, we do not see obvious evidence for the same mechanisms of adaptation described by Rudolph et al. In their case, proteomics revealed a striking change in the expression of heat shock proteins, such as GroEL, and intriguingly lysyl-tRNA synthetase, yet we do not see any changes indicating a similar adaptation. On the surface, this suggests that different pathways to adaptation may exist. However, it is important to note that proteomic and genomic analyses tell different stories, and similar genotypes may have arisen in the two experiments. For example, it may be possible that genetic changes in a global transcriptional regulator like rpoD (our results) might lead to changes in the expression of GroEL (their results). Indeed, rpoD has been shown to be a multicopy suppressor of defects in heat shock proteins in E. coli (56). In the future, it will be important to combine various high-throughput approaches (genomics, transcriptomics, proteomics, etc.) to more thoroughly understand the adaptive process.
Using the Evolugator, the entire process of adaptation from beginning to end took approximately 8 months. It is intriguing to note that Rudolph et al. achieved nearly identical results using traditional serial batch culture in 2 years. While there was no deliberate intention on either part to test which methodology could achieve these results faster—the Evolugator or traditional serial transfer—the fact that these studies were done in parallel and without knowledge of each others' efforts suggests that the Evolugator methodology may be capable of facilitating experimental evolution more rapidly. Since evolution is a stochastic process, it is difficult to ascribe a definitive reason for this enhanced performance. Indeed, it may be that our experiment randomly hit upon a faster pathway to thermoadaptation. However, we propose that the improvement may lie in the automation of the Evolugator and its ability to continuously monitor growth in real time, which provides several advantages.
The effectiveness of serial batch culture for selecting for adaptive traits depends on the number of generations per unit of time, the population size, and the bottleneck severity (59, 60). It works best when cells are maintained in late log phase, when short generation times are accompanied by large population sizes and when dilution is achieved with low dilution ratios so that a large sample of the population is transferred. Without knowing where a culture is on the growth curve, it is difficult to meet these requirements. With real-time growth rate monitoring, the Evolugator allows consistent dilution in late log phase based on cell density, even when the growth rate changes from cycle to cycle, thereby maximizing the number of generations per unit of time. On the other hand, the only way to reduce the laborious nature of traditional serial transfer to a manageable level is to initiate dilution during stationary phase or to rely on high dilution ratios (1:1,000) to maintain cells in log phase for longer periods of time. The former approach suffers from longer generation times and fewer generations per unit of time, while the latter approach results in severe genetic bottlenecks (60). Rudolph et al. chose to dilute in stationary phase after 48 h of growth despite the longer generation times, ostensibly because mutation rates are higher in this phase of growth. However, it is unclear if the higher mutation rate offsets the impact of the reduced number of generations per day. In addition, while the population size in our experiment is approximately the same as that employed by Rudolph et al. (7 ml of culture), they report a dilution ratio of 1:8, while the dilution ratio in the Evolugator is fixed at 1:2. Thus, the Evolugator imposed a significantly less severe bottleneck with the same population size, resulting in stronger selective pressure.
Moreover, both this study and Rudolph et al. reported instances where increases in selective pressure resulted in an immediate decrease in cell density and sometimes the death of the culture, requiring a restart of the adaptation process from a frozen stock. Real-time monitoring of growth allowed us to quickly reduce the temperature upon encountering such events, allowing the cells to recover and adaptive mutations to arise without having to restart the culture. Indeed, we only killed the culture outright twice in 8 months, while we successfully recovered the culture after excessive increases in temperature dozens of times, saving valuable time.
Finally, it should also be noted that traditional serial transfer is highly prone to contamination upon exposure of the culture to the outside world during dilution. Rudolph et al. addressed the problem in two ways. First, they reduced the frequency of dilution to every 48 h. Second, they used a kanamycin-resistant (kan+) E. coli strain as the starting point to control for contamination throughout the experiment. While it is unclear if kanamycin was added to the medium during the entire adaptation process, even periodic selection for kanamycin resistance to control for contamination could inadvertently eliminate adaptations for temperature, further slowing down the process. Moreover, it is unclear how the kan+ genotype (and perhaps the addition of kanamycin to the culture) may have influenced the generation time and the overall trajectory of the adaptation process. On the other hand, the Evolugator tubing is sealed at inception, meaning that the only way for contamination to enter the experiment is through the initial inoculum or when the tubing is changed after many rounds of dilution.
While none of the limitations in the methodology used by Rudolph et al. is particularly severe in isolation, together they likely combine to make the selection for complex phenotypes like improved Topt/Tmax exceedingly difficult, making what Rudolph et al. achieved all the more remarkable. The Evolugator circumvents these problems and, as a result, is capable of producing strains faster than conventional methods and would be of tremendous economic value for industrial strain development.
ACKNOWLEDGMENTS
I.K.B., B.J.L., E.W.-H., T.P.P, G.C.F.P., T.J.L., V.D.C.-L., and E.D.C. were funded by Evolugate, LLC, and conflicts of interest must be reported for these authors. S.A.B. and S.G.C. acknowledge ongoing support from the NHGRI.
We thank Ellen Dickstein, Matt Gitzendammer, Brandon Walts, Eric Triplett, Lauren McIntyre, John Rice, Chythanaya Rajanna, Tamara Revazishvili, and the Oregon Core Facility for help.
Footnotes
Published ahead of print 21 October 2011
REFERENCES
- 1. Angelov A, Mientus M, Liebl S, Liebl W. 2009. A two-host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst. Appl. Microbiol. 32:177–185 [DOI] [PubMed] [Google Scholar]
- 2. Antal M, Bordeau V, Douchin V, Felden B. 2005. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 280:7901–7908 [DOI] [PubMed] [Google Scholar]
- 3. Araya CL, Payen C, Dunham MJ, Fields S. 2010. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrick JE, et al. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 [DOI] [PubMed] [Google Scholar]
- 6. Bausum HT, Matney TS. 1965. Boundary between bacterial mesophilism and thermophilism. J. Bacteriol. 90:50–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buckling A, Craig Maclean R, Brockhurst M, Colegrave N. 2009. The Beagle in a bottle. Nature 457:824–829 [DOI] [PubMed] [Google Scholar]
- 8. Christ D, Chin JW. 2008. Engineering Escherichia coli heat resistance by synthetic gene amplification. Protein Eng. Des. Sel. 21:121–125 [DOI] [PubMed] [Google Scholar]
- 9. Cooper VS, Bennett AF, Lenski RE. 2001. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evolution 55:889–896 [DOI] [PubMed] [Google Scholar]
- 10. Dallinger WH. 1878. On the life history of a minute septic organism: with an account of experiments made to determine its thermal death point. Proc. R. Soc. Lond. 27:332–350 [Google Scholar]
- 11. de Crécy E, Jaronski S, Lyons B, Lyons TJ, Keyhani NO. 2009. Directed evolution of a filamentous fungus for thermotolerance. BMC Biotechnol. 9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Crécy E, et al. 2007. Development of a novel continuous culture device for experimental evolution of bacterial populations. Appl. Microbiol. Biotechnol. 77:489–496 [DOI] [PubMed] [Google Scholar]
- 13. Di Giulio M. 2003. The universal ancestor and the ancestor of bacteria were hyperthermophiles. J. Mol. Evol. 57:721–730 [DOI] [PubMed] [Google Scholar]
- 14. Duo M, Hou S, Ren D. 2008. Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl. Microbiol. Biotechnol. 81:731–741 [DOI] [PubMed] [Google Scholar]
- 15. Elena SF, Cooper VS, Lenski RE. 1996. Punctuated evolution caused by selection of rare beneficial mutations. Science 272:1802–1804 [DOI] [PubMed] [Google Scholar]
- 16. Ells TC, Speers RA, Hansen LT. 2009. Insertional mutagenesis of Listeria monocytogenes 568 reveals genes that contribute to enhanced thermotolerance. Int. J. Food Microbiol. 136:1–9 [DOI] [PubMed] [Google Scholar]
- 17. Feist AM, et al. 2010. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng. 12:173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feofilova EP, Tereshina VM, Khokhlova NS, Memorskaya AS. 2000. Different mechanisms of the biochemical adaptation of mycelial fungi to temperature stress: changes in the cytosol carbohydrate composition. Microbiology 69:504–508 [PubMed] [Google Scholar]
- 19. Gaucher EA, De Kee DW, Benner SA. 2006. Application of DETECTER, an evolutionary genomic tool to analyze genetic variation, to the cystic fibrosis gene family. BMC Genomics 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grogan DW, Cronan JE., Jr 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herendeen SL, van Bogelen RA, Neidhardt FC. 1979. Levels of major proteins of Escherichia coli during growth at different temperatures. J. Bacteriol. 139:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishchuk OP, Voronovsky AY, Abbas CA, Sibirny AA. 2009. Construction of Hansenula polymorpha strains with improved thermotolerance. Biotechnol. Bioeng. 104:911–919 [DOI] [PubMed] [Google Scholar]
- 23. Kajiwara Y, Ogawa K, Takashita H, Omori T. 2000. Enhanced glycerol production in Shochu yeast by heat-shock treatment is due to prolonged transcription of GPD1. J. Biosci. Bioeng. 90:121–123 [DOI] [PubMed] [Google Scholar]
- 24. Kitagawa M, et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 25. Lee DH, Palsson BO. 2010. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, l-1,2-propanediol. Appl. Environ. Microbiol. 76:4158–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315–1341 [Google Scholar]
- 27. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li WH. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96–99 [DOI] [PubMed] [Google Scholar]
- 29. Lindsay JA. 1995. Is thermophily a transferrable property in bacteria? Crit. Rev. Microbiol. 21:165–174 [DOI] [PubMed] [Google Scholar]
- 30. Madigan MT, Martino JM. 2006. Brock biology of microorganisms, 11th ed. Pearson/Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- 31. Magnuson K, Jackowski S, Rock CO, Cronan JE. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57:522–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malmberg RL, Mauricio R. 2005. QTL-based evidence for the role of epistasis in evolution. Genet. Res. 86:89–95 [DOI] [PubMed] [Google Scholar]
- 33. Meyer V. 2008. Genetic engineering of filamentous fungi: progress, obstacles and future trends. Biotechnol. Adv. 26:177–185 [DOI] [PubMed] [Google Scholar]
- 34. Montero-Barrientos M, Cardoza RE, Gutiérrez S, Monte E, Hermosa R. 2007. The heterologous overexpression of hsp23, a small heat-shock protein gene from Trichoderma virens, confers thermotolerance to T. harzianum. Curr. Genet. 52:45–53 [DOI] [PubMed] [Google Scholar]
- 35. Nichols RJ, et al. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Notley-McRobb L, Ferenci T. 1999. The generation of multiple coexisting mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1:45–52 [DOI] [PubMed] [Google Scholar]
- 37. Omori T, et al. 1996. Enhancement of glycerol production by brewing yeast (Saccharomyces cerevisiae) with heat shock treatment. J. Ferment. Bioeng. 82:187–190 [Google Scholar]
- 38. Park KS, et al. 2003. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol. 21:1208–1214 [DOI] [PubMed] [Google Scholar]
- 39. Patnaik R. 2008. Engineering complex phenotypes in industrial strains. Biotechnol. Prog. 24:38–47 [DOI] [PubMed] [Google Scholar]
- 40. Pena MI, Davlieva M, Bennett MR, Olson JS, Shamoo Y. 2010. Evolutionary fates within a microbial population highlight an essential role for protein folding during natural selection. Mol. Syst. Biol. 6:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Philippe N, Crozat E, Lenski RE, Schneider D. 2007. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays 29:846–860 [DOI] [PubMed] [Google Scholar]
- 42. Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. 2010. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 44. Richey DP, Lin EC. 1972. Importance of facilitated diffusion for effective utilization of glycerol by Escherichia coli. J. Bacteriol. 112:784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Riehle MM, Bennett AE, Long AD. 2005. Differential patterns of gene expression and gene complement in laboratory-evolved lines of E. coli. Integ. Comp. Biol. 45:532–538 [DOI] [PubMed] [Google Scholar]
- 46. Riehle MM, Bennett AF, Long AD. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 98:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rivera EC, et al. 2006. Evaluation of optimization techniques for parameter estimation: application to ethanol fermentation considering the effect of temperature. Process Biochem. 41:1682–1687 [Google Scholar]
- 48. Rozes N, Peres C. 1998. Effects of phenolic compounds on the growth and the fatty acid composition of Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 49:108–111 [Google Scholar]
- 49. Rudolph B, Gebendorfer KM, Buchner J, Winter J. 2010. Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem. 285:19029–19034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Serina S, et al. 2004. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res. Microbiol. 155:692–701 [DOI] [PubMed] [Google Scholar]
- 52. Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shehata TE, Marr AG. 1975. Effect of temperature on the size of Escherichia coli cells. J. Bacteriol. 124:857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi B, Xia X. 2005. Genetic variation in clones of Pseudomonas pseudoalcaligenes after ten months of selection in different thermal environments in the laboratory. Curr. Microbiol. 50:238–245 [DOI] [PubMed] [Google Scholar]
- 55. Shi DJ, Wang CL, Wang KM. 2009. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 36:139–147 [DOI] [PubMed] [Google Scholar]
- 56. Shiozawa T, Ueguchi C, Mizuno T. 1996. The rpoD gene functions as a multicopy suppressor for mutations in the chaperones, CbpA, DnaJ and DnaK, in Escherichia coli. FEMS Microbiol. Lett. 138:245–250 [DOI] [PubMed] [Google Scholar]
- 57. Unell M, Kabelitz N, Jansson JK, Heipieper HJ. 2007. Adaptation of the psychrotroph Arthrobacter chlorophenolicus A6 to growth temperature and the presence of phenols by changes in the anteiso/iso ratio of branched fatty acids. FEMS Microbiol. Lett. 266:138–143 [DOI] [PubMed] [Google Scholar]
- 58. van Derlinden E, Bernaerts K, van Impe JF. 2008. Dynamics of Escherichia coli at elevated temperatures: effect of temperature history and medium. J. Appl. Microbiol. 104:438–453 [DOI] [PubMed] [Google Scholar]
- 59. Wahl LM, Gerrish PJ. 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55:2606–2610 [DOI] [PubMed] [Google Scholar]
- 60. Wahl LM, Gerrish PJ, Saika-Voivod I. 2002. Evaluating the impact of population bottlenecks in experimental evolution. Genetics 162:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. 2010. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat. Biotechnol. 28:856–862 [DOI] [PubMed] [Google Scholar]
- 62. Yokotani N, et al. 2009. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229:1065–1075 [DOI] [PubMed] [Google Scholar]
- 63. Yuk HG, Marshall DL. 2003. Heat adaptation alters Escherichia coli O157:H7 membrane lipid composition and verotoxin production. Appl. Environ. Microbiol. 69:5115–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao G, Winkler ME. 1994. An Escherichia coli K-12 tktAtktB mutant deficient in transketolase activity requires pyridoxine (vitamin B6) as well as the aromatic amino acids and vitamins for growth. J. Bacteriol. 176:6134–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]