Abstract
This study compared the three most commonly used assays for detecting Cryptosporidium sp. infections in cell culture: immunofluorescent antibody and microscopy assay (IFA), PCR targeting Cryptosporidium sp.-specific DNA, and reverse transcriptase PCR (RT-PCR) targeting Cryptosporidium sp.-specific mRNA. Monolayers of HCT-8 cells, grown in 8-well chamber slides or 96-well plates, were inoculated with a variety of viable and inactivated oocysts to assess assay performance. All assays detected infection with low doses of flow cytometry-enumerated Cryptosporidium parvum oocysts, including infection with one oocyst and three oocysts. All methods also detected infection with Cryptosporidium hominis. The RT-PCR assay, IFA, and PCR assay detected infection in 23%, 25%, and 51% of monolayers inoculated with three C. parvum oocysts and 10%, 9%, and 16% of monolayers inoculated with one oocyst, respectively. The PCR assay was the most sensitive, but it had the highest frequency of false positives with mock-infected cells and inactivated oocysts. IFA was the only infection detection assay that did not produce false positives with mock-infected monolayers. IFA was also the only assay that detected infections in all experiments with spiked oocysts recovered from Envirochek capsules following filtration of 1,000 liters of treated water. Consequently, cell culture with IFA detection is the most appropriate method for routine and sensitive detection of infectious Cryptosporidium parvum and Cryptosporidium hominis in drinking water.
INTRODUCTION
Cryptosporidium spp. are intracellular parasites that infect the epithelial cells lining the luminal surfaces of the digestive and respiratory tracts of a wide variety of animal hosts. The genus contains over 20 named species, which infect a variety of vertebrates, and over 60 genotypes (16, 29). Cryptosporidium parvum and Cryptosporidium hominis are the species most often responsible for human infections. Other species that are infrequently isolated from humans include C. meleagridis, C. canis, C. felis, C. suis, C. ubiquitum, C. muris, and a few wildlife genotypes (28). The disease is usually self-limiting in otherwise healthy humans, but persistent infection can contribute to mortality in individuals with weakened immune systems.
Almost 2 decades after the largest waterborne disease outbreak in the United States resulted in over 400,000 Cryptosporidium infections and contributed to the deaths of approximately 150 people (15), public health and regulatory agencies are still trying to determine the risk from Cryptosporidium oocysts in drinking water. An important element in accurately assessing this risk is measuring the prevalence of infectious Cryptosporidium oocysts in drinking water. According to one study, 1.4% of conventionally treated water samples contained infectious oocysts and 27% of surface water treatment plants in the United States released infectious oocysts in their treated water at least once during the 3-year study (1). The results translated to an annual cryptosporidiosis risk of 52 infections per 10,000 people, which is much higher than the 1 in 10,000 goal for annual risk of infection adopted by the U.S. Environmental Protection Agency (USEPA). A previous study detected infectious oocysts using in vitro cell culture coupled with a PCR assay targeting a Cryptosporidium-specific heat shock protein gene (hsp70) (14). Other assays have also been used for detecting Cryptosporidium infection in cell culture. These include an immunofluorescence microscopy assay (IFA) (23) and reverse transcriptase PCR (RT-PCR) targeting hsp70 mRNA (21). The various methods have been used to detect infectious Cryptosporidium spp. in filter backwash water, untreated source waters, and reclaimed wastewater (5, 8, 14), to demonstrate equivalency of cell culture with mouse assays (21, 24), and to evaluate UV inactivation of oocysts (10, 18).
If a cell culture method is used to assess the prevalence of infectious oocysts in drinking water and the resulting data are used to make operational, treatment, or regulatory decisions, it is imperative that the method be sensitive to low, environmentally relevant oocyst concentrations with minimal false-negative or false-positive results. However, there has been no rigorous comparison of the various infectivity detection assays and relatively little standardization of assay conditions or procedures. Therefore, the objective of this study was to compare the three most commonly used assays to detect Cryptosporidium infection in HCT-8 cell culture (IFA, PCR, and RT-PCR). Sensitivity, reproducibility, and frequency of false positives were evaluated with low doses of flow cytometry-enumerated viable oocysts and with oocysts exposed to a variety of inactivating agents. The assays were evaluated with two isolates of C. parvum (Iowa and Moredun) and a C. hominis isolate and were compared in two laboratories (Metropolitan Water District of Southern California [MWDSC] and Texas AgriLife Research Center [AgriLife]) using large numbers of replicate cell monolayers. In addition, the infectivity assays were evaluated with oocysts that were recovered from large volumes of drinking water using USEPA Method 1623 (26).
MATERIALS AND METHODS
Sources and quality control of Cryptosporidium sp. oocysts.
Mouse-propagated C. parvum oocysts (Iowa isolate) were obtained from Waterborne, Inc. (New Orleans, LA). Oocysts from the same lot were shipped to both analysis laboratories at the same time. Oocysts of the C. hominis TU728 isolate propagated in gnotobiotic pigs were obtained from Giovanni Widmer (Tufts University, North Grafton, MA). Sheep-propagated oocysts of the C. parvum Moredun isolate were supplied by Steve Wright (Moredun Research Institute, Penicuik, Scotland).
The quality of each lot of oocysts was tested prior to their use. This included microscopic observation of wet mounts and Gram-stained samples, inoculation of nutrient broth, Sabouraud dextrose plates, and m-Endo plates to assess the level of bacteria and fungi in oocyst preparations, staining with fluorescein isothiocyanate (FITC)-labeled anti-Cryptosporidium antibody (Cellabs, Brookvale, Australia) to assess gross physical structure, and inoculating RPMI 1640 cell culture medium containing penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin B (0.625 μg/ml), and 2% fetal bovine serum (FBS) to ensure that bacteria and fungi in oocyst preparations would not contaminate cell cultures. The majority of oocysts used for infectivity assays (>80%) were less than 4 weeks old (postshedding).
Enumerating oocysts.
For enumeration using well slides, 10 aliquots of an oocyst suspension were placed on 2-well Superstick slides (Waterborne) and allowed to dry overnight at room temperature. FITC-labeled anti-Cryptosporidium antibody (Cellabs) was applied to the wells, and the slides were incubated at 37°C in a humidified chamber for 30 min. Following rinsing in phosphate-buffered saline (PBS) and application of antifade mounting medium (Waterborne), slides were examined by epifluorescence microscopy with 485-nm excitation and 510-nm dichroic and 520-nm emission filters. The coefficient of variation (CV) for all microscopic oocyst enumerations in this study was ≤16%.
Oocysts were also enumerated and sorted by flow cytometry at the Wisconsin State Laboratory of Hygiene (WSLH; Madison, WI). Oocysts were sorted into individual microcentrifuge tubes for each experiment. Once returned to the analysis laboratories, oocysts were inoculated onto cell monolayers without any further dilution. The average relative standard deviation for flow cytometry-enumerated oocyst doses of 1 to 100 was 3.7% (n = 182 tubes of oocysts). A trip control tube of oocysts was included in every shipment of oocysts sent to WSLH for sorting by flow cytometry. These oocysts were then shipped back to the analysis laboratories and used alongside the flow cytometry-enumerated oocysts in infectivity assays to determine whether the shipping process affected oocyst infectivity.
Inactivated oocysts.
Gamma-irradiated (0.5 kGy) EasySeed oocysts were obtained from BTF (North Ryde, Australia), and some lots of C. parvum Iowa oocysts were also gamma irradiated (0.5 kGy) by Food Technology Services, Inc. (Mulberry, FL). Oocysts were also irradiated with approximately 60 mJ/cm2 of low-pressure UV using previously described collimated beam equipment and procedures (18). For heat inactivation, oocysts were incubated at 70°C for 30 min. Oocysts were also disrupted by three cycles of freezing in liquid nitrogen for 2 min followed by thawing at 95°C for 1 min. Inactivated oocysts were enumerated and sorted into individual tubes by flow cytometry (WSLH) before being used for infectivity assays.
Cell culture procedures.
Monolayers of the human ileocecal adenocarcinoma HCT-8 cell line (ATCC CCL-244; American Type Culture Collection, Manassas, VA) were maintained at both analysis laboratories. Stock cells were maintained in 150-cm2 flasks and passaged twice a week in RPMI 1640 cell culture medium with GlutaMAX (Invitrogen, Carlsbad, CA) containing 5% heat-inactivated fetal bovine serum (FBS; HyClone, Logan, UT), penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin B (0.25 μg/ml), and 20 mM HEPES buffer. All stock cells were maintained in this medium regardless of the assay eventually used to detect infections. Cells were passaged by adding 10 ml of Hanks' balanced salt solution (HBSS; Sigma, St. Louis, MO) containing 0.25% trypsin and 0.02% EDTA to the monolayer and incubating for 5 min at 37°C to release the cell monolayer from the flask. Trypsin was inactivated and removed by adding an equal volume of cell culture medium, centrifuging the cells at 160 × g for 5 min, and resuspending the cells in fresh cell culture medium. Cells were enumerated using a hemocytometer, and 4 × 106 cells per flask were inoculated into a 150-cm2 flask containing 50 ml fresh medium. The cells were maintained in a humidified incubator at 37°C and 5% CO2. Cells were not used beyond passage 30. Separate biological safety cabinets and incubators were used for uninfected stock cells and infected monolayers.
All newly thawed HCT-8 cell stocks were tested for the presence of contaminating Mycoplasma spp. by a direct DNA fluorochrome staining technique (Bionique Testing Laboratories, Saranac Lake, NY). Prior to testing, new batches of HCT-8 cells were passaged twice in maintenance medium without antibiotics to allow for maximum growth of Mycoplasma spp., if present. All cells used in this study were certified Mycoplasma negative before being used for infectivity assays and comparison experiments.
To prepare monolayers for infectivity assays, a stock flask of HCT-8 cells was split into two 150-cm2 flasks of fresh maintenance medium. One flask (the assay flask, seeded with 5 × 106 cells) was used to set up 96-well plates (Corning Inc., Corning, NY) for the PCR and RT-PCR detection assays and 8-well Lab-Tek II chamber slides (Thermo Scientific, Rochester, NY) for IFA detection, while the second flask was used for subsequent cell passages. The assay flask was incubated for 42 to 52 h to achieve 80 to 100% confluence, and the monolayer was then removed from the flask by trypsinization as described above. Ninety-six-well plates were seeded with 100 μl of RPMI maintenance medium containing 5 × 105 cells/ml, while 8-well slides were seeded with 500 μl of RPMI maintenance medium containing 4 × 105 cells/ml. After 42 to 52 h of incubation at 37°C, the maintenance medium was removed and monolayers were inoculated with oocysts suspended in the inoculation medium specific to each detection assay (described below).
Oocyst pretreatment.
Individual aliquots of oocysts were pretreated prior to infection of the HCT-8 monolayers. Oocysts were incubated in acidified HBSS, pH 2.0, containing 1% trypsin (AHBSS/T) for 1 h at 37°C (6). Tubes were vortexed vigorously every 15 min. The oocysts were washed twice by adding fresh cell culture inoculation medium, centrifuging at 13,000 × g for 2 min, and then discarding the supernatant as previously described (22). The final pellet of oocysts was resuspended in cell culture inoculation medium and used to inoculate monolayers.
For oocysts that were eluted from Envirochek HV capsules (described below), the oocysts were removed from water concentrates by immunomagnetic separation (IMS; Dynabeads, Invitrogen). Oocysts were detached from the magnetic beads prior to inoculating monolayers by incubating in AHBSS/T for 1 h at 37°C with vigorous vortexing every 15 min. The sample was then placed on the magnet, and the supernatant containing the oocysts was transferred to a fresh tube. An aliquot of fresh AHBSS/T was added to the magnetic beads, and the sample was incubated at 37°C for an additional 5 min. The tube was placed on the magnet, and the supernatant was transferred to the tube containing the rest of the sample. The sample was then washed twice in fresh cell culture inoculation medium to remove all traces of trypsin before inoculating monolayers.
IFA-based infectivity detection assay.
Cell culture maintenance medium was removed from HCT-8 monolayers grown to at least 80% confluence in 8-well chamber slides, and 100 μl of inoculation medium was added to each well to prevent the monolayers from drying out during the inoculation procedure. Following incubation in AHBSS/T as described above, oocysts were resuspended in inoculation medium and added to the well in a final volume of 400 μl. The inoculated chamber slides were then incubated at 37°C for 64 to 72 h in a 5% CO2 humidified incubator. Cell culture inoculation medium for the IFA was RPMI 1640 plus GlutaMAX containing 10% heat-inactivated FBS, 20 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.625 μg/ml amphotericin B, and 100 μg/ml kanamycin.
After incubation, inoculation medium was removed from the wells and the monolayers were immediately fixed with methanol for 10 min. At MWDSC, slide chambers were removed and the monolayers were incubated in blocking buffer (2% goat serum and 0.002% Tween 20 in PBS) for 30 min at room temperature. After removal of blocking buffer, rat anti-Cryptosporidium sporozoite antibody (SporoGlo; Waterborne) diluted 1:500 in 1× PBS was added to monolayers. Slides were incubated in a humidified chamber for 45 min at room temperature. After four washes in 1× PBS, goat anti-rat IgG FITC-labeled antibody (Sigma) diluted 1:150 in 1× PBS was added to monolayers, and slides were incubated for an additional 45 min. The antibody was removed with four washes in 1× PBS, slides were dried at room temperature, coverslips were applied over mounting medium (Waterborne), and slides were examined using epifluorescence microscopy (485-nm excitation, 520-nm emission).
At the AgriLife laboratory, slide chambers were left intact on the slide for staining and microscopy as previously described (22). Stained monolayers with the chambers still in place were examined on an inverted epifluorescence microscope.
Infection detected by IFA was defined as a monolayer that contained at least one focus of life stages. A focus of life stages was defined as at least three life stages within an area ≤175 μm in diameter. An individual life stage was defined as an intracellular life cycle stage ≥1 μm and ≤10 μm in diameter, with the correct apple green color and intensity of fluorescence, and not an obvious fluorescent artifact. This definition was based on measurement of 80 infectious foci generated by control oocysts at the start of the study.
PCR-based infectivity detection assay.
Cell culture maintenance medium was removed from HCT-8 monolayers grown to at least 80% confluence in 96-well plates, and 50 μl fresh inoculation medium was immediately added to each well. Following incubation in AHBSS/T, oocysts were resuspended in inoculation medium and added to the well in a final volume of 100 μl. Inoculated monolayers were then incubated at 37°C for 64 to 72 h in a 5% CO2 humidified incubator. Cell culture inoculation medium for the PCR assay was RPMI 1640 plus GlutaMAX containing 10% heat-inactivated FBS, 15 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 12.5 μg/ml tetracycline, 0.5 μg/ml amphotericin B, 50 mM glucose, 35 μg/ml ascorbic acid, 1 μg/ml folic acid, 4 μg/ml 4-aminobenzoic acid, and 2 μg/ml calcium pantothenate (adapted from reference 27).
Following incubation, cell culture medium was removed from the wells, monolayers were washed five times with 1× PBS, and DNA was extracted using a QIAamp DNA minikit by following the manufacturer's instructions (Qiagen, Valencia, CA). Extracted DNA was eluted off the column in 50 μl of 100 μM Tris and 10 μM EDTA, pH 8, at 70°C.
The entire 50 μl of DNA was used in a 100-μl amplification reaction mixture. DNA was amplified by either conventional PCR at MWDSC or quantitative PCR (qPCR) at AgriLife using previously described procedures (14, 6). Conventional PCR used AmpliTaq Gold DNA polymerase with GeneAmp 10× PCR Gold buffer (Applied Biosystems, Foster City, CA), 2.5 mM MgCl2, 200 μM each dATP, dCTP, and dGTP, 800 μM dUTP, 1.5 mg/ml bovine serum albumin (BSA; New England BioLabs, Ipswich, MA), 0.01 U/μl uracil DNA glycosylase (UDG; New England BioLabs), and 0.20 μM (each) forward and reverse CPHSPT2 primers targeting the Cryptosporidium heat shock protein 70 gene (hsp70) (14). Amplification conditions were as follows: 10 min at 25°C; 10 min at 95°C; 55 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s; and a final extension for 5 min at 72°C. The amplification reagents and conditions were the same for qPCR with the addition of the TaqMan probe (6). Amplification products were detected by real-time TaqMan PCR and gel electrophoresis at the AgriLife laboratory and by electrophoresis only at MWDSC. Only gel electrophoresis results were used for method comparisons.
RT-PCR infectivity detection assay.
Cell culture maintenance medium was removed from HCT-8 monolayers grown to at least 80% confluence in 96-well tissue culture plates, and fresh growth medium was added to each well. Following incubation in AHBSS/T, oocysts were resuspended in growth medium and added to the well in a final volume of 100 μl. Inoculated monolayers were then incubated at 37°C for 64 to 72 h in a 5% CO2 humidified incubator. Cell culture growth medium for the RT-PCR assay was RPMI 1640 plus GlutaMAX containing 2% heat-inactivated FBS, 20 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.625 μg/ml amphotericin B, and 100 μg/ml kanamycin.
Following incubation, growth medium was removed from monolayers, which were then washed twice with 1× PBS. Cells were lysed, and RNA was extracted using an RNeasy 96 RNA extraction kit by following the manufacturer's instructions (Qiagen). Residual DNA that may have carried over in the RNA extraction was removed by treating the extraction filters twice with 80 μl DNase I (1,800 kU/ml) for 20 min at 37°C. The RNA was eluted in 80 μl of RNase-free water.
Reverse transcription and PCR were performed as described previously (21) using murine leukemia virus (MuLV) reverse transcriptase (2.5 U/μl), RNase inhibitor (1 U/μl), oligo(dT)16 primers (2.5 μM), and 10 μl of RNA in a 20-μl reaction mixture. The entire 20 μl reverse transcription reaction mixture was used as the template for the amplification reaction. The amplification reaction mixture consisted of Platinum Taq 10× PCR buffer, 1.5 mM MgCl2, 200 μM (each) dATP, dCTP, dGTP, and dUTP, 0.25 μM forward and reverse CPHSP2 primers targeting the Cryptosporidium heat shock protein 70 gene (hsp70) (19), 0.01 U/μl UDG, and 0.025 U/μl Platinum Taq polymerase (Invitrogen). Amplification conditions were as follows: 10 min at 25°C; 10 min at 95°C; 45 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 60 s; and a final extension for 5 min at 72°C. To detect potential false positives due to DNA carryover in the RNA extraction, a second RT-PCR for each RNA sample omitted MuLV reverse transcriptase. Amplification products were visualized by gel electrophoresis.
Control infections.
The infectivity of each lot of oocysts was assessed prior to their use in comparison experiments by inoculating six HCT-8 monolayers in 8-well chamber slides with 1,000 oocysts each. These oocysts were enumerated by fluorescent staining and microscopy according to USEPA Method 1623 (26) with an acceptable CV of ≤16% (n = 10). Inoculated cell cultures were incubated at 37°C for 64 to 72 h and stained using the IFA, and the infectious foci were enumerated by fluorescence microscopy. Oocysts of C. parvum were used for assay comparison experiments only if their mean infectivity in six replicate monolayers was ≥5% (≥50 infectious foci per monolayer inoculated with 1,000 oocysts).
Mock infection controls.
Precise numbers of flow cytometry-sorted oocysts were added to previously uninoculated cell monolayers after the washing steps of the PCR and RT-PCR detection assays and immediately prior to methanol fixation of monolayers for the IFA. These monolayers were then processed using each of the three detection assays. This allowed for the evaluation of false positives due to known numbers of oocysts remaining on monolayers after the initial assay sample processing steps. An incubation period to allow oocysts to attach to monolayers was not necessary because lysis reagents were immediately added to the monolayers for the PCR and RT-PCR infection detection assays. This approach may have given a slight advantage to IFA detection, since false positives with that assay depend on oocyst and sporozoite attachment. However, it is unlikely that unexcysted oocysts and sporozoites will cause significant false positives with the cell culture-IFA (CC-IFA) due to the distinctive microscopic characteristics of infectious foci.
Spiked water samples.
Treated drinking water (990 liters) from a conventional water treatment plant was filtered through Envirochek HV capsules by following the procedure described in USEPA Method 1623 (26). Sodium thiosulfate was added upstream of the filter using a proportioning injector to inactivate residual chlorine. In each laboratory, 10 liters of drinking water spiked with viable flow cytometry-enumerated oocysts was then filtered through the same capsules. Filter capsules were pretreated with 5% (wt/vol) sodium hexametaphosphate to remove mineral deposits that can interfere with oocyst elution (4). Oocysts were eluted according to Method 1623, dissociated from the magnetic beads with AHBSS/T, and inoculated onto HCT-8 monolayers. Infectivity assays were performed as described above. Similar filters were also spiked with 50 and 100 flow cytometry-sorted viable and gamma-irradiated C. parvum oocysts. These filters were blind-spiked by an independent third party (CH Diagnostics, Berthoud, CO), and the spiking details were not released to the analysis laboratories until after the infectivity assay was completed and results were recorded.
RESULTS
IFA, PCR, and RT-PCR were compared for detecting infectious Cryptosporidium sp. oocysts in HCT-8 cell culture. Comparisons included sensitivity with C. parvum and C. hominis, frequency of false positives with inactivated oocysts, reliability for detecting low numbers of infectious oocysts, and detection of infectious oocysts recovered from large volumes of drinking water. Most of the comparisons used flow cytometry-enumerated and -sorted oocysts, which required shipping oocysts to WSLH. Trip controls included with all oocyst shipments indicated that 48 h of transit did not affect their infectivity.
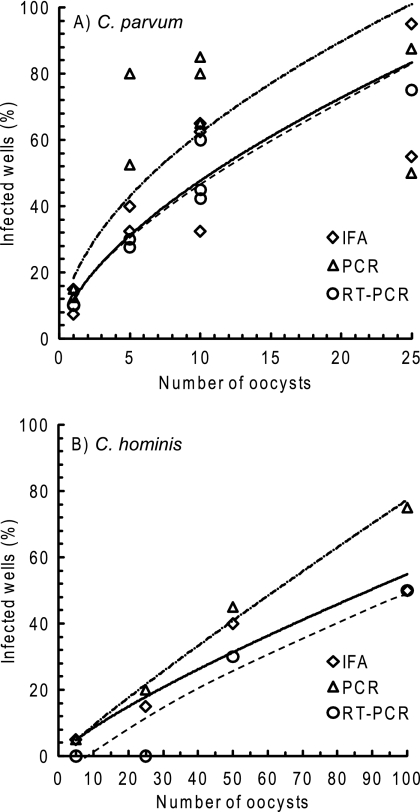
All three methods detected infection with the Iowa isolate of C. parvum and the C. hominis TU728 isolate (Fig. 1). All methods also detected infection with the Moredun isolate of C. parvum (results not shown). Infection was quantified as the percentage of cell culture wells that developed infection (W%) when multiple wells (Wn) were inoculated with the same dose of flow cytometry-enumerated and -sorted oocysts. The highest W% values for both C. parvum and C. hominis were obtained using the PCR detection assay (Fig. 1).
Fig 1.
Comparison of three assays for detecting infection of HCT-8 cells with low numbers of Cryptosporidium parvum (A) and Cryptosporidium hominis (B) oocysts. Infectivity detection assays were immunofluorescence microscopy assay (IFA; solid lines), PCR targeting hsp70 DNA (dot-dashed lines), and reverse transcriptase PCR (RT-PCR) targeting hsp70 mRNA (dashed lines). Oocysts were enumerated by flow cytometry, and replicate doses were inoculated into 10 cell culture wells. Each data point is the average of two experiments replicated in two laboratories for C. parvum and one experiment replicated in two laboratories for C. hominis, so a total of 20 to 40 wells were inoculated with each oocyst dose. Lines are best-fit curves to illustrate overall similarities and differences between assays.
The percentage of cell culture wells that developed infection was less than 100% because not all oocysts in each lot are capable of initiating infection, even when freshly shed. The infectivity of each oocyst lot was measured prior to its use for assay comparison. One thousand oocysts were inoculated onto six replicate HCT-8 monolayers, and, following 72 h of incubation, infection was detected using the IFA and infectious foci were enumerated by epifluorescence microscopy. Infectivity was calculated as the percentage of infectious foci per inoculum oocyst. Oocysts of C. parvum were used for comparing assays only if their initial infectivity (averaged across six monolayers) was ≥5% (≥50 infectious foci per monolayer). In this study, initial oocyst infectivity in control infections was 7 to 19% for C. parvum and 1 to 11% for C. hominis.
With all three detection assays, C. hominis was less infectious than C. parvum, with only <10 to 20% of cell culture wells developing infection when inoculated with 25 C. hominis oocysts compared to 33 to 88% with the same dose of C. parvum oocysts. However, although C. hominis oocysts produced fewer infectious foci, there was no significant difference between the number of life stages per C. hominis focus (23.5 ± 11.6 life stages/focus, mean ± standard deviation [SD]; n = 48) and the number per C. parvum focus (30 ± 11.7 life stages/focus; n = 39). This indicates that individual C. hominis oocysts are capable of developing cell culture infection foci similar to C. parvum oocysts but that fewer C. hominis oocysts in each lot are capable of initiating an infection, possibly due to inefficient production of fully mature oocysts by the pig propagation model. Previous studies also indicated that C. hominis is less infectious than C. parvum in HCT-8 cells (21).
The frequency of false positives was assessed with a variety of inactivated oocysts (Table 1). Potential false-positive results were generated by the cell culture-IFA (CC-IFA) in 2 out of 10 cell culture wells inoculated with 100 freeze/thaw-inactivated oocysts in one of the analysis laboratories. These objects did not have the appearance of typical infectious foci but still met the definition of infection (correct size and fluorescence characteristics). These foci could have resulted from dead, intact, or partial sporozoites released by inactivated oocysts that were unable to initiate infection but were stained with the antisporozoite antibody. The CC-RT-PCR assay produced false-positive detection only for mock infections but had the highest overall frequency of positives from these controls (8 out of 70 mock infections). In contrast, the CC-PCR assay generated false positives with oocysts inactivated by freeze/thaw, UV, and gamma irradiation, in addition to 4 out of 70 wells that were mock infected with 25 or 100 oocysts (Table 1). These results indicate that the CC-PCR assay is the most prone to false positives due to oocysts that stick to cell monolayers but do not initiate infection and may explain the higher W% values for the PCR assay (Fig. 1).
Table 1.
False-positive detection of infectivity with inactivated Cryptosporidium parvum oocysts
| Inactivation method | No. of oocysts inoculated | Wna | Cell culture wells positiveb (%) by: |
||
|---|---|---|---|---|---|
| IFA | PCR | RT-PCR | |||
| 70°C, 30 min | 10 | 20 | 0 | 0 | 0 |
| 100 | 20 | 0 | 0 | 0 | |
| 3×(LiqN2, 2 min/ 95°C, 1 min)c | 10 | 20 | 0 | 0 | 0 |
| 100 | 20 | 10 | 5 | 0 | |
| UV irradiation, ∼60 mJ/cm2 | 10 | 20 | 0 | 25 | 0 |
| 100 | 20 | 0 | 60 | 0 | |
| 0.5 kGy gamma irradiation | 100 | 20 | 0 | 55 | 0 |
| 100 | 15 | 0 | 45 | 0 | |
| 100 | 20 | 0 | 10 | 0 | |
| None (mock infectionsd) | 100 | 20 | 0 | 5 | 15 |
| 100 | 20 | 0 | 10 | 13e | |
| 25 | 10 | 0 | 0 | 0 | |
| 25 | 10 | 0 | 10 | 0 | |
| 25 | 10 | 0 | 0 | 20 | |
| None (unseeded wells) | 0 | 20 | 0 | 0 | 0 |
| None (untreated viable oocysts) | 10 | 40 | 62 | 65 | 45 |
Number of cell culture wells inoculated with each dose of oocysts.
Average of replicate experiments performed in two laboratories.
Three cycles of freezing in liquid nitrogen for 2 min followed by thawing at 95°C for 1 min.
Mock infections involved inoculating oocysts onto cell monolayers and then immediately processing the monolayers according to each of the detection assays.
Wn = 15.
To assess the performance of each method with low oocyst doses, viable C. parvum oocysts were enumerated and sorted by flow cytometry and inoculated onto cell monolayers at doses of 1 oocyst to 5 oocysts per monolayer. Although there was variability in the proportion of wells that developed infection, aggregate data from both laboratories demonstrated that CC-IFA detected infection in 43% and 25% of wells inoculated with five and three oocysts, respectively, compared to 60% and 51% for PCR and 32% and 23% for RT-PCR (Table 2). However, the CC-PCR assay again had the highest frequency of false positives with mock-infected monolayers. For monolayers that were inoculated with a single flow cytometry-sorted oocyst, the IFA, RT-PCR assay, and PCR assay detected infection in 9%, 10%, and 16% of cell monolayers, respectively (Wn = 100 for IFA and PCR and 89 for RT-PCR).
Table 2.
Reproducibility in detecting low doses of infectious Cryptosporidium parvum
| Infection type | No. of oocysts/wella | Wn | Labb | Cell culture wells positive (%) by: |
||
|---|---|---|---|---|---|---|
| IFA | PCR | RT-PCR | ||||
| Viable oocysts | 5 | 50 | A | 48 | 60 | 33† |
| 5 | 50 | B | 38 | 72 | 30 | |
| Mean | 43 | 60 | 32 | |||
| 3 | 40 | A | 42 | 70 | 15 | |
| 3 | 40 | A | 25 | 48 | 30 | |
| 3 | 40 | B | 12 | 40 | 18 | |
| 3 | 40 | B | 20 | 48 | 30 | |
| Mean | 25 | 51 | 23 | |||
| 1 | 50 | A | 10 | 18 | 13c | |
| 1 | 50 | B | 8 | 14 | 6 | |
| Mean | 9 | 16 | 10 | |||
| Mock | 3 | 40 | A | 0 | 48 | 3 |
| 3 | 40 | A | 0 | 12 | 0 | |
| 3 | 40 | B | 0 | 5 | 0 | |
| 3 | 40 | B | 0 | 0 | 0 | |
| Mean | 0 | 16 | 1 | |||
Oocysts were enumerated by flow cytometry and inoculated into multiple cell culture wells (Wn) in both laboratories.
Lab A, MWDSC; lab B, AgriLife.
Wn = 39 wells.
An advantage of CC-IFA detection is the ability to directly visualize and quantify infections by enumerating the number of infectious foci that develop on each monolayer assuming a single infectious oocyst will generate one focus of life stages. Both the proportion of wells that developed infection (InfW) and the average number of infectious foci on infection-positive wells increased approximately 10-fold as the inoculum dose increased from 1 oocyst to 100 oocysts (Table 3). Infectivity based on the number of infectious foci per inoculum oocyst remained relatively constant (mean = 10.9%), regardless of the size of the inoculum dose.
Table 3.
Quantification of Cryptosporidium parvum infections detected by IFA
| No. of oocysts/well | No. of inoculated wells (Wn) | Total no. of oocysts (O) | No. of positive wells (Wpos) | InfWa (%) | Mean no. of infectious foci in positive wells ± SD | Total no. of infectious foci (C) | InfOb (%) |
|---|---|---|---|---|---|---|---|
| 1 | 40 | 40 | 4 | 10 | 1.0 ± 0 | 4 | 10 |
| 3 | 160 | 480 | 40 | 25 | 1.3 ± 0.5 | 51 | 10.6 |
| 5 | 40 | 200 | 18 | 45 | 1.3 ± 0.5 | 23 | 11.5 |
| 10 | 80 | 800 | 50 | 63 | 1.6 ± 0.9 | 80 | 10 |
| 25 | 20 | 500 | 19 | 95 | 3.3 ± 2.1 | 63 | 12.6 |
| 100 | 40 | 4,000 | 40 | 100 | 10.8 ± 5.3 | 431 | 10.8 |
Infectivity based on number of cell culture wells that developed infection according to the formula (Wpos/Wn) × 100.
Infectivity based on the total number of infectious foci as a proportion of the total number of oocysts inoculated at each dose according to the formula (C/O) × 100.
The three cell culture detection assays were also compared using oocysts recovered from spiked water samples. Following filtration of 990 liters of treated drinking water through Envirochek HV capsules, a further 10 liters of water spiked with 56 viable flow cytometry-enumerated oocysts was filtered through the same capsule. Oocysts were recovered using Method 1623 with the addition of sodium hexametaphosphate, and following dissociation from magnetic beads in AHBSS/T, the entire sample was inoculated onto a single HCT-8 monolayer. The average infectivity of these oocysts prior to spiking Envirochek HV capsules was 11% based on the number of infectious foci detected by CC-IFA per inoculum oocyst. Therefore, only 6 out of the 56 spiked oocysts were predicted to be infectious. All three assays in both laboratories detected infection with these oocysts (Table 4). In a separate experiment, Envirochek HV capsules were blind-spiked with 12 and 6 flow cytometry-enumerated infectious oocysts (100 and 50 total oocysts, respectively) by an independent laboratory and shipped to both analysis laboratories for processing using modified Method 1623 and cell culture. The entire recovered sample was inoculated onto a single monolayer. The average infectivity of these oocysts prior to spiking Envirochek HV capsules was 12.6% based on the number of infectious foci per inoculum oocyst. The CC-IFA detected infection with samples spiked with 12 and 6 infectious oocysts in both laboratories. However, only one of the two analysis laboratories detected infection by CC-PCR with the same number of oocysts, and neither laboratory detected infection with 6 infectious oocysts using the CC-RT-PCR assay (Table 4). None of the methods detected infections from filters spiked with 100 oocysts inactivated by gamma radiation.
Table 4.
Detection of infectious Cryptosporidium parvum oocyst spikes recovered from 1,000 liters of drinking water using modified USEPA Method 1623
| No. of infectious oocystsa | Oocyst lot | Lab | Wnb | Detection of infection by: |
||
|---|---|---|---|---|---|---|
| IFA | PCR | RT-PCR | ||||
| 12 (100) | KE | A | 1 | + | −c | + |
| 12 (100) | KE | B | 1 | + | + | + |
| 6 (56) | KU | A | 1 | + | + | + |
| 6 (56) | KU | B | 1 | + | + | + |
| 6 (50) | KE | A | 1 | + | −c | −c |
| 6 (50) | KE | B | 1 | + | + | −c |
| 100 gamma irradiated | KE | A | 1 | − | − | − |
| 100 gamma irradiated | KE | B | 1 | − | − | − |
| 0 | A | 1 | − | − | − | |
| 0 | B | 1 | − | − | − | |
Based on CC-IFA infectivity assessment of the oocyst stock prior to seeding filters (total numbers of oocysts are in parentheses).
Number of cell culture wells inoculated with each dose of oocysts.
False-negative result.
DISCUSSION
Many cell lines, medium formulations, assay formats (chamber slides, coverslips, 24-well and 96-well plates), oocyst inoculation procedures, and methods for detecting infection have been used to assess and quantify infectivity of Cryptosporidium sp. oocysts. However, water industry applications have focused primarily on HCT-8 cells, and the three most common assays for detecting or quantifying infections in cell culture are IFA, PCR, and RT-PCR. Each of these detection assays has been used for a variety of applications, all using the HCT-8 cell line. Equivalency between CD-1 mice and cell culture infectivity was demonstrated using the RT-PCR detection assay targeting hsp70 mRNA for fresh and UV-exposed oocysts (18, 21). Detection of infections in HCT-8 cells using quantitative PCR (qPCR) targeting the 18S rRNA gene was also equivalent to a mouse assay for measuring oocyst inactivation by pulsed UV (7). In addition, there was a statistically significant correlation between CC-IFA and a mouse assay for assessing the efficacy of chlorine dioxide and UV inactivation of oocysts (24). The effects of chemical and UV disinfection, alum coagulation, and dissolved air flotation were evaluated using qPCR targeting the 18S rRNA gene to detect infections in HCT-8 cells (11, 12). Infectious oocysts were detected in 1.4% of treated drinking water samples using a CC-PCR assay targeting hsp70 DNA (1), and the same assay detected infectious oocysts in 4.5% of raw water samples and 7.4% of treatment plant filter backwash water samples (5, 14). Infectious oocysts were also detected in 40 to 50% of reclaimed water effluents using CC-IFA following recovery of oocysts by method 1623 (8, 17).
However, acceptance and adoption of the methods have been limited because there has been no rigorous comparison of the various assays used to detect and quantify infections in HCT-8 cell culture. A comparison of IFA and PCR for detecting infections in HCT-8 cells found that the PCR assay was less reproducible than IFA, particularly at low oocyst concentrations (9). However, the authors used oocysts with very low infectivity (0.1 to 2%, based on the number of infectious foci per inoculum oocyst). In the current study, the mean infectivity for oocysts used to compare assays was 14%. Other studies using the same definition of infectivity have reported C. parvum infectivity values of 8 to 9.5% (2), 10 to 22% (22), and 5 to 14% (20).
The CC-PCR assay had the highest frequency of false positives with inactivated oocysts and mock infections. It has previously been reported that this assay underestimates UV inactivation of oocysts (3), probably due to amplification of DNA in irradiated oocysts or sporozoites that remain on the cell monolayer but do not initiate infection. The qPCR version of the PCR assay used in the present study was also not recommended for disinfection studies because of the likely background signal obtained at the high oocyst inoculum densities typically used (6). Consequently, the apparently higher infection rates generated by the PCR detection assay may be attributed in part to its relatively high rate of false positives.
The intent of this study was to compare the three most commonly used infection detection assays as published. The published methods each used different oocyst inoculation medium formulations, and the assays were optimized using these formulations. Therefore, these same assay-specific medium formulations were used in the current study. In particular, the PCR detection assay used inoculation medium containing glucose and vitamin supplements. These supplements have been shown to increase Cryptosporidium parvum cell culture infection (25), which may partly explain the high detection levels observed for the PCR assay.
This study represents the first rigorous comparison of the three most commonly used assays for detecting Cryptosporidium sp. infection in HCT-8 cell culture. The results demonstrated that the CC-IFA is the most appropriate for use on a more widespread basis by the water industry. It generated the fewest false positives with inactivated oocysts and mock infections and detected infection with low numbers of oocysts (1 oocyst to 3 oocysts per monolayer). It also performed better than the other assays with oocysts recovered from spiked filters using Method 1623. In addition, it was the simplest method to perform, with the fewest processing steps. In an earlier study, CC-IFA accurately predicted the number of infectious oocysts in blind coded samples of purified oocyst suspensions (2). The authors suggested that water utilities with high oocyst prevalence in their source waters would benefit from assessing the infectivity of recovered oocysts to better ascertain the threat to public health from these waterborne oocysts. Importantly, both freshly confluent and aged HCT-8 cell monolayers can be used for the CC-IFA, making it logistically feasible for water quality and utility laboratories (22). Further, a variation of the CC-IFA used in the present study for the detection of both total and infectious oocysts in individual water samples was recently described (13).
Cell culture methods require the use of specialized equipment and materials that are not usually available in water utility laboratories. In addition, there is inherent variability in cell culture-based infectivity assays for Cryptosporidium (21). Nevertheless, the standardized cell culture-IFA described in this paper appears to be suitable for more widespread use by the water industry. Data on the prevalence of infectious Cryptosporidium in drinking water will be useful in refining assessments of the public health risk from waterborne Cryptosporidium.
ACKNOWLEDGMENTS
This work was funded by the Water Research Foundation.
We are grateful to Rebecca Hoffman of the Wisconsin State Laboratory of Hygiene for enumerating and sorting oocysts by flow cytometry and Greg Sturbaum at CH Diagnostics for spiking oocysts into filter capsules. We thank Joe Hernandez and Nicholas Garcia at Texas AgriLife Research Center and Katrin Hanley, Patty Huang, and Melinda Tan at the Metropolitan Water District of Southern California for assistance in sample processing. We also thank Theresa Slifko, John Albert, Ricardo De Leon, Mark LeChevallier, Marilyn Marshall, Jennifer Clancy, and Christobel Ferguson for helpful discussions throughout the study.
Footnotes
Published ahead of print 28 October 2011
REFERENCES
- 1. Aboytes A, et al. 2004. Detection of infectious Cryptosporidium in filtered drinking water. J. Am. Water Works Assoc. 96:88–98 [Google Scholar]
- 2. Bukhari Z, Holt DM, Ware MW, Schaefer FWIII. 2007. Blind trials evaluating in vitro infectivity of Cryptosporidium oocysts using cell culture immunofluorescence. Can. J. Microbiol. 53:656–663 [DOI] [PubMed] [Google Scholar]
- 3. Bukhari Z, LeChevallier M. 2003. Assessing UV reactor performance for treatment of finished water. Water Sci. Technol. 47:179–184 [PubMed] [Google Scholar]
- 4. Clancy JL, McCuin RM, Hargy TM. 2003. Recovery of Cryptosporidium oocysts from high-volume water samples. American Water Works Association, Denver, CO [Google Scholar]
- 5. Di Giovanni GD, et al. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture PCR. Appl. Environ. Microbiol. 65:3427–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Giovanni GD, LeChevallier MW. 2005. Quantitative-PCR assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 71:1495–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garvey M, Farrell H, Cormican M, Rowan N. 2010. Investigations of the relationship between use of in vitro culture-quantitative PCR and a mouse-based bioassay for evaluating critical factors affecting the disinfection performance of pulsed UV light for treating Cryptosporidium parvum oocysts in saline. J. Microbiol. Methods 80:267–273 [DOI] [PubMed] [Google Scholar]
- 8. Gennaccaro AL, McLaughlin MR, Wuintero-Betancourt W, Huffman DE, Rose JB. 2003. Infectious Cryptosporidium parvum oocysts in final reclaimed effluent. Appl. Environ. Microbiol. 69:4983–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Joachim A, Eckert E, Petry F, Bialek R, Daugschies A. 2003. Comparison of viability assays for Cryptosporidium parvum oocysts after disinfection. Vet. Parasitol. 111:47–57 [DOI] [PubMed] [Google Scholar]
- 10. Johnson AM, et al. 2005. UV inactivation of Cryptosporidium hominis as measured in cell culture. Appl. Environ. Microbiol. 71:2800–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keegan A, Daminato D, Saint CP, Monis PT. 2008. Effect of water treatment processes on Cryptosporidium infectivity. Water Res. 42:1805–1811 [DOI] [PubMed] [Google Scholar]
- 12. Keegan AR, Fanok S, Monis PT, Saint CP. 2003. Cell culture-TaqMan PCR assay for evaluation of Cryptosporidium parvum disinfection. Appl. Environ. Microbiol. 69:2505–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lalancette C, Di Giovanni GD, Prevost M. 2010. Improved risk analysis by dual direct detection of total and infectious Cryptosporidium oocysts on cell culture in combination with immunofluorescence assay. Appl. Environ. Microbiol. 76:566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. LeChevallier MW, et al. 2003. Comparison of Method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Appl. Environ. Microbiol. 69:971–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. MacKenzie WR, et al. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161–167 [DOI] [PubMed] [Google Scholar]
- 16. Nichols RAB, Conelly L, Sullivan CB, Smith HV. 2010. Identification of Cryptosporidium species and genotypes in Scottish raw and drinking waters during a one-year monitoring period. Appl. Environ. Microbiol. 76:5977–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quintero-Betancourt W, Gennaccaro AL, Scott TM, Rose JB. 2003. Assessment of methods for detection of infectious Cryptosporidium oocysts and Giardia cysts in reclaimed effluents. Appl. Environ. Microbiol. 69:5380–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rochelle PA, et al. 2004. Irreversible UV inactivation of Cryptosporidium spp. despite the presence of UV repair genes. J. Eukaryot. Microbiol. 51:553–562 [DOI] [PubMed] [Google Scholar]
- 19. Rochelle PA, et al. 1997. An assay combining cell culture with reverse transcriptase-PCR to detect and determine infectivity of waterborne Cryptosporidium parvum. Appl. Environ. Microbiol. 63:2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rochelle PA, Ferguson DM, Johnson AM, De Leon R. 2001. Quantitation of Cryptosporidium parvum infection in cell culture using a colorimetric in situ hybridization assay. J. Eukaryot. Microbiol. 48:565–574 [DOI] [PubMed] [Google Scholar]
- 21. Rochelle PA, et al. 2002. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 68:3809–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sifuentes LY, Di Giovanni GD. 2007. Aged HCT-8 cell monolayers support Cryptosporidium parvum infection. Appl. Environ. Microbiol. 73:7548–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slifko TR, Huffman DE, Rose JB. 1999. A most-probable-number assay for enumeration of infectious Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 65:3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Slifko TR, et al. 2002. Comparison of tissue culture and animal models for assessment of Cryptosporidium parvum infection. Exp. Parasitol. 101:97–106 [DOI] [PubMed] [Google Scholar]
- 25. Upton SJ, Tilley M, Brillhart DB. 1995. Effects of select medium supplements on in vitro development of Cryptosporidium parvum in HCT-8 cells. J. Clin. Microbiol. 33:371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. U.S. Environmental Protection Agency 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 815-R-05-002. Office of Research and Development, Government Printing Office, Washington, DC [Google Scholar]
- 27. Woods KM, Nesterenko MV, Upton SJ. 1995. Development of a microtitre ELISA to quantify development of Cryptosporidium parvum in vitro. FEMS Microbiol. Lett. 128:89–94 [DOI] [PubMed] [Google Scholar]
- 28. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]
- 29. Xiao L, Ryan UM. 2008. Molecular epidemiology, p 119–163 In Fayer R. (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, FL [Google Scholar]