Abstract
Efficient biosynthesis of l-tyrosine from glucose is necessary to make biological production economically viable. To this end, we designed and constructed a modular biosynthetic pathway for l-tyrosine production in E. coli MG1655 by encoding the enzymes for converting erythrose-4-phosphate (E4P) and phosphoenolpyruvate (PEP) to l-tyrosine on two plasmids. Rational engineering to improve l-tyrosine production and to identify pathway bottlenecks was directed by targeted proteomics and metabolite profiling. The bottlenecks in the pathway were relieved by modifications in plasmid copy numbers, promoter strength, gene codon usage, and the placement of genes in operons. One major bottleneck was due to the bifunctional activities of quinate/shikimate dehydrogenase (YdiB), which caused accumulation of the intermediates dehydroquinate (DHQ) and dehydroshikimate (DHS) and the side product quinate; this bottleneck was relieved by replacing YdiB with its paralog AroE, resulting in the production of over 700 mg/liter of shikimate. Another bottleneck in shikimate production, due to low expression of the dehydroquinate synthase (AroB), was alleviated by optimizing the first 15 codons of the gene. Shikimate conversion to l-tyrosine was improved by replacing the shikimate kinase AroK with its isozyme, AroL, which effectively consumed all intermediates formed in the first half of the pathway. Guided by the protein and metabolite measurements, the best producer, consisting of two medium-copy-number, dual-operon plasmids, was optimized to produce >2 g/liter l-tyrosine at 80% of the theoretical yield. This work demonstrates the utility of targeted proteomics and metabolite profiling in pathway construction and optimization, which should be applicable to other metabolic pathways.
INTRODUCTION
The traditional approach to improve microbial production of natural products, such as amino acids and antibiotics, consists of altering key structural or regulatory genes of the biosynthetic pathway, followed by measuring the amount of desired product that is produced. Each change then reveals the presence or absence of a bottleneck, and based on those results, the next gene is deleted or overexpressed, and the cycle repeats until product titers/yields can no longer be improved substantially. Although this stepwise approach can yield improvements in flux through these pathways, it is a tedious and time-consuming strategy, given that metabolic pathways tend to be well balanced, and rarely does a single change increase flux dramatically. Indeed, some bottlenecks are not revealed until others are relieved. This process typically leads to the identification of local yield maxima, but not the global optimal yield.
These challenges are particularly evident in efforts to engineer Escherichia coli to produce high yields of aromatic amino acids. With advances in metabolic engineering and the discovery of novel biosynthetic pathways in plants, aromatic amino acids, which have been important commodities used as animal feeds, food additives, and supplements, can also serve as precursors to a variety of commercially valuable molecules and pharmaceutical drugs (13, 42). Recently, several publications used l-tyrosine-overproducing strains of E. coli grown on glucose to produce biopolymer starting materials, such as p-hydroxycinnamic acid and p-hydroxystyrene (39), and drug precursors, such as reticuline, an important intermediate in the biosynthesis of benzylisoquinoline alkaloids (29, 40). However, of the three aromatic amino acids derived from the shikimate (SHIK) pathway, the l-tyrosine yield is the lowest, ranging from 0.10 to 0.15 g per g glucose (Table 1). Though Patnaik et al. recently reported l-tyrosine titers of over 50 g/liter using E. coli in a 200-liter bioreactor by improving the fermentation and isolation steps (35), the production strain yielded only 0.09 g of l-tyrosine per gram of glucose (33), which is less than 20% of the theoretical yield (Table 1). Further improvement in the yield is needed to make the process as economically competitive as the processes used to synthesize other amino acids, such as l-lysine, l-glutamate, and l-alanine (17, 21).
Table 1.
l-Tyrosine production yields from various E. coli strains engineered within the past 10 years
| Straina | Genotype | Titer (g/liter)b | Ytyr/glu (g/g)b | % Ytc | References |
|---|---|---|---|---|---|
| DPD4193 | K-12; aroH367 tyrR366 tna-2 lacY5 malT384 aroG397(fbr)dtrpE382 Ptrc-tyrA::kanR ΔpheLA | 0.18 (55) | 0.09 | 16 | 33, 35 |
| T2 | K-12; ΔtyrR pCL1920::PLtetO-1-aroGfbr-tyrAfbr-ppsA-tktA | 0.62 (9.7) | 0.124 | 22 | 26 |
| T2-YK | K-12; ΔtyrR pCL1920::PLtetO-1-aroGfbr-tyrAfbr-ppsA-tktA pBR322::PLtetO-1-aroK-ydiB | 0.70 | 0.14 | 25 | 25 |
| PB12CP | JM101; Δ(ptsHI crr) Glc+ pJLB::aroGfbr pTrc::tyrC-pheACM | 0.18 (3) | 0.064 | 12 | 6 |
| ygdT KO | K-12; ΔpheA ΔtyrR ygdT::kan pCL1920::tyrAfbr-aroGfbr | 0.59 | 0.118 | 21 | 38 |
KO, knockout.
The reported titers and yields (Ytyr/glu, yields of l-tyrosine and l-glucose) were results from the shake flask cultures, except those in parentheses, which were from bioreactors. All cultures grown were grown at 37°C, except PB12CP, which was grown at 30°C.
The theoretical yield (% Yt) was calculated based on the maximum value of 0.55 g of l-tyrosine per g of glucose (43).
fbr, feedback resistant.
Despite a vast wealth of literature accumulated over the past 30 years pertaining to the enzymatic activities and expression properties of the shikimate pathway, it remains difficult to engineer (4, 13, 14, 17, 42). Previous l-tyrosine engineering work has most often focused on the transcriptional deregulation of the tyrR and/or trpR regulons, followed by removing the feedback inhibition on two key enzymes, 3-deoxy-d-arabino-heptulosonate (DAHP) synthase (AroG), which catalyzes the first step committed to the shikimate pathway, and the dual-function chorismate mutase/prephenate dehydrogenase (TyrA), which catalyzes the first two steps in l-tyrosine biosynthesis from chorismate (26, 33). Coexpression of the rate-limiting enzymes, shikimate kinase (AroK or AroL) and quinate (QUIN)/shikimate dehydrogenase (YdiB), and deletion of the l-phenylalanine branch of the aromatic amino acid biosynthetic pathway have been shown to increase l-tyrosine production (12, 25, 33). Furthermore, overexpression of phosphoenolpyruvate synthase (PpsA) and transketolase A (TktA), altering glucose transport and the use of other carbon sources, such as xylose and arabinose, have also been shown to increase the pools of precursors to the shikimate pathway (1, 9, 22, 26, 34, 47, 48).
In these previous studies, gene expression was modified for only a few candidates of the l-tyrosine pathway at a time, and a large number of strains had to be screened to circumvent bottlenecks. We hypothesized that, using these approaches, one pathway bottleneck might be eliminated while a new bottleneck might be introduced somewhere else along the pathway. In this study, we constructed a strain that harbored all of the genes necessary for the production of l-tyrosine from erythrose-4-phosphate (E4P) and phosphoenolpyruvate (PEP) on two plasmids (11 genes in total, 9 for the tyrosine biosynthetic pathway plus 2 for overexpressing E4P and PEP) and analyzed it for l-tyrosine production, pathway enzyme levels, and intermediates. The results of these analyses were used to identify multiple bottlenecks and to engineer subsequent strains for improved production of specific enzymes. By applying these techniques over several rounds of engineering, we were able to significantly improve l-tyrosine production from 20% to 80% of the theoretical yield (0.44 g l-tyrosine/g glucose) without resorting to time- and resource-consuming characterization of the complete pathway gene expression landscape.
MATERIALS AND METHODS
PCR amplification of genes and construction of plasmids.
All genes (open reading frames [ORFs]) were amplified by PCR from the genomic DNA of E. coli MG1655 and extended with 5′-AAAGGAGGCCATCC-3′ at the 5′ end and with the corresponding endonuclease restriction sites at the 5′ and 3′ ends of the fragments. The feedback-resistant mutants, aroG* [D146N] and tyrA* [M53I; A354V] (26), were obtained using the technique of rapid PCR site-directed mutagenesis (45) (the asterisks in aroG* and tyrA* refer to the feedback-resistant variants of aroG and tyrA, respectively). To improve the expression of AroB, rare codons found within the first 15 codons were optimized, as shown in boldface (ATG GAG CGT ATT GTC GTT ACT CTG GGC GAA CGT AGC TAC CCA ATT), yielding aroBop, the codon-optimized variant of aroB. The codon usage analysis was performed using the Web-based software E. coli Codon Usage Analyzer 2.1 by Morris Maduro (http://www.faculty.ucr.edu/∼mmaduro/codonusage/usage.htm).
The l-tyrosine biosynthetic pathway (central metabolites to l-tyrosine) was encoded in several variants of two modules: the shikimate module and the tyrosine module. The shikimate modules encode the enzymes that transform pyruvate, fructose 6-phosphate (F6P), and glyceraldehyde 3-phosphate (G3P) into shikimate, and versions of this module containing various plasmid origins, gene variants, promoters, and transcription terminators are referred to as S0 to S5 and are harbored on plasmids pS0 to pS5 (Fig. 1 and 2). The tyrosine modules encode the enzymes that transform shikimate into l-tyrosine, and versions of this module containing various gene variants, promoters, and transcription terminators are referred to as Y0 to Y3 and are harbored on plasmids pY0 to pY3 (Fig. 1 and 3). For construction of the initial shikimate module, S0 (Table 2), the fragment containing ydiB, aroD, and aroB, was constructed using splicing by overlapping extension (SOE)-PCR (15), with an EcoRI restriction site at the 5′ end and combined NheIXXXBamHI restriction sites at the 3′ end, where XXX are random nucleotides functioning as a spacer; this ydiB-to-aroB fragment was then cloned into pZS21 (27) between the EcoRI and BamHI restriction sites. The genes that encode the enzymes that produce DAHP from pyruvate, F6P, and G3P (aroG*, ppsA, and tktA) were cloned into pPro33 (20). The fragment containing 5′-aroG*-XbaI-ppsA-NdeI-tktA-3′, a DAHP-expressing operon, was amplified and extended by PCR with the NheI and BamHI restriction sites at the 5′ and 3′ ends, respectively; it was inserted at the 3′ end of aroB between the NheI and BamHI sites, creating plasmid pS0.
Fig 1.
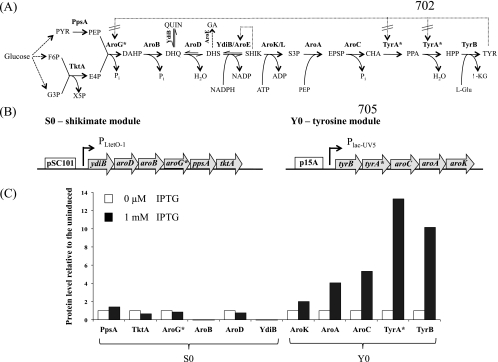
(A) The biosynthetic pathway of l-tyrosine (TYR) in E. coli from glucose. X5P, xylulose 5-phosphate; PYR, pyruvate; EPSP, 5-enolpyruvoylshikimate 3-phosphate; CHA, chorismate; PPA, prephenate; HPP, 4-hydroxyphenlypyruvate; l-Glu, glutamic acid; and α-KG, α-ketoglutarate. The enzymes (in boldface) are as follows: PpsA, phosphoenolpyruvate synthase; TktA, transketolase A; AroG, DAHP synthase; AroB, DHQ synthase; AroD, DHQ dehydratase; YdiB, quinate/shikimate dehydrogenase; AroE, shikimate dehydrogenase; AroK/L, shikimate kinase I/II; AroA, EPSP synthase; AroC, chorismate synthase; TyrA, chorismate mutase/prephenate dehydrogenase; and TyrB, tyrosine aminotransferase. QUIN and gallic acid (GA) are side products. QUIN is formed by YdiB from DHQ (18), while GA is formed by AroE from DHS (18, 30). The dashed lines indicate where feedback inhibitions occur. Allosteric regulation of AroG and TyrA were removed in this study by employing their respective feedback-resistant mutants, AroG* (D146N) and TyrA* (M53I;A354V), respectively. (B) Structures of the initial modules, S0 and Y0, for production of shikimate and l-tyrosine, respectively. The open blocks indicate the origins of replication, the shaded arrows represent the genes, and the angled arrows indicate the promoters. Note that for each operon, the genes are placed in the reverse order relative to the reaction pathway. Using the shikimate module as an example, ydiB, which catalyzes the last step in the formation of shikimate, was placed next to the promoter, and so on. (C) SRM analysis of the protein production levels from S0 and Y0 in strain A when induced or uninduced with IPTG. The protein levels shown are ratios relative to the uninduced levels.
Fig 2.
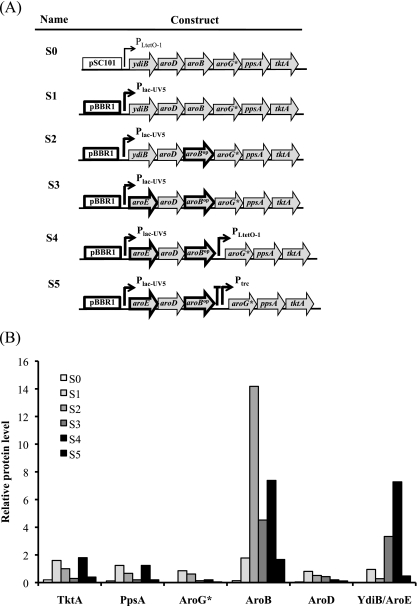
(A) Stepwise improvements of the shikimate module by changing the origin of replication from pSC101 to pBBR1 and the promoter from PLtetO-1 to Plac-UV5 (S1), followed by codon optimization of aroB (S2), replacement of ydiB with aroE (S3), and inserting a second promoter PLtetO-1 5′ of aroG* (S4). In S5, a combination of the rrnB terminator T1 (large T) and Ptrc was used to substitute PLtetO-1 5′ of aroG*, which resulted in significant reductions in protein and shikimate production. (B) The SRM results indicate the relative levels of TktA through YdiB/AroE as a consequence of the various modifications to the shikimate module. All cultures were performed under the conditions described in Table 3.
Fig 3.
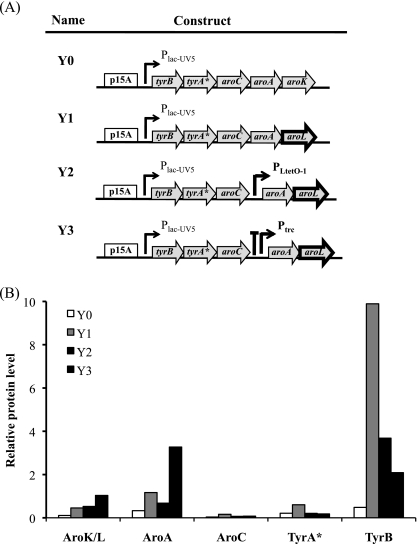
(A) Stepwise improvements of the tyrosine module by replacement of aroK with aroL (Y1) and inserting a second promoter, either PLtetO-1 (Y2) or a combination of the rrnB terminator T1 (large T) and Ptrc (Y3), 5′ of aroA. (B) The SRM results indicate the relative levels of TyrB through AroL as a consequence of the various modifications to the tyrosine module. All cultures were performed under the conditions described in Table 3.
Table 2.
List of plasmids used in this study
| Plasmida | Descriptionb |
|---|---|
| Base plasmids | |
| pZS21 | pSC101; Kanr PLtetO-1 |
| pZA31 | p15A; Cmr PLtetO-1 |
| pBbB5c | pBBR1; CmrlacI Plac-UV5 |
| pRBS01 | p15A; Ampr Plac-UV5 |
| pBbA5a | p15A; AmprlacI Plac-UV5 |
| Shikimate plasmids | |
| pS0 | pZS21::ydiB-aroD-aroB-aroG*-ppsA-tktA |
| pS1 | pBbB5c::ydiB-aroD-aroB-aroG*-ppsA-tktA |
| pS2 | pBbB5c::ydiB-aroD-aroBop-aroG*-ppsA-tktA |
| pS3 | pBbB5c::aroE-aroD-aroBop-aroG*-ppsA-tktA |
| pS4 | pBbB5c::aroE-aroD-aroBop PLtetO-1-aroG*-ppsA-tktA |
| pS5 | pBbB5c::aroE-aroD-aroBop T1-Ptrc-aroG*-ppsA-tktA |
| Tyrosine plasmid | |
| pY0 | pRBS01::tyrB-tyrA*-aroC-aroA-aroK |
| pY1 | pRBS01::tyrB-tyrA*-aroC-aroA-aroL |
| pY2 | pBbA5a::tyrB-tyrA*-aroC PLtetO-1-aroA-aroL |
| pY3 | pBbA5a::tyrB-tyrA*-aroC T1-Ptrc-aroA-aroL |
| Other plasmids | |
| pDHQ | pZA31::aroB-aroG*-ppsA-tktA |
| pDHS | pZA31::aroD-aroB-aroG*-ppsA-tktA |
The parental plasmids pZS21 and pZA31 are described elsewhere (27). Descriptions of plasmids pRBS01 and pBb are available at the registry (http://registry.jbei.org). The copy numbers determined for pBBR1 and p15A ori in pBbB5c and pBbA5a are listed in the registry; they range from 8 to 10 and from 17 to 20 copies per cell, respectively.
aroBop is the codon-optimized variant of aroB. T1 is the E. coli rrnB terminator t1. It precedes Ptrc to prevent run through by the first promoter.
Because some of the intermediates in the biosynthetic pathway cannot be purchased to use as standards for analysis, we needed to engineer strains that could produce these intermediates. Two plasmids, pDHQ and pDHS, were constructed using pZA31 (27), whose only differences from pZS21 are its replication origin (p15A versus pSC101) and resistance marker (chloramphenicol versus kanamycin) (Table 2), to produce standards for the measurement of the metabolic intermediates involved in the hydroaromatic equilibrium. The dehydroquinate (DHQ) operon is essentially S0 without ydiB-aroD; when pDHQ is transformed into cells, those cells accumulate DHQ. The dehydroshikimate (DHS) operon is essentially S0 without ydiB; when pDHS is transformed into cells, those cells accumulate DHS.
For construction of the plasmids harboring the tyrosine modules Y0 (pY0) and Y1 (pY1) (Table 2), plasmid pRBS01 (http://registry.jbei.org) was used as the backbone. Two fragments, the first containing tyrB and tyrA* and the second containing aroC and aroA, were assembled using SOE-PCR (15). They were cloned between BglII and HindIII of the plasmid as 5′-BglII-tyrB-tyrA*-XhoI-aroC-aroA-KpnI-HindIII-3′. Subsequently, either aroK or aroL was cloned between the KpnI and HindIII sites to produce pY0 or pY1, respectively (Table 2).
Construction of biobrick operons.
The remaining shikimate and tyrosine plasmids (pS1 to pS5, pY2, and pY3) were constructed using the Bglbrick standard and plasmids pBbB5c and pBbA5a (3), which are described in the Joint BioEnergy Institute (JBEI) registry (http://registry.jbei.org). For each ORF to be cloned into these BglBrick plasmids, all EcoRI, BglII, BamHI, and XhoI restriction sites within the sequence were removed by codon substitution. The ORFs were then amplified by PCR with primers that extended the 5′ and 3′ ends with EcoRIXXBglII and BamHIXXXhoI, respectively. Positions XX are the adenylate dinucleotides (AA) but can be any random sequence. Similar to the other ORFs in this study, all genes contained the consensus 5′-AGGAGG-3′ ribosome binding site (RBS) followed by a spacer sequence, 5′-CCATCC-3′ (41). Prior to cloning, all PCR fragments were digested with BglII and XhoI and then inserted into the corresponding plasmid stepwise, from the 5′ end to the 3′ end, replacing the original insert, gfp or rfp, respectively.
In brief, pS1 replaces the pSC101 origin and the promoter PLtetO-1 on pS0 with the pBBR1 origin and Plac-UV5, respectively. pS2 replaces aroB on pS1 with its codon-optimized variant, aroBop. pS3 replaces ydiB on pS2 with aroE. pS4 adds to pS3 an additional promoter, PLtetO-1, between aroBop and aroG*. pS5 adds to pS3 a transcription terminator and Ptrc between aroBop and aroG*. For the tyrosine plasmids, pY2 adds the promoter PLtetO-1 between aroC and aroA on pY1. pY3 adds a transcription terminator and Ptrc between aroC and aroA on pY1.
E. coli strains and culture conditions.
All plasmid manipulations were performed using E. coli strain DH10B unless otherwise stated. For shikimate and l-tyrosine production data, E. coli MG1655 was used and cultured in 50 ml MOPS (morpholinepropanesulfonic acid)-M9 minimal medium containing 0.5% glucose (31) and supplemented with the appropriate amounts of antibiotics: carbenicillin at 100 μg/ml, chloramphenicol at 30 μg/ml, and/or kanamycin at 50 μg/ml. All cultures were grown at 37°C, which is the optimal temperature for production, in a 250-ml shake flask shaking at 200 rpm. For induction, 50 μM to 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the culture after 3 h of incubation time. Samples used to analyze l-tyrosine and shikimate levels were collected at 24 h; except for strain F, samples were also collected at 48 h (Table 3).
Table 3.
l-Tyrosine production of the various strains constructed in the studya
| Strain | Plasmids | Titer (mg/liter) | mgTyr/gCDWb | % Yt |
|---|---|---|---|---|
| A | pS0 and pY0 | 746 ± 18 | 921 ± 22 | 27 |
| B | pS0 and pY1 | 897 ± 28 | 944 ± 29 | 33 |
| C | pS4 and pY1 | 1,150 ± 28 | 1,307 ± 32 | 42 |
| D | pS5 and pY1 | 1,219 ± 24 | 1,451 ± 29 | 44 |
| E | pS4 and pY2 | 908 ± 5 | 987 ± 5 | 33 |
| F | pS4 and pY3 | 2,169 ± 382 | 2,645 ± 466 | 79 |
| G | pS5 and pY3 | 1,310 ± 75 | 1,617 ± 93 | 48 |
The basal strain was E. coli MG1655, which was transformed with the various shikimate and l-tyrosine plasmids (Table 2). The l-tyrosine titer (in mg/liter) reported was the final production obtained within 24 h when glucose had been completely consumed, with the exception of strain F (which was 48 h), in a 50-ml MOPS-M9 minimal medium shake flask culture containing 5 g/liter glucose, shaken at 200 rpm and 37°C. For strain F, the l-tyrosine titer at 24 h was 1,512 ± 27 mg/liter. All cultures were induced with 50 μM IPTG, except for that of strain A. For strain A, the production was maximized at 1 mM IPTG.
The average cell dry weight for all of the strains was 0.38 g/liter per optical-density (OD) unit of culture; it is consistent with the value reported previously (6).
HPLC measurements for l-tyrosine production.
l-Tyrosine titers were measured using high-performance liquid chromatography (HPLC) with UV detection. An aliquot (500 μl) of culture was drawn and diluted into 1 N HCl, followed by incubation at 55°C for 30 min with occasional vortexing. The sample was then centrifuged, and the collected supernatant was diluted further with the appropriate amount of water prior to injection into an Agilent 1200 Series HPLC system equipped with a photodiode array detector set at 210, 254, and 280 nm (Agilent Technologies, Santa Clara, CA). The separation was achieved with a reverse-phase C18 column (Inertsil; 2.1 by 250 mm, 3.5 μm; GL Sciences, Inc., Torrance, CA) at a flow rate of 0.15 ml/min. l-Tyrosine was eluted with a linear gradient of water (A) and methanol (B) as follows: 5% B from 0 to 8 min, 5 to 40% B from 8 to 13 min, holding at 40% B from 13 to 16 min, 40 to 5% B from 16 to 21 min, and finally allowing the column to equilibrate at 5% B for 10 min. l-Tyrosine from E. coli extracts was quantified using a five-point calibration curve ranging from 14 mg/liter to 448 mg/liter. The R2 coefficient for the l-tyrosine calibration curve was 0.99.
Targeted proteomics analysis.
The levels of enzymes in the l-tyrosine biosynthetic pathway were determined using single-reaction monitoring (SRM) mass spectrometry (MS). After 24 h of cultivation, cells were pelleted by centrifugation, and the supernatant was discarded. Protein extraction, alkylation, digestion, and analysis were performed as described elsewhere (36). Briefly, the protein was extracted from the cell pellet by using chloroform-methanol precipitation and resuspended in 10% methanol for total protein quantification via the DC Protein reagent (Bio-Rad, Hercules, CA). Fifty micrograms of protein was reduced with 5 mM tris(2-carboxyethyl)phosphine, subsequently alkylated with 200 mM iodoacetic acid, and digested overnight at 37°C by using trypsin at a ratio of 1:50 (trypsin-sample). Prior to LC-MS analysis, bovine serum albumin digest was added at a concentration of 17 fmol/μl to serve as an internal standard.
Protein samples were analyzed using an Eksigent Tempo nanoLC-2D coupled to an AB Sciex 4000 Q-Trap mass spectrometer running with Analyst 1.5 operating in SRM mode. Samples were loaded onto a PepMap100 μ-guard column (Dionex-LC Packings) and washed (20 min; 15 μl/min) with buffer A (2% [vol/vol] acetonitrile, 0.1% [vol/vol] formic acid, and the balance H2O). Samples were eluted over a Pepmap100 analytical column (75-μm inside diameter [i.d.], 150-mm length, 100-Å packing pore size, 3-μm bead size) with a 15-min gradient from 5% to 30% buffer B (98% [vol/vol] acetonitrile, 0.1% formic acid, and the balance H2O). Following peptide elution, the column was washed at 80% buffer B for 10 min and allowed to equilibrate for 13 min at 5% buffer B prior to the next analysis.
Three unique peptide transitions were chosen and optimized for each protein encoded on the tyrosine and shikimate modules. Each peptide transition was verified by using full tandem mass spectrometry (MS-MS) scans and subsequent database searching to confirm that the correct peptide was selected. MultiQuant version 1.2 and 2.0 software (AB Sciex) was used to determine the peak area for each transition. Sample load variations were normalized by using the antibiotic markers specific to each plasmid and a bovine serum albumin (BSA) internal standard.
Analysis of pathway intermediates.
All pathway intermediates were quantified using HPLC–electrospray ionization (ESI)–time-of-flight (TOF) MS. For quantification of anionic, nonphosphorylated metabolites, 1 ml of culture was mixed with ice-cold methanol (1:1 [vol/vol]), and deproteinated by filtration (YM-3 centrifuge filter; Millipore Inc., Billerica, MA). For phosphorylated intracellular metabolites, cells from the 50-ml culture were collected by centrifugation and extracted with 0.5 ml of ice-cold methanol, followed by 0.5 ml of ice-cold water. The samples were then dried by lyophilization (Labconco Co., Kansas City, MO) and reconstituted in 0.5 ml water-methanol (1:1 [vol/vol]), and protein was removed as described above.
All chemical standards were purchased from Sigma-Aldrich and prepared as 100-μM stock solutions in methanol-water (50:50 [vol/vol]). The standards for DHQ and DHS, which were produced using E. coli engineered with the pathway ending at these metabolic intermediates, were purified via an Agilent 1200 Series preparative LC system and a Carbomix H-NP 10%-8% preparative column (21.2 by 300 mm, 10 μm; Sepax Technologies, Inc., Newark, DE).
The separation of metabolites was conducted on the fermentation-monitoring HPX-87H column with 8% cross-linkage (150-mm length, 7.8-mm inside diameter, and 9-μm particle size; Bio-Rad, Richmond, CA) using an Agilent Technologies 1100 Series HPLC system. A sample injection volume of 10 μl was used throughout. The sample tray and column compartment were set to 4 and 50°C, respectively. Metabolites were eluted isocratically with a mobile-phase composition of 0.1% formic acid in water at a flow rate of 0.5 ml/min.
The HPLC system was coupled to an Agilent Technologies 6210 series time-of-flight mass spectrometer (for LC-TOF MS) via a MassHunter workstation (Agilent Technologies, CA). Drying and nebulizing gases were set to 13 liters/min and 30 lb/in2, respectively, and a drying-gas temperature of 330°C was used throughout. ESI was conducted in the negative ion mode and a capillary voltage of −3,500 V was utilized. All other MS conditions were described previously (11). Metabolites from E. coli extracts were quantified via seven-point calibration curves ranging from 625 nM to 50 μM. The R2 coefficients for the calibration curves were ≥0.99.
RESULTS
Initial pathway construction.
The l-tyrosine biosynthetic genes (Fig. 1A) were assembled in two modules, one consisting of six genes for the production of shikimate from E4P and PEP and another consisting of five genes for the final production of l-tyrosine from shikimate. E. coli MG1655 was selected as the production host, rather than strains that have been engineered to overproduce l-tyrosine, because the genetically unmodified strain is better suited for analyzing the performance of the synthetic system we engineered in this study. We had determined that the basal levels of proteins and metabolites in the l-tyrosine biosynthetic pathway in E. coli MG1655 were negligible compared to those produced by our modular system. We chose to divide the l-tyrosine production pathway into two modules at the intermediate shikimate, rather than at chorismate, which is the natural branch point in the shikimate pathway (8), for several reasons. First, chorismate is unstable at 37°C, which would make optimization of a partial pathway ending at chorismate more difficult. Moreover, splitting the pathway at chorismate would require the first nine genes to be cloned into one plasmid, while splitting at shikimate required the cloning of only the first six genes into the first plasmid, which was easier and faster to accomplish. Additionally, shikimate is stable in liquid culture and is commercially valuable as a precursor for the synthesis of Tamiflu (19).
Previous studies had made advances in engineering the l-tyrosine pathway (24–26), providing us with useful information from which to build the modular system described here. In their initial constructs, Lütke-Eversloh and Stephanopoulos achieved l-tyrosine production of 621 ± 26 mg/liter (26) by overexpressing aroG* (D146N), tyrA* (M53I; A354V), ppsA, and tktA on a pSC101 plasmid under the control of a constitutive promoter (PLtetO-1 with no tetR) in the tyrR knockout strain; following combinatorial analysis, production was increased by ∼26% when the isozymes YdiB and AroK were additionally coexpressed on a pBR322 plasmid (25). Utilizing this work as a starting point, we selected pZS21 (27), consisting of the pSC101 origin of replication and PLtetO-1, to express the genes of the first shikimate module (S0), creating plasmid pS0, and pRBS01 (http://registry.jbei.org), a medium-copy-number plasmid with a p15A origin of replication and the IPTG-inducible Plac-UV5, to express the genes of the first tyrosine module (Y0), creating plasmid pY0. Moreover, we selected YdiB (instead of AroE) as the dehydrogenase for production of shikimate and AroK (instead of AroL) as the shikimate kinase in our first set of operons (Fig. 1B and Table 2). All of the genes in the operons were initially ordered so that the last gene in the metabolic pathway was placed closest to the promoter, and so on. According to our experimental observations that genes close to the promoter are usually induced at much higher levels than those distal from the promoter, we hypothesized that this reverse arrangement would create a metabolic flux pull toward the product by increasing the protein concentration of the enzymes occurring in the latter part of the pathway. Under our production conditions, strain A, harboring pS0 and pY0, yielded 746 ± 18 mg/liter l-tyrosine (Table 3), consistent with the previous studies (25, 26). When we reversed the orientation of the genes in the module S0 or Y0, the production dropped significantly to ∼180 mg/liter in either case (data not shown). Changes of this magnitude in the product titer resulting from reversing the order of genes in the operon have been reported previously (2). In their construction of the taxadiene biosynthetic pathway, Ajikumar et al. (2) observed that when the order of the genes of geranyl geranyl pyrophosphate synthase (GGPS) and taxadiene synthase (TS) was reversed in the operon, opposite to the sequence of the reaction mechanism, the production increased by 2- to 3-fold.
Analysis of pathway enzyme and intermediate levels.
In order to rapidly identify pathway bottlenecks in strain A, we performed LC-MS-based analyses of pathway enzymes and intermediates. By targeted proteomics analysis, the protein levels in the shikimate module were unchanged between the induced and uninduced samples because expression was constitutive, under the control of PLtetO-1 with no tetR (Fig. 1C). In contrast, the genes in the tyrosine module were expressed under the control of Plac-UV5, and strong induction of protein production by IPTG was observed (Fig. 1C). However, the size of the increase in protein level decreased the further away the ORF was from the promoter (i.e., a 14-fold increase in TyrB but only a 2-fold increase in AroK, the protein product of the final gene in the operon). Consistent with the plasmid copy number, enzymes in the shikimate module were produced at lower levels than those in the tyrosine module. The targeted proteomics results indicated that YdiB and AroB were produced at very low levels, suggesting that they would be good targets for subsequent engineering efforts.
To complement the targeted proteomics analysis, we analyzed the levels of each of the pathway intermediates in strain A plus the precursors E4P and PEP. We used an LC-ESI-TOF MS approach to identify the intermediates and precursors based on their retention times and accurate mass measurements. Results from the metabolite analysis (Table 4) showed an accumulation of intermediates preceding AroK in the pathway: SHIK, DHS, DHQ, and the side product QUIN (Fig. 1A), which occurred due to the quinate dehydrogenase activity of YdiB (28). Knop and colleagues reported the hydroaromatic equilibrium between these intermediates and quinate (18), thus reducing flux to shikimate and downstream metabolites. There was no other significant accumulation of pathway intermediates. These data, along with the proteomic analysis, point to at least one bottleneck between AroB and AroK in the pathway.
Table 4.
Hydroaromatic equilibrium and intermediate levels accumulated in E. coli MG1655 harboring various shikimate modules constructed in the studya
| Module | Level (mg/liter) |
|||
|---|---|---|---|---|
| QUIN | DHQ | DHS | SHIK | |
| S0 | 106 ± 1 | 36 ± 1 | 588 ± 13 | 79 ± 2 |
| S1 | 165 ± 1 | 51 ± 1 | 1,029 ± 6 | 194 ± 2 |
| S2 | 242 ± 4 | 56 ± 1 | 1,227 ± 65 | 278 ± 28 |
| S3 | 2 ± 0 | 5 ± 0 | 70 ± 6 | 510 ± 18 |
| S4 | 3 ± 1 | 8 ± 0 | 113 ± 3 | 759 ± 39 |
| S5 | 11 ± 1 | 4 ± 0 | 46 ± 4 | 273 ± 20 |
Equilibrium exists between SHIK and the two preceding intermediates, DHQ and DHS, and the side product QUIN, whose formation is catalyzed by YdiB (28). It occurs immediately after the formation of DAHP and prior to the formation of shikimate 3-phosphate, the first intermediate in the tyrosine module (Fig. 1A). The hydroaromatic levels were determined under the conditions described in Table 3.
Optimization of the shikimate module.
From the initial proteomic analysis, significant improvement to flux through the pathway was made by making improvements to the protein levels encoded on the shikimate plasmid. Since the levels of YdiB and AroB were very low and intermediates produced or consumed by these enzymes accumulated, we targeted them for further engineering. To increase protein levels and hopefully to address the bottlenecks leading to the conversion of shikimate to shikimate-3-phosphate (S3P) catalyzed by AroK, a new plasmid (pS1) was constructed with the shikimate biosynthetic genes from pS0 cloned into pBbB5c (http://registry.jbei.org), which has a higher-copy-number origin of replication (Fig. 2A and Table 2). Metabolite analysis of the shikimate pathway intermediates (Table 4) for the strain harboring pS1 showed 194 ± 2 mg/liter shikimate, an increase of greater than 100% in shikimate production relative to the original construct, pS0. The precursors DHQ and DHS, as well as the side product QUIN, increased to a lesser extent, by factors of 42, 75, and 56%, respectively. However, when SRM analysis was performed to determine protein production from the new plasmid, neither modification to the plasmid (i.e., increasing the copy number) improved YdiB or AroB levels significantly (Fig. 2B), which only compounded the problems associated with metabolic flux to shikimate.
Since multiple attempts to modify the plasmid construct did not improve YdiB and AroB levels, we modified the gene sequences directly. Analysis of the aroB nucleotide sequence revealed several rare codons at the beginning of the gene. In particular, its third codon, AGG, has a frequency of occurrence of 1.6 out of 1,000 (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=83333); no other gene in this study has a rare codon with an occurrence frequency lower than 1.6 that is close to the initiation start site. To improve aroB expression, the first 15 codons were optimized by removing rare codons (16, 44), generating aroBop, which was used to construct pS2 (Fig. 2A and Table 2). Targeted proteomic analysis indicated that codon optimization of aroB improved the production of AroB (Fig. 2B), and shikimate pathway metabolite analysis showed increased accumulation of the downstream metabolites DHQ, DHS, and SHIK and the side product QUIN (Table 4); however, the bottleneck at YdiB remained. In place of YdiB, the isoenzyme AroE (5) was substituted in pS3 (Fig. 2A). Subsequently, protein analysis indicated that aroE produced much more protein than ydiB (Fig. 2B). Moreover, metabolite analysis indicated that AroE did not favor the formation of quinate (Table 4), consistent with its specificity for shikimate (5, 28). Compared to the original shikimate module (in pS0), shikimate production using pS3 increased approximately 5-fold (Table 4). Nevertheless, the total flux through S3, as indicated by the molar sum of DHQ, DHS, SHIK, and QUIN, decreased compared to those of YdiB-containing modules S1 and S2, which could be due to the feedback inhibition on AroE by shikimate (7, 18) and recently proposed AroE-catalyzed production of gallic acid from dehydroshikimate (30).
To determine if any new bottlenecks were created by the preceding engineering efforts, we individually expressed the genes of the shikimate module (aroE, aroD, aroB, aroG*, ppsA, and tktA) on pBbB5c in a strain also harboring the shikimate module S0 (on pS0) and the tyrosine module Y1 (on pY1) (strain B) (Table 3). In this case, l-tyrosine production relative to the two-plasmid strain was measured (Table 5). The results indicated that the first three enzymes in the pathway were rate-limiting steps and that increasing even one of these enzymes at a time yielded greater than 14% improved l-tyrosine production. Interestingly, these three genes (aroG*, ppsA, and tktA) catalyze the formation of the pathway precursors PEP and E4P and the first committed intermediate in the pathway, DAHP (Fig. 1A). In order to further increase the expression of aroG*, ppsA, and tktA on the shikimate module S3, two different promoters were inserted 5′ of aroG* to increase the levels of the three genes furthest from the promoter (Fig. 2A). When the constitutive PLtetO-1 was inserted 5′ of aroG* (S4), shikimate production increased by 50% (Table 4). However, proteomic analysis indicated that only the production of PpsA and TktA increased, but not that of AroG*, compared to expression from pS3 (Fig. 2B). Strangely, when a regulated Ptrc preceded by an E. colirrnB T1 terminator, to prevent readthrough from the first promoter, was inserted 5′ of aroG* (pS5), the shikimate titer decreased by ∼46% ± 4% (Table 4). This most likely occurred because protein production from the module decreased uniformly by a factor of ∼4 (Fig. 2B). Even more, expression decreased with increasing IPTG concentration (data not shown).
Table 5.
Combinatorial analyses of the shikimate module for improving l-tyrosine production in strain B
| Plasmid coexpresseda | % Improvementb |
|---|---|
| pBbB5c::ydiB | 4.4 ± 0.2 |
| pBbB5c::aroE | 9.9 ± 0.6 |
| pBbB5c::aroD | 10.9 ± 0.7 |
| pBbB5c::aroBop | 3.4 ± 0.2 |
| pBbB5c::aroG | 33.5 ± 1.6 |
| pBbB5c::tktA | 20.2 ± 1.1 |
| pBbB5c::ppsA | 14.5 ± 1.0 |
The description of plasmid pBbB5c can be found in Table 1. The genes ydiB through tktA were individually cloned into pBbB5c and transformed into strain B.
The percent improvement in l-tyrosine production was calculated based on the l-tyrosine titer of strain B.
Optimization of the tyrosine module and production analysis.
Based on the improvements observed with increasing protein levels for the shikimate module, a similar approach was used to improve expression of AroA and AroK at the end of the operon in the tyrosine module (Fig. 3A). We first increased the expression of AroK, since its protein level was relatively low compared to the other four enzymes in the operon (Fig. 1C) and because it catalyzes the conversion of shikimate to S3P, which is known to be a rate-limiting step (25). However, repeated attempts to insert a second promoter 5′ of the gene or to modify the RBS of aroK failed consistently, which might have been be due to plasmid instability, as reported previously for an aroK-containing plasmid (37).
Consequently, we reconsidered using AroL, which has higher affinity for shikimate than does AroK (46), despite the earlier observations regarding AroK (25). Substitution of aroL for aroK in the original tyrosine plasmid (pY1 for pY0) yielded ∼900 mg/liter of l-tyrosine, an increase of 20% when paired with pS0 (strain B) (Table 3). The substitution also resulted in higher cell density, and as such, the l-tyrosine production/gram of dry cell weight ratios (mgTyr/gCDW) for strains A and B did not significantly differ (Table 3). Interestingly, the use of AroL in strain B eliminated the accumulation of shikimate and other metabolites involved in the hydroaromatic equilibrium (data not shown). Moreover, the proteomic analysis indicated that protein levels in the aroL-containing pY1 were higher than those in the aroK-containing pY0 (Fig. 3B). These results suggest that the major bottleneck existed at AroK in the original l-tyrosine-producing strain. However, when pY1 was cotransformed with the best shikimate-producing plasmid, pS4, the l-tyrosine titer increased by 28% to 1.15 g/liter (Table 3), indicating that improvements in metabolic flux to shikimate yielded a significant impact on l-tyrosine production. To improve the l-tyrosine production further, two different promoters were inserted after the third ORF, aroC, on the tyrosine plasmid to increase expression of aroA and aroL. As with the shikimate plasmid, first PLtetO-1 was inserted between aroC and aroA, creating pY2. Interestingly, when pY2 was transformed into cells harboring pS4, only 908 mg/liter of l-tyrosine was produced after 24 h (Table 3), indicating that addition of the second promoter reduced l-tyrosine production compared to the strain harboring pS4 and pY1. Proteomic analysis suggests that this decreased production occurred due to reduction in AroA production, despite the additional promoter, and also TyrB, TyrA*, and AroC in the preceding operon (Fig. 3B). A second double-operon tyrosine plasmid (pY3) was constructed to incorporate an E. coli rrnB T1 terminator 5′ of Ptrc, which is 5′ of aroA. Over twice as much AroL and more than five times as much AroA were produced by cells harboring pY3 than by cells harboring pY2 (Fig. 3B); however, AroC levels remained quite low, and TyrB and TyrA* decreased even further. When this plasmid was transformed into cells harboring pS4, we obtained a titer of 1.5 g/liter after 24 h of growth and 2.2 ± 0.4 g/liter after 48 h, significantly higher than that of the single-operon plasmid (Table 3). This titer corresponds to an overall yield of 0.44 g l-tyrosine/g glucose fed, which is 80% of the theoretical yield (Table 3). The increase in l-tyrosine production caused by the increase in AroA and AroL expression in pY3, despite smaller amounts of AroC, TyrA*, and TyrB, indicated that both AroA and AroL are rate-limiting enzymes in the lower half of the l-tyrosine biosynthetic pathway.
DISCUSSION
We have engineered an efficient l-tyrosine production system using E. coli engineered with two modules that convert central metabolic intermediates into l-tyrosine. Expression of the genes in the tyrosine modules was optimized using two LC-MS technologies to elucidate the levels of enzymes and metabolic intermediates in the biosynthetic pathway and various replication origins, promoters, transcription terminators, and gene and enzyme variants. Unlike previous studies, we expressed all of the genes in the pathway instead of a selective few targets. Using LC-MS-based targeted proteomics and metabolite profiling, we were able to monitor the levels of all of the pathway enzymes and intermediates simultaneously, allowing us to quickly identify the bottlenecks and to adjust the gene expression to optimize the metabolic flux for production of l-tyrosine. The result of our work is a modular production system comprised of two dual-operon plasmid-based modules optimized for expression of shikimate and l-tyrosine, respectively. Compared to previous strategies, which selectively chose one or two genes to modify on the chromosome or plasmid, the modular system we developed here is optimized for each half of the tyrosine biosynthetic pathway, which could be readily used for expression in different E. coli host strains without a need for chromosomal mutations, i.e., deletion of tyrR, pheA, or pheL from the host strain. Because the system was constructed in two pieces (e.g., divided at shikimate), it is also possible to use the modules independently.
Our analyses unambiguously revealed several bottlenecks of the shikimate pathway not previously known and led to engineering efforts that significantly increased l-tyrosine production to 80% of the theoretical yield. Metabolite profiling indicated that DHQ, DHS, and SHIK accumulated in the initial strain, suggesting that the main bottleneck in the first part of the pathway was due to YdiB. Efforts to increase YdiB levels improved shikimate production; however, the levels of DHQ and DHS increased proportionally. These data support observations in the literature that YdiB possesses both quinate and shikimate dehydrogenase activities (10, 23) and that the hydroaromatic equilibrium between DHQ, DHS, SHIK, and the side product quinate limits production of shikimate and downstream metabolites. The molar ratios of DHQ, DHS, SHIK, and QUIN, calculated from the amounts expressed by all of the shikimate modules containing YdiB, pS0 to pS3, were 1.0:21.4:4.0:3.4. When aroE was substituted for ydiB, quinate could not be detected significantly in the culture, and DHS was efficiently converted to SHIK. The molar ratio between DHS and SHIK calculated from pS3 through pS5 was 1:7.
Potential loss in the metabolic flux due to the feedback inhibition of AroE by shikimate and formation of gallic acid (7, 30) was avoided by replacing aroK with aroL on the plasmids containing the l-tyrosine pathway. Previously, it has been shown that overexpression of AroK is better than that of AroL to increase l-tyrosine production (25); however, limitations from YdiB could have precluded any improvements from AroL. The metabolite analysis presented above suggests the highest l-tyrosine production from that system (∼700 mg/liter) could be limited due to inefficient conversion to shikimate even if high levels of YdiB are present.
SRM analysis of protein levels provided valuable insight into gene expression from both plasmids. The SRM data for the initial constructs (Fig. 1C) are consistent with our experimental observation that genes farther away from the promoter are induced to a lesser extent by IPTG than those closer to the promoter. Thus, insertion of a second promoter in the plasmid following the first three genes improved protein production from the genes 3′ of the promoter. However, inserting PLtetO-1 alone, without a terminator 5′ of the promoter, did not increase production of the protein encoded by the gene directly following that promoter; the second and third genes following the promoter were expressed at a higher level, as is evident in both the shikimate and tyrosine modules S4 and Y1. Inserting a trc promoter, with an E. coli rrnB T1 terminator 5′ of the promoter to prevent readthrough, increased production of AroA and AroL in the tyrosine module but, oddly, resulted in a general repression of the genes in the shikimate module S5.
It is interesting that our production data and SRM analysis are consistent with a previous study in which pulse-feeding experiments and statistical analysis identified AroB, AroA, and AroL as promising metabolic engineering targets to alleviate flux control in l-phenylalanine-producing strains (32). In our study, we demonstrated, using the shikimate operons, that AroB expression remained relatively low unless its first 15 codons were optimized. Additionally, using the tyrosine production module, we demonstrated that the large increase in production only occurred when expressions of AroA and AroL were upregulated by insertion of T1-Ptrc 5′ of aroAL in the tyrosine module Y3. Compared to the tyrosine module without the second promoter (Y1), l-tyrosine production doubled in strains harboring pY3 when used in conjunction with the plasmid (pS4) that harbors shikimate module S4. Nevertheless, the AroC level was relatively low in all constructs tested, suggesting that further improvements could be achieved by increasing AroC levels. In the end, with all the improvements we have made to the shikimate pathway in the two bioperon modules, we successfully achieved an l-tyrosine yield of 0.44 g/g glucose, which is 80% of the theoretical yield (43). The techniques we used, targeted proteomics and metabolite profiling, to optimize the l-tyrosine biosynthesis pathway can be readily applied to the biosynthesis of l-phenylalanine, l-tryptophan, and other complex metabolic pathways.
ACKNOWLEDGMENTS
This work was supported in part by the Synthetic Biology Engineering Research Center, which is funded by National Science Foundation award no. 0540879, and by the Joint BioEnergy Institute, which is funded by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231.
We thank James M. Carothers (QB3, CA) for his comments on the manuscript and Lisa Zhang (Agilent Technologies, Santa Clara, CA) for carrying out the purification of dehydroshikimate and dehydroquinate. D.J. thanks Nanyang Technological University, Singapore, for funding his 2-year stay in J.D.K.'s laboratory and Heng-Phon Too at the National University of Singapore, Singapore, for a brief rotation in his laboratory prior to engagement in this study.
Footnotes
Published ahead of print 21 October 2011
REFERENCES
- 1. Ahn JO, et al. 2008. Exploring the effects of carbon sources on the metabolic capacity for shikimic acid production in Escherichia coli using in silico metabolic predictions. J. Microbiol. Biotechnol. 18:1773–1784 [PubMed] [Google Scholar]
- 2. Ajikumar PK, et al. 2010. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson JC, et al. 2010. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bongaerts J, Kramer M, Muller U, Raeven L, Wubbolts M. 2001. Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3:289–300 [DOI] [PubMed] [Google Scholar]
- 5. Chaudhuri S, Coggins JR. 1985. The purification of shikimate dehydrogenase from Escherichia coli. Biochem. J. 226:217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chavez-Bejar MI, et al. 2008. Metabolic engineering of Escherichia coli for l-tyrosine production by expression of genes coding for the chorismate mutase domain of the native chorismate mutase-prephenate dehydratase and a cyclohexadienyl dehydrogenase from Zymomonas mobilis. Appl. Environ. Microbiol. 74:3284–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dell KA, Frost JW. 1993. Identification and removal of impediments to biocatalytic synthesis of aromatics from d-glucose: rate-limiting enzymes in the common pathway of aromatic amino acid biosynthesis. J. Am. Chem. Soc. 115:11581–11589 [Google Scholar]
- 8. Dosselaere F, Vanderleyden J. 2001. A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Crit. Rev. Microbiol. 27:75–131 [DOI] [PubMed] [Google Scholar]
- 9. Draths KM, et al. 1992. Biocatalytic synthesis of aromatics from d-glucose—the role of transketolase. J. Am. Chem. Soc. 114:3956–3962 [Google Scholar]
- 10. Duncan K, Chaudhuri S, Campbell MS, Coggins JR. 1986. The overexpression and complete amino acid sequence of Escherichia coli 3-dehydroquinase. Biochem. J. 238:475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eudes A, et al. 2011. Production of tranilast [N-(3′,4′-dimethoxycinnamoyl)-anthranilic acid] and its analogs in yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 89:989–1000 [DOI] [PubMed] [Google Scholar]
- 12. Gavini N, Pulakat L. 1991. Role of translation of the pheA leader peptide coding region in attenuation regulation of the Escherichia coli pheA gene. J. Bacteriol. 173:4904–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gosset G. 2009. Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 20:651–658 [DOI] [PubMed] [Google Scholar]
- 14. Herrmann KM, Weaver LM. 1999. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:473–503 [DOI] [PubMed] [Google Scholar]
- 15. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 16. Humphreys DP, et al. 2000. High-level periplasmic expression in Escherichia coli using a eukaryotic signal peptide: importance of codon usage at the 5′ end of the coding sequence. Protein Expr. Purif. 20:252–264 [DOI] [PubMed] [Google Scholar]
- 17. Ikeda M. 2006. Towards bacterial strains overproducing l-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 69:615–626 [DOI] [PubMed] [Google Scholar]
- 18. Knop DR, et al. 2001. Hydroaromatic equilibration during biosynthesis of shikimic acid. J. Am. Chem. Soc. 123:10173–10182 [DOI] [PubMed] [Google Scholar]
- 19. Kramer M, et al. 2003. Metabolic engineering for microbial production of shikimic acid. Metab. Eng. 5:277–283 [DOI] [PubMed] [Google Scholar]
- 20. Lee SK, Keasling JD. 2005. A propionate-inducible expression system for enteric bacteria. Appl. Environ. Microbiol. 71:6856–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leuchtenberger W, Huthmacher K, Drauz K. 2005. Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69:1–8 [DOI] [PubMed] [Google Scholar]
- 22. Li K, Frost JW. 1999. Microbial synthesis of 3-dehydroshikimic acid: a comparative analysis of d-xylose, l-arabinose, and d-glucose carbon sources. Biotechnol. Prog. 15:876–883 [DOI] [PubMed] [Google Scholar]
- 23. Lindner HA, et al. 2005. Site-directed mutagenesis of the active site region in the quinate/shikimate 5-dehydrogenase YdiB of Escherichia coli. J. Biol. Chem. 280:7162–7169 [DOI] [PubMed] [Google Scholar]
- 24. Lutke-Eversloh T, Santos CN, Stephanopoulos G. 2007. Perspectives of biotechnological production of l-tyrosine and its applications. Appl. Microbiol. Biotechnol. 77:751–762 [DOI] [PubMed] [Google Scholar]
- 25. Lutke-Eversloh T, Stephanopoulos G. 2008. Combinatorial pathway analysis for improved l-tyrosine production in Escherichia coli: identification of enzymatic bottlenecks by systematic gene overexpression. Metab. Eng. 10:69–77 [DOI] [PubMed] [Google Scholar]
- 26. Lutke-Eversloh T, Stephanopoulos G. 2007. l-Tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75:103–110 [DOI] [PubMed] [Google Scholar]
- 27. Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michel G, et al. 2003. Structures of shikimate dehydrogenase AroE and its paralog YdiB. A common structural framework for different activities. J. Biol. Chem. 278:19463–19472 [DOI] [PubMed] [Google Scholar]
- 29. Minami H, et al. 2008. Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U. S. A. 105:7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muir RM, et al. 2011. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 75:555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oldiges M, Kunze M, Degenring D, Sprenger GA, Takors R. 2004. Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnol. Prog. 20:1623–1633 [DOI] [PubMed] [Google Scholar]
- 33. Olson MM, et al. 2007. Production of l-tyrosine from sucrose or glucose achieved by rapid genetic changes to phenylalanine-producing Escherichia coli strains. Appl. Microbiol. Biotechnol. 74:1031–1040 [DOI] [PubMed] [Google Scholar]
- 34. Patnaik R, Liao JC. 1994. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl. Environ. Microbiol. 60:3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patnaik R, Zolandz RR, Green DA, Kraynie DF. 2008. l-Tyrosine production by recombinant Escherichia coli: fermentation optimization and recovery. Biotechnol. Bioeng. 99:741–752 [DOI] [PubMed] [Google Scholar]
- 36. Redding-Johanson AM, et al. 2011. Targeted proteomics for metabolic pathway optimization: application to terpene production. Metab. Eng. 13:194–203 [DOI] [PubMed] [Google Scholar]
- 37. Rood JI, Sneddon MK, Morrison JF. 1980. Instability in tyrR strains of plasmids carrying the tyrosine operon: isolation and characterization of plasmid derivatives with insertions or deletions. J. Bacteriol. 144:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santos CN, Stephanopoulos G. 2008. Melanin-based high-throughput screen for l-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 74:1190–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sariaslani FS. 2007. Development of a combined biological and chemical process for production of industrial aromatics from renewable resources. Annu. Rev. Microbiol. 61:51–69 [DOI] [PubMed] [Google Scholar]
- 40. Sato F, Inui T, Takemura T. 2007. Metabolic engineering in isoquinoline alkaloid biosynthesis. Curr. Pharm. Biotechnol. 8:211–218 [DOI] [PubMed] [Google Scholar]
- 41. Shine J, Dalgarno L. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 71:1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sprenger GA. 2007. From scratch to value: engineering Escherichia coli wild type cells to the production of l-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 75:739–749 [DOI] [PubMed] [Google Scholar]
- 43. Varma A, Boesch BW, Palsson BØ. 1993. Biochemical production capabilities of Escherichia coli. Biotechnol. Bioeng 42:59–73 [DOI] [PubMed] [Google Scholar]
- 44. Wang H, O'Mahony DJ, McConnell DJ, Qi SZ. 1993. Optimization of the synthesis of porcine somatotropin in Escherichia coli. Appl. Microbiol. Biotechnol. 39:324–328 [DOI] [PubMed] [Google Scholar]
- 45. Weiner MP, Costa GL. 1994. Rapid PCR site-directed mutagenesis. PCR Methods Appl. 4:S131–S136 [DOI] [PubMed] [Google Scholar]
- 46. Whipp MJ, Pittard AJ. 1995. A reassessment of the relationship between aroK- and aroL-encoded shikimate kinase enzymes of Escherichia coli. J. Bacteriol. 177:1627–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yi J, Draths KM, Li K, Frost JW. 2003. Altered glucose transport and shikimate pathway product yields in E. coli. Biotechnol. Prog. 19:1450–1459 [DOI] [PubMed] [Google Scholar]
- 48. Yi J, Li K, Draths KM, Frost JW. 2002. Modulation of phosphoenolpyruvate synthase expression increases shikimate pathway product yields in E. coli. Biotechnol. Prog. 18:1141–1148 [DOI] [PubMed] [Google Scholar]