Abstract
Leaf-nodulated plants are colonized by vertically inherited bacterial endosymbionts, which maintain symbioses throughout host generations. The permanent character of the interaction implies phylogenetic congruence between the host and the endosymbiont. However, the present population genetic study of Psychotria leptophylla provides evidence for a mixed symbiont transmission involving both vertical inheritance and horizontal transfers from the environment.
TEXT
Bacterial endophytes of leaf-nodulated plants are accommodated in specialized leaf nodule structures, and it is generally accepted that the bacteria are vertically transmitted to seeds (14, 16). To ensure passage of the symbiont from the host to the progenitors, a bacterial colony is maintained within the shoot tip, operating as an inoculum for infection of new developing reproductive structures and embryos (16). Generally, the establishment of vertical symbiont transmission has been suggested as a general key mechanism toward the evolution of a mutualistic relationship (5, 6). Consequently, heritable symbionts have been commonly found in many lineages that are involved in a wide variety of beneficial effects conferred on their host (1, 17, 18). The exact benefit of leaf-nodulated endosymbionts for the host still needs to be addressed. Most studies, however, have proposed that the endophytes are involved in phytohormone production, since they are essential for normal host development (16). All attempts to cultivate the bacterial partner failed, suggesting that the endosymbionts are absolutely dependent on the host for growth and survival (9, 16, 24). Recent studies using culture-independent PCR methods revealed the identity of the endophytes of all leaf-nodulated plants as Burkholderia (10–12, 24). More interestingly, it was also demonstrated that high host specificity occurs as a result of vertical symbiont transfer and that frequent host switches are observed, rejecting a long-term cospeciation (13). These results suggest a diffuse bacterium-plant interaction as an ancestral state of bacterial leaf symbiosis, followed by an evolution toward a more specific one-to-one interaction, where the bacterial associate became an obligate and vertically inherited endosymbiont of the host. The presumed transition from a free-living state to an obligate and heritable lifestyle of the bacterial partner will be investigated by analyzing the cospeciation pattern of the host and symbiont at a lower taxonomic level. In the present article, we focus on different populations of Psychotria leptophylla to provide novel and valuable information about the host specificity of the interaction at an intraspecific level and the transmission mode of the endosymbionts.
This study includes eight populations (A to H) of Psychotria leptophylla sampled at four localities in Cameroon: the Southwest, Littoral, Center, and South provinces. For all populations, we sampled two individuals at different localities with the exception of population G (Fig. 1; see also Table S1 in the supplemental material). Psychotria brachyanthoides, P. camerunensis, and P. darwiniana were used as an outgroup. All plant species with voucher and GenBank accession numbers are listed in Table S1. DNA extraction, PCR amplification, and sequencing follow the method of Lemaire et al. (13). Primer and temperature profiles are listed in Table S2. Purified PCR products were sent to Macrogen for sequencing (Macrogen Inc., Seoul, South Korea). Sequences were edited, assembled, and aligned in Geneious Pro v.5.1.7 (3), and phylogenetic analyses were done with maximum-likelihood (ML) and Bayesian inference (BI) methods. Data partitions were analyzed separately and combined. The congruency between DNA markers was tested with the parsimony-based incongruence length difference test (ILD test) (4) as implemented in the software program PAUP* v.4b10 (23). ML and BI analyses were conducted as described in previous studies (12, 13). For the BI analyses, the software program MrModeltest v.3.06 (20) selected an HKY+I model for the gltB, recA, and external transcribed sequence (ETS) data sets, a GTR+I model for the gyrB, lepA, and internal transcribed spacer (ITS) data sets, and a GTR+G model for the trpB data set. The cospeciation analyses between host and endosymbiont lineages were investigated with the software program TreeMap v.3.0β (2) as described previously (12), and the reconciliation analysis was performed under the default setting of event cost (codivergence = 0; duplication = host switch = 1). The significance of observed event types were estimated using a randomization test of 1,000 randomly generated trees. Quantitative PCR (qPCR) was conducted in a StepOne Plus apparatus (Applied Biosystems, Foster City, CA) using Fast SYBR green master mix (Applied Biosystems, Foster City, CA). The qPCR primers (see Table S2) were designed based on an alignment of gyrB clonal sequences of Psychotria kirkii var. hirtella and related Psychotria species generated from the study of Lemaire et al. (12). Primer specificity and quality were checked in silico using the software program Primer Express v.3.0 (Applied Biosystems). The reliability, specificity, and evaluation of the negative controls were done by melt curve analyses in the software program StepOne v.2.1 (Applied Biosystems). The relative abundance of the bacterial gyrB gene was measured in the vegetative (leaves and shoot tips) and reproductive (flowers and seeds) parts from Psychotria kirkii var. hirtella (plant accession no. BR 2000103624; National Botanic Garden of Belgium Living Plant Collections Database), which were sampled from fresh material and frozen in liquid nitrogen. Negative controls of corresponding vegetative and reproductive material were obtained from endosymbiont-free Psychotria carthagenensis (plant accession no. BR 19842833). The actin gene of the host plant was used for normalizing gyrB expression.
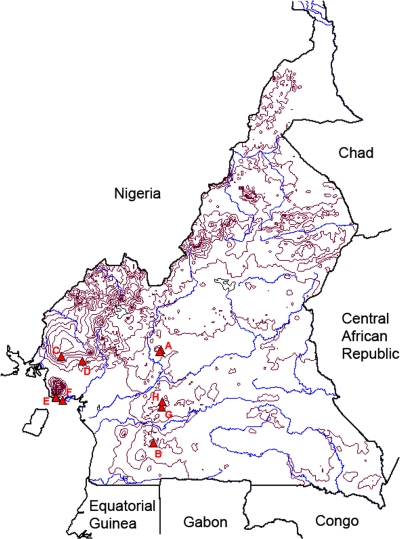
Fig 1.
Map of Cameroon with height contour lines (red) and rivers (blue). Red triangles show the geographical positions of the sampled individuals of the populations (A to H) of Psychotria leptophylla. A, Ngoro; B, Ngoakélé; C, Rumpi Hills; D, Mount Kupe; E, Bimbia-Bonadikombo; F, Mount Etinde; G, Nkolbisson; H, Mbam Minkom.
Leaf-nodulated plants are known to maintain the symbiont through all stages of the host's life cycle. This balanced association results in a complex developmental adaptation, which is morphologically well documented (14, 16). However, much work is still required to elucidate the mechanism of symbiont transmission in the seed-borne stage of the host plant; in leaf-nodulated Psychotria species, the exact position where the endosymbionts reside in the seeds remains unknown (16). To verify the hypothesis that the symbionts are present in the entire life cycle of the host plants, we examined their presence in all vegetative and reproductive stages of Psychotria kirkii var. hirtella with qPCR. The relative abundance of Burkholderia gyrB DNA is high in the leaves, shoot tips, and flower but low in the seeds (Table 1). This result supports findings of previous morphological studies and the hypothesis that the symbionts are vertically transmitted. In Psychotria leptophylla, the presence of endosymbionts in every plant part is also expected because different populations of either P. kirkii or P. leptophylla are characterized by a host-specific endosymbiont (13). More important, the small amount of bacterial DNA within the seeds might explain why it remains so difficult to observe endosymbionts within the seed stage of the plant life cycle (16).
Table 1.
Relative abundance of the endosymbiont gyrB gene in leaves, shoot tips, flowers, and seeds of leaf-nodulated Psychotria kirkii (two replicates) and nonnodulated Psychotria carthagenensis (negative control)a
| Sample type | Plant structure | Value of target abundance | Range |
|---|---|---|---|
| P. kirkii var. hirtella (replicate 1) | Leaf | 126.60 | 24.82–30.88 |
| Shoot | 24.03 | 94.58–152.51 | |
| Flower | 201.54 | 14.37–15.47 | |
| Seed | 1.00 | 0.09–0.10 | |
| P. kirkii var. hirtella (replicate 2) | Leaf | 160.61 | 125.84–136.53 |
| Shoot | 117.72 | 26.74–34.61 | |
| Flower | 9.76 | 0.02–0.03 | |
| Seed | 1.50 | 0.19–0.21 | |
| P. carthaginensis (control) | Leaf | 0.00 | 0.00–0.00 |
| Shoot | 0.00 | 0.00–0.00 | |
| Flower | 0.00 | 0.00–0.00 | |
| Seed | 0.00 | 0.00–0.00 |
The gyrB gene was normalized against the plant actin gene. The data presented here are three technical replicates from two biological replicates and were determined by the delta CT method.
In general, heritable and mutualistic endosymbionts are often found to exhibit cocladogenesis with their hosts (15, 27). A previous study (13), however, rejected a history of strict cospeciation in leaf-nodulated Psychotria species and suggested a recent shift toward a host-specific association and ongoing cospeciation. To test this hypothesis, we analyzed a five-gene endosymbiont and two-gene host data set, including 15 and 8 lineages and populations of Psychotria leptophylla, respectively. The observation of topologically similar gene trees and the absence of statistical incongruencies between the different data sets (for host DNA markers, the ILD test indicated congruent ITS and ETS data sets [P = 1.0]; Table 2 shows results for endosymbiont DNA markers) allowed for data combination. The combined phylogenetic analyses (Fig. 2) show that all Psychotria leptophylla populations harbored their own specific Burkholderia lineage, indicating vertical transmission of the endosymbionts. However, we found one population sampled on Mount Etinde (population F; Fig. 1) with different endosymbiont lineages, suggesting a horizontally acquired symbiont. Further cogent evidence for horizontal transfer was demonstrated by comparing the host and endosymbiont phylogeny; phylogenetic incongruencies were detected between the populations A, B, C, D, and F. A strict cospeciation pattern between the host and the endosymbiont was observed only for populations A, G, and H (Fig. 2). The topological differences observed between hosts and their endosymbionts were statistically confirmed by the cospeciation analysis in TreeMap v.3.0β. The reconciliation analysis introduced 16 cospeciation events, 12 duplications, and 6 host switches (total event cost of 18) (P < 0.001).
Table 2.
Results of ILD test
| Endosymbiont DNA marker | Congruency |
||||
|---|---|---|---|---|---|
| gltB | gyrB | lepA | recA | trpB | |
| gltB | 0.121 | 0.066 | 0.456 | 0.433 | |
| gyrB | 1.000 | 1.000 | 1.000 | ||
| lepA | 1.000 | 1.000 | |||
| recA | 1.000 | ||||
| trpB | |||||
Fig 2.
Comparison of the maximum-likelihood phylogenies of Psychotria host plants (left) and the Burkholderia endosymbiont (right). Maximum-likelihood bootstrap values and Bayesian posterior probabilities are shown above branches. Branch lengths represent the numbers of substitutions per site.
Several potential mechanisms might explain the observation of topological incongruencies between the host and the endosymbiont as a result of horizontally acquired symbionts. Horizontal symbiont transmission between populations of Psychotria leptophylla might occur via pollinators, acting as vectors of endosymbionts. During pollination, the developing pollen tube could deposit an “immigrant endosymbiont” into the ovule of a new host plant, thereby replacing the indigenous endosymbiont. However, this hypothesis may not explain symbiont/pollen transfer over long distances, as observed in this study (Fig. 2). An alternative hypothesis to explain horizontal transmission reflected by phylogenetic incongruencies is an intraspecific gene flow followed by repeated backcrossing, resulting in chloroplast introgression (21). Chloroplast introgression has been considered to be a major cause of differential transfer of cytoplasmic and nuclear genetic elements. In this case, the discrepancy between the two data sets (i.e., biparental inherited nuclear DNA of the host and maternal inherited DNA from the endosymbionts) might be the result of a constant pollen transfer from the indigenous population to an invaded population, which originated with a seed dispersal event. Our current results may be explained by this introgression hypothesis but fail to support previously observed intergenic symbiont transmission by hybridization of species from different leaf-nodulated Psychotria, Pavetta, and Sericanthe genera (13). Finally, external infections by soil bacteria provide the best scenario to explain the horizontal symbiont transmissions at intraspecific, interspecific, and intergenic taxonomic levels. Direct evidence for this hypothesis is the close relationship between some soil Burkholderia species and leaf-nodulated Burkholderia species (8, 13). Under this hypothesis, symbiont replacement by soil bacteria may occur when the host plant fails to transfer its original bacteria through the seeds. Indeed, a low number of endosymbionts was measured in the seeds (see above), which might occasionally be insufficient as a threshold for a vital endosymbiont colony. An ineffective symbiont inoculation of the seed during floral development results in bacterium-free dwarf plants, known as cripples, which occur naturally in a portion of every batch of seeds (16). In this study, natural crippled plants were obtained by germinating hundreds of seeds of Psychotria kirkii. Bacterium-free seedlings develop normally until the fourth pair of leaves, but then growth and differentiation cease and shoot tips degenerate to callus (Fig. 3). This static condition will persist for several years, after which seedlings will eventually perish (7, 25). Given the uncertain survival of crippled hosts, external reinfection by beneficial soil bacteria is required to restore normal growth and development. The observation of external reinfection of soil Burkholderia implies that the endosymbionts also occur freely in the soil and are not strictly obligate or dependent on their host. Consequently, the strict obligatory status of the endosymbionts needs to be revised.
Fig 3.
(A) Comparison of a nodulated (left) and crippled (right) phenotype of Psychotria kirkii. (B) Shoot tip of a nodulated plant with leaf nodules. (C) Degenerated shoot tip of a crippled plant showing a distorted leaf formation and missing leaf nodules.
Supplementary Material
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189 [DOI] [PubMed] [Google Scholar]
- 2.Charleston M, Page R.Treemap 2.0. Macintosh program for co-phylogenetic analysis. 1998. http://sydney.edu.au/engineering/it/∼mcharles/.
- 3. Drummond A, et al. 2010. Geneious v5.1. Biomatters, Auckland, New Zealand. [Google Scholar]
- 4. Farris JS, Kallersjo M, Kluge AG, Bult C. 1994. Testing significance of incongruence. Cladistics 10:315–319 [Google Scholar]
- 5. Frank SA. 1996. Host-symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 263:339–344 [DOI] [PubMed] [Google Scholar]
- 6. Genkai-Kato M, Yamamura N. 1999. Evolution of mutualistic symbiosis without vertical transmission. Theor. Popul. Biol. 55:309–323 [DOI] [PubMed] [Google Scholar]
- 7. Gordon JF. 1963. The nature and distribution within the plant of the bacteria associated with certain leaf-nodulated species of the families Myrsinaceae and Rubiaceae. Thesis. University of London, London, United Kingdom [Google Scholar]
- 8. Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5:446–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lebard S, Belin-Depoux M. 2003. Structure and ontogeny of foliar bacterial nodules of Ardisia crenata Sims (Myrsinaceae). Acta Bot. Gallica 150:19–33 [Google Scholar]
- 10. Lemaire B, et al. 2011. Identification, origin and evolution of leaf nodulating symbionts of Sericanthe (Rubiaceae). J. Microbiol. doi:10.1007/s12275011-1163-5. [DOI] [PubMed]
- 11. Lemaire B, Smets E, Dessein S. 2011. Bacterial leaf symbiosis in Ardisia (Myrsinoideae, Primulaceae): molecular evidence for host specificity. Res. Microbiol. doi:10.1016/j.resmic.2011.04.003. [DOI] [PubMed]
- 12. Lemaire B, et al. 2011. Identification of the bacterial endosymbionts in leaf nodules of Pavetta (Rubiaceae). Int. J. Syst. Evol. Microbiol. doi:10.1099/ijs.0.028019-0. [DOI] [PubMed]
- 13. Lemaire B, Vandamme P, Merckx VS, Smets E, Dessein S. 2011. Bacterial leaf symbiosis in angiosperms: host specificity without co-speciation. PLoS One 6:e24430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lersten NR, Horner HT. 1976. Bacterial leaf nodule symbiosis in angiosperms with emphasis on Rubiaceae and Myrsinaceae. Bot. Rev. 42:145–214 [Google Scholar]
- 15. Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. 2003. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol. Biol. Evol. 20:907–913 [DOI] [PubMed] [Google Scholar]
- 16. Miller IM. 1990. Bacterial leaf nodule symbiosis. Adv. Bot. Res. 17:163–234 [Google Scholar]
- 17. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 18. Moran NA, Plague GR, Sandstrom JP, Wilcox JL. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 100:14543–14548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817. [DOI] [PubMed] [Google Scholar]
- 21. Rieseberg LH, Wendel JF. 1993. Introgression and its consequences in plants, p 70–114. In Harrison R. (ed.), Hybrid zones and the evolutionary process. Oxford University Press, New York, NY [Google Scholar]
- 22. Reference deleted.
- 23. Swofford D. 2002. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 24. Van Oevelen S, De Wachter R, Vandamme P, Robbrecht E, Prinsen E. 2004. ‘Candidatus Burkholderia calva’ and ‘Candidatus Burkholderia nigropunctata’ as leaf gall endosymbionts of African Psychotria. Int. J. Syst. Microbiol. 54:2237–2239 [DOI] [PubMed] [Google Scholar]
- 25. von Faber FC. 1912. Das erbliche Zusammenleben von Bakterien und tropischen Pflanzen. Jahrb. Wiss. Bot. 51:285–375 [Google Scholar]
- 26. Reference deleted.
- 27. Wu D, et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.