Abstract
Activated sludge plants suffer frequently from the operational problem of stable foam formation on aerobic reactor surfaces, which can be difficult to prevent. Many foams are stabilized by mycolic acid-containing Actinobacteria, the mycolata. The in situ biocontrol of foaming using phages is an attractive strategy. We describe two polyvalent phages, GTE5 and GRU1, targeting Gordonia terrae and Gordonia rubrupertincta, respectively, isolated from activated sludge. Phage GRU1 also propagates on Nocardia nova. Both phages belong to the family Siphoviridae and have similar-size icosahedral heads that encapsulate double-stranded DNA genomes (∼65 kb). Their genome sequences are similar to each other but markedly different from those of other sequenced phages. Both are arranged in a modular fashion. These phages can reduce or eliminate foam formation by their host cells under laboratory conditions.
INTRODUCTION
Many activated sludge plants suffer from the generation of stable foam on the surfaces of the aerated reactors (13, 43, 44). This foam causes acute operational problems and may also pose environmental and health hazards (13, 43, 44). The foam is stabilized by highly abundant hydrophobic bacteria, including the mycolic acid-containing Actinobacteria, the mycolata (13, 23, 41). Many control measures have been described to eliminate these foams, but none are successful in all cases, which probably reflects the poor understanding of foam microbial ecology (13). The more frequently reported mycolata in foam include members of the genus Gordonia (13), and among those cultured from foam is Gordonia terrae (26, 27). One environmentally attractive approach to prevent foaming is to apply lytic phages to reduce the numbers of the causative organisms below the threshold required for stable foam formation (33, 47, 49). A similar philosophy has been proposed, and in some cases adopted, to treat antibiotic-resistant organisms in clinical infections (24) and to eliminate pathogenic bacteria during food processing (30).
Such phages are obtained readily from activated sludge. Thomas et al. (47) isolated 17 mycolata phages, 7 of which lysed Gordonia species. Each displayed the attractive feature of targeting a broad range of hosts. The characterization data they presented were restricted to descriptions of virion morphology and genome type, but it has been suggested that for applications like the one discussed here it is important to understand phage genome composition (30). Consequently, we have characterized the genome sequences of two of the Gordonia phages (GTE5 and GRU1) isolated originally by Thomas et al. (47) and have assessed their abilities to control the stabilization of foam caused by their host bacteria.
MATERIALS AND METHODS
Bacterial strains used in the study.
The mycolata bacterial strains used and the methods for their growth are listed by Petrovski et al. (34).
Phage purification, host range determination, and characterization.
The GTE5 and GRU1 phages were isolated from the Carrum (Victoria, Australia) and Loganholme (Queensland, Australia) treatment plants, respectively, as detailed by Thomas et al. (47). Phage recovery and purification, achieved with their respective hosts, G. terrae and Gordonia rubropertincta, were described by Petrovski et al. (34), as were the methods used for determining their host ranges and phage morphologies by transmission electron microscopy (TEM). Single-step phage growth experiments were performed as described previously (1, 34).
DNA isolation and sequencing.
Prior to DNA isolation, the two phages were precipitated separately using NaCl/polyethylene glycol (PEG) 8000, and phage DNA was isolated using SDS/proteinase K, as previously described (34). The genomes of GTE5 and GRU1 were sequenced by Genoseq (University of California—Los Angeles, Los Angeles, CA), and pyrosequencing reads were assembled separately, as described previously (34). The two resulting single contigs obtained for each phage had a minimum of 50 times sequence coverage.
Genome annotation.
The genomes of GTE5 and GRU1 were annotated using the Integrative Services for Genomic Analysis (http://isga.cgb.indiana.edu; 22) interface with Egatis (31), followed by manual inspection of all gene predictions.
Putative open reading frames (ORFs) longer than 90 bases were predicted using Glimmer3 (12) with the iterative process described by Delcher et al. (11) to enhance predictive accuracy. All predicted start codons were inspected manually for the presence of putative ribosomal binding sites and adjusted as required.
Sequence similarity searches were performed using BLAST X against a nonredundant database, including data sourced from the NCBI, Swiss-Prot, and Protein Data Bank (PDB) databases using a significance value of 1e−04. The BLAST X results were used as input for the BLAST-Extend-Repraze algorithm (http://sourceforge.net/projects/ber/) to identify potential frameshifts or point mutations. Protein domain searches were performed using hmmpfam (http://hmmer.janelia.org/) against the PFAM (2) and TIGRFAM HMM (19) databases to identify protein family or domain matches. Each ORF was also checked manually using the conserved domain database (CDD) (16). Transmembrane domains were predicted using DAS (dense alignment surface method)-transmembrane prediction (9; http://www.sbc.su.se/∼miklos/DAS/). ORFs were also screened for the presence of lipoprotein motifs (3).
The presence of tRNA and transfer mRNA (tmRNA) was screened for using RNAmmer (25) and tRNAscan-SE (29, 40). Putative rho-independent transcriptional terminators were identified with TransTerm (15). Global alignment of the genomes was performed using LAGAN (5).
Mass spectroscopy of phage proteins.
For the identification of structural proteins, the purified phages (∼1010 PFU) were precipitated from concentrated stocks using ZnCl2 (39) to remove PEG. The pellet was reduced using 100 mM dithiothreitol (DTT) and heat denatured (100°C for 5 min). Samples were loaded into 12% SDS-polyacrylamide gels for electrophoresis prior to staining with Coomassie brilliant blue. All visible proteins were excised from the gel and pooled. The excised proteins were trypsin digested (42), followed by analysis on electrospray ionization-time of flight mass spectrometry (ESI-TOF MS) by the Australian Proteome Analysis Facility (APRF) Sydney.
Effects of GTE5 and GRU1 phages on foam stability.
Triplicate 20-ml cultures of each bacterial host (with the A600 adjusted to 1.0) were incubated at room temperature overnight with or without the addition of either the GTE5 or GRU1 phage (multiplicity of infection [MOI] = 0.3). The foaming potential was assessed using the laboratory scale foaming apparatus described by Stratton et al. (45). The foaming apparatus consists of a glass cylinder with a sintered glass disc fitted to its base and connected to a rotameter. A 20-ml aliquot of the bacterial broth culture (with the A600 adjusted to 1.0) was added to the cylinder and aerated at 100 ml/min for 1 min. Foaming abilities were assessed using the criteria of Petrovski et al. (33).
Nucleotide sequence accession numbers.
The nucleotide sequences for the GTE5 and GRU1 phages have been deposited in GenBank under accession numbers JF923796 and JF923797, respectively.
RESULTS AND DISCUSSION
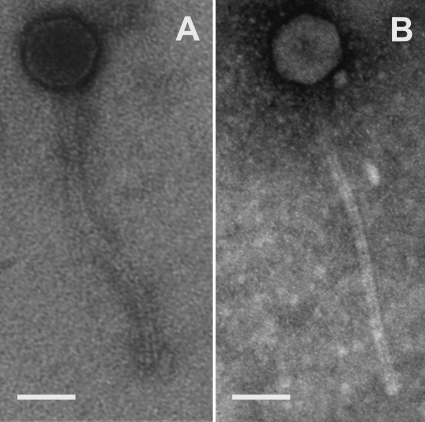
Phages GTE5 and GRU1 were isolated originally on lawn plates of G. terrae Gter34 and G. rubropertincta Grub38, respectively, from samples collected at the Carrum (Victoria) and Loganholme (Queensland) activated sludge plants (47). Southern hybridization studies revealed that the two phages shared extensive DNA sequence similarity (46). Examination of plaque morphologies revealed that GRU1 phage plaques were ∼1 mm in diameter, while those of the GTE5 phage were slightly larger. TEM of the GTE5 and GRU1 phages revealed that both were Siphoviridae, sharing similar morphological dimensions and comprising long noncontractile tails (∼250 nm) with B1 (1) isometric capsids (∼55 nm) (Fig. 1). The burst sizes were determined to be 85 ± 5 PFU (GTE5) and 76 ± 5 PFU (GRU1) per infective center, with a latency period of 4 h (data not shown). Both GTE5 and GRU1 propagate on G. terrae (strains Gter34 and G232) and G. rubropertincta (strain Grub38). Additional host range studies revealed that GRU1 alone formed plaques on Nocardia nova (strain Nnov47).
Fig 1.
Electron micrographs of GTE5 (A) and GRU1 (B). Scale bars, 50 nm.
Genome sequencing and general features of GTE5 and GRU1 phages.
Several restriction endonucleases (i.e., PstI, EcoRI, and HindIII) failed to digest the genomic DNA of either phage. However, when NotI, SacI, and ScaI were applied, it became clear that the two phage genomes were different and circularly permuted (data not shown). The genomes of GTE5 and GRU1 were sequenced twice independently with an average of ∼20,000 reads for each replicate. The assembled sequences showed that they possessed genomes of 65,839 bp and 65,766 bp, respectively. The GC content of GTE5 DNA was 65.0 mol%, and it was 65.5 mol% for GRU1. Both fall within the ranges of their host bacterial genome DNA GC contents of 63 to 69 mol% (18).
Excluding the unpublished partial sequence of phage GTE5 deposited by J. Thomas in GenBank (accession no. AAY16491), the genomes of both GTE5 and GRU1 are novel but related at the DNA level (Fig. 2). Each genome can be divided into regions sharing high levels of sequence similarity separated by small regions unique to each phage (Fig. 2A). Analysis of the GTE5 and GRU1 genomes revealed 93 and 95 putative ORFs, respectively, but no tRNA or tmRNA sequences were detected. The ORFs in GTE5 and GRU1 are numbered consecutively in Fig. 2, except for the small and large terminase genes (terS and terL). A total of 35 ORFs from GTE5 and 36 ORFs from GRU1 show high levels of similarity to known ORFs, but of these, only 18 and 15 ORFs, respectively, could be assigned putative functions (see Tables S1 and S2 in the supplemental material). Both phage genomes are modularly organized, with regions for DNA packaging, DNA replication, and capsid and tail assembly (Fig. 2B and C).
Fig 2.
Pairwise alignment and genetic maps of the GTE5 and GRU1 phage genomes. (A) Pairwise LAGAN alignment of GTE5 and GRU1. (B and C) Genetic maps of GTE5 (B) and GRU1 (C). The lines at the bottom indicate the modular regions. The arrows representing genes are shaded similarly if their protein products have similar functions.
Sequence repeats.
Sequence repeats were identified in both phage genomes, as reported for other circularly permuted phages (34). The GTE5 genome contains 78 inverted repeat sequences ranging from 16 bp to 47 bp in length (see Table S3 in the supplemental material). GRU1 contains 69 inverted repeat sequences that range from 16 bp to 110 bp, with the largest sharing identity with two smaller repeat sequences (see Table S4 in the supplemental material). The functional roles of these repeats, if any, remain unclear. Six palindromic sequences were also identified in GTE5 and 10 in GRU1, four of which are shared between the two genomes (Table 1). In GTE5, most occur within coding sequences, but this is not the case in GRU1. It is possible that these sequences may act as rho-independent terminators (Table 1), although this remains to be determined.
Table 1.
Palindrome sequences found within GTE5 and GRU1 phage genomes
| Palindrome | Coordinates | Sequence | Gene affected |
|---|---|---|---|
| P1-GTE5 | 58990–59005 | ATTGAAACGTTTCAAT | Intergenic orf8-orf81 |
| P2-GTE5 | 43588–43605 | GTCATCGACGTCGATGAC | orf53 |
| P3-GTE5 | 52704–52723 | ACCGGTTGACGTCAACCGGT | orf67 |
| P4-GTE5 | 63953–63974 | CGCGGTGCGGATCCGCACCGCG | orf88 |
| P5-GTE5 | 50428–50459 | CGTGCAGCACGGCCGAGTGACCGTGCTGCACG | orf63 |
| P6-GTE5 | 43282–43320 | GAAGCGGGTGGACCGACCCCTCATCGGCCCACCCGCTTC | Intergenic orf52-orf53 |
| P1-GRU1 | 58637–58652 | ATTGAAACGTTTCAAT | Intergenic orf83-orf84 |
| P2-GRU1 | 61605–61620 | GACCGAGATCTCGGTC | orf86 |
| P3-GRU1 | 12754–12771 | ACGCCTCGCGCGAGGCGT | orf20 |
| P4-GRU1 | 52027–52046 | GCATGTCAGCGCTGACATGC | Intergenic orf68-orf69 |
| P5-GRU1 | 51941–51960 | ACCGGTTGACGTCAACCGGT | Intergenic orf68-orf69 |
| P6-GRU1 | 63058–63079 | CGCGGTGCGGATCCGCACCGCG | Intergenic orf89-orf90 |
| P7-GRU1 | 56419–56442 | ATCTGACAACGCGCGTTGTCACAT | Intergenic orf77-orf78 |
| P8-GRU1 | 21892–21917 | CCGGCCTACGCCGCGGCGTTGGCGGG | orf29 |
| P9-GRU1 | 57326–57354 | CGGGTGGCCGCTGCACCGGCGGCCACCCG | Intergenic orf79-orf80 |
| P10-GRU1 | 42256–42294 | GAAGCGGGTGGACCGACCCCTCATCGGCCCACCCGCTTC | Intergenic orf52-orf53 |
Analysis of the GTE5 and GRU1 phage genomes.
The predicted amino acid sequences of the genes orf1 to orf10 in phages GRU1 and GTE5 have no significant identity with any other amino acid sequences in GenBank, although the sequences of these ORFs are highly similar between the phages. orf11 (975 bp) in GRU1 appears to be a truncated version of orf11 (1,974 bp) in GTE5. The first 197 and the last 739 bp of both genes share identities of 90% and 66%, respectively, but the GTE5 version contains an additional region of 1,050 bp that has no identity with any DNA sequence present in GRU1. This suggests two possibilities: (i) orf11 in phage GTE5 has acquired a DNA insertion of >1 kb or (ii) orf11 in phage GRU1 has undergone a deletion event. To resolve these possibilities, the predicted amino acid sequences of both orf11s were investigated. Homologues of the GRU1 Orf11 occur in other genomes, and it shares significant sequence identity with a hyaluronoglucosaminidase in the Streptococcus phage phi3396 genome that is responsible for hyaluronic acid hydrolysis (10). No predicted function could be assigned to Orf11 in GTE5, although the COG5434 conserved motif was recognized within its amino acid sequence. The same motif is found in plant cell wall-degrading endopyalacturonase enzymes. Interestingly, this conserved motif is absent from the GRU1 Orf11, suggesting that they may have different functions.
DNA packaging and structural proteome genes.
Terminase enzymes are involved in packaging phage DNA into capsids and thus are essential components for replication in Siphoviridae phages (7, 17, 38). Both the small (terS) and large (terL) terminase genes were identified in GTE5 and GRU1 and were organized in the expected order. The large terminase was identified from the presence of the conserved motif pfam03354 and its sequence similarity to other large terminases. The gene encoding the small terminase, terS, was located upstream of terL in both genomes and contained the pfam01844 motif typical of HNH endonucleases and small terminases.
The cluster of genes from the terL gene through orf34 (Fig. 2) appear to encode the structural proteomes of both phages. GTE5 contains at least eight structural proteins, based on earlier published SDS-PAGE profiles (46). Furthermore, in silico analysis, N-terminal sequencing, and mass spectroscopy together show that the genes encoding the structural proteins are located within this cluster. For example, the first gene (orf16 in GTE5 and its homologue orf15 in GRU1) is predicted to encode the phage portal protein, on the basis of the characteristic pfam05133 motif and its high sequence similarity to the portal protein of phage P1201 (8). A portal vertex protein encoded by orf43 (GTE5) and orf41 (GRU1) appears to be located outside this module and was identified based on the presence of the conserved PHA02531 domain.
Two major structural proteins from GTE5 (∼40 kDa and ∼30 kDa) were N-terminally sequenced by Thomas (46) (PINRDYVDPAEITRQVRVAL and PSFQTLAKRQGELI, respectively). These sequences are identical to those of the proteins encoded by orf20 and orf26 in GTE5, corresponding to orf19 and orf25 in GRU1. When shotgun mass spectroscopy was performed on purified whole GTE5 phage, six peptide fragments were identified from three genes: Orf19 (GTE5) and Orf18 (GRU1) (GGTPLGQITAAGATK), Orf20 (GTE5) and Orf19 (GRU1) (IPLMEEDRIR, LVFVGNDQNFEVPFGR, and AFDAELPLANDEALGQMR), and Orf26 (GTE5) and Orf25 (GRU1) (PSFQTLAK and KPLAGVIATAPEDFVLDAEFK). These results confirm that orf19, orf20, and orf26 and orf18, orf19, and orf25 each encode structural proteins in GTE5 and GRU1, respectively.
This genome region is further divided into two modules, the head morphogenesis module and tail morphogenesis module (Fig. 2B and C). It is common for head morphogenesis genes to cluster together and precede the tail protein genes (6). The same arrangement is observed in both GTE5 and GRU1. Genes orf16 to orf20 (GTE5) and orf15 to orf19 (GRU1) are organized into an operon-like structure and appear to include the head morphogenesis genes. The genes orf20 (GTE5) and orf19 (GRU1) appear to encode the main capsid protein, while the putative genes orf17 (GTE5) and orf16 (GRU1) encode a protein sharing high similarity with the predicted head protein in phage P1201 (8).
Immediately downstream of this region is the putative tail morphogenesis region. Based on amino acid identity and mass spectroscopy data, we propose that the major tail protein subunit is encoded by orf26 (GTE5) and orf25 (GRU1) and predict that additional structural proteins involved in tail assembly are encoded by orf27 to orf33 (GTE5) and orf26 to orf32 (GRU1). The protein encoded by orf30 in GTE5 and orf29 in GRU1 is predicted to encode the tape measure protein (TMP), since the genes encode the largest protein (∼173 kDa), which contains the pfam06737 (lytic transglycosylase) motif typical of tape measure proteins. The C terminus of this protein contains a lytic transglycosylase domain and a peptidoglycan hydrolase domain, also seen in Mycobacterium phage TM4 (37) and Tsukamurella phage TPA2 (34).
Putative genes located between these two regions (i.e., orf28-orf29 in GTE5 and orf27-orf28 in GRU1) seem to be expressed using a programmed translational frameshift (50). The two resulting proteins are also thought to be involved in tail assembly (50). This expression mechanism is found in a wide range of seemingly unrelated phages, but its functional purpose is unclear (50). Genes encoding other minor tail structural proteins were also identified based on sequence identity. In both GTE5 and GRU1, the predicted amino acid sequences of genes following the tape measure protein gene (orf31 to orf33 and orf30 to orf32, respectively) show significant sequence identity with those of phage genes suspected of encoding minor proteins, including the tail fibers (Tables 1 and 2).
Table 2.
Influence of GTE5 and GRU1 phages on production of stable foams by selected mycolata strains under laboratory conditions
| Culture | Foaming scorea |
||
|---|---|---|---|
| Without phage | In the presence of GTE5 | In the presence of GRU1 | |
| Gordonia aichiensis (Raic22)b | 6 | 6 | 6 |
| G. terrae (Gter34) | 2 | 0 | 0 |
| G. terrae (G232) | 2 | 0 | 0 |
| G. rubropertincta (Grub38) | 1 | 0 | 0 |
| N. nova (Nnov47) | 1a | 1a | 1a |
Foaming scores are in accordance with the modified scale of Blackall and Marshall (4) illustrated in Petrovski et al. (33, 35). The scale is as follows: 0, as for pure water with no foam; 1, 1.0 to 3.0 cm of foam with fragile, ill-formed bubbles; 1a, flotation of clumped bacterial cells to the surface of the air-water interface; 2, intermittent, sufficiently stable films; 3, substantial foaming (i.e., bubbles about 10 cm in diameter) to 3 to 8 cm high; 4, initially 8 to 15 cm of foam (about 1-cm-diameter bubbles) with stable films formed at regular intervals; 5, stable foam 5 to 10 cm high in 2 min, after which it collapses to 3 to 5 cm high (foam is stable when aeration ceases); 6, stable foam 15 to 30 cm high with no films.
Used as a negative control. GTE5 and GRU1 do not lyse this host.
Host cell lysis genes.
The genes orf39 and orf40 in GTE5 and their equivalents in GRU1 (i.e., orf37 and orf38) encode proteins sharing high sequence similarities with a chitinase from Rhodococcus equi and more distantly with a lysis-encoding gene in phage P1201 (8). Since these gene products appear to share identity with a single protein in other systems, perhaps they were once encoded by a single gene in GTE5/GRU1 or, alternatively, fused in other systems. Despite the high similarity between the Orf39 sequence in GTE5 and the Orf37 sequence in GRU1 (94% similar), the predicted protein in GRU1 encodes a pfam03412 motif, while that from GTE5 does not. This motif belongs to the peptidase C39 family, also found in bacteriocins (14).
The genes orf40 in GTE5 and orf38 in GRU1 encode a protein with the conserved motif pfam00182 found in chitinases (48). The same motif also characterizes the lysozyme-like family of proteins and some phage lysins. Thus, these two genes resemble the lysA and lysB systems described in other phages (32). The subsequent genes encoding the putative lysin proteins (orf41 in GTE5 and orf39 in GRU1) may encode holin proteins, with two transmembrane domains characteristic of some holins. However, they show no close sequence identity with any currently known proteins.
The gene product of orf44 in GTE5 and orf42 in GRU1 is similar to the primase from phage P1201 and other Mycobacterium phages (20, 21). This protein contains two conserved domains (pfam08706 and pfam09250), which characterize the primase protein in double-stranded DNA viruses (28, 51). The gene products of orf47 (GTE5) and orf46 (GRU1) share no significant similarity with any known protein sequences, but Orf47 and Orf46 contain the cd04762 conserved domain of a helix-turn-helix motif, suggesting they are possible DNA binding proteins. A DNA polymerase is expressed from orf49 in both phages, a conclusion based on its high level of similarity to the DNA polymerase III alpha subunit from Mycobacterium phages and the presence of the conserved domain pfam07733. DNA helicases are predicted to be encoded by orf60 (GTE5) and orf61 (GRU1). The corresponding ORF protein sequences are closely related to those in the Corynebacterium phage P1201 and contain conserved domains characteristic of helicases (8).
Proteins encoded by GRU1 genes that have no match to any known sequences but possess conserved motifs are found in proteins Orf54 and Orf85 of phages of Gram-positive bacteria. Orf54 has no known function but contains a cd00569 domain, characteristic of helix-turn-helix motifs, suggesting some regulatory role. Similarly, the predicted sequence of Orf85 contains a PRK00409 conserved domain associated with proteins that inhibit DNA recombination (36).
GTE5 and GRU1 phage genomes are related evolutionarily.
Most of the predicted proteins encoded by GTE5 and GRU1 genes could not be assigned functions (see Tables S1 and S2 in the supplemental material). Many of the encoding genes show a mosaic structure, with subregions of both high sequence conservation and divergence, consistent with genetic exchange. A similar gene level organization was reported for Tsukamurella phage TPA2 (34), supporting the hypothesis that phage evolution may come about by DNA exchange events between different phages and/or their hosts. Comparing the GTE5 and GRU1 phage genomes on a global level (Fig. 2A) suggests that the level of sequence conservation is module dependent. Structural-functional modules (i.e., the DNA-packaging, capsid, tail, and lysis modules) are more highly conserved at the DNA level than those involved in DNA replication or those with unknown functions. Large regions of low DNA sequence similarity separate the functional modules. The reason for this genomic structure is unknown, but it suggests that each phage is based on a conserved core genome with a variable accessory genome aiding replication and host specificity.
GTE5 and GRU1 as phage biocontrol agents.
The isolation of GTE5 and GRU1 was undertaken originally for the purpose of developing biocontrol agents for activated sludge foaming (47). The two phages appear to be similar, although GRU1 can propagate on N. nova (Nnov47) while GTE5 cannot. Any successful use of phages for the biocontrol of foaming will require that a cocktail of different phages be used to minimize problems with inherent or acquired host resistance. These two phages may prove to be of value in such a cocktail.
We used GTE5 and GRU1 in laboratory scale experiments to determine if they reduced the foam stability provided by their host bacteria, as demonstrated for the actinobacterial phage GTE2 (35). The data show that the stability of foaming of G. terrae and G. rubropertincta decreased markedly in the presence of GTE5 and GRU1, and with some strains, no stable foam was generated (Table 2). Whether these phages can be used in large-scale activated sludge plants remains to be demonstrated, but the results so far are promising.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by an Australian Research Council (ARC) Linkage Grant (LP0774913), together with Melbourne Water and South East Water, who we thank for their financial support. S.P. was funded by ARC Linkage and La Trobe University grants.
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ackermann HW. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bateman A, et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Billion A, Ghai R, Chakraborty T, Hain T. 2006. Augur—a computational pipeline for whole genome microbial surface protein prediction and classification. Bioinformatics 22:2819–2820 [DOI] [PubMed] [Google Scholar]
- 4. Blackall LL, Marshall KC. 1989. The mechanism of stabilization of actinomycete foams and the prevention of foaming under laboratory conditions. J. Ind. Microbiol. 4:181–188 [Google Scholar]
- 5. Brudno M, et al. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brüssow H, Desiere F. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213–222 [DOI] [PubMed] [Google Scholar]
- 7. Catalano CE. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol. Life Sci. 57:128–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen CL, et al. 2008. Genome sequence of the lytic bacteriophage P1201 from Corynebacterium glutamicum NCHU 87078: evolutionary relationships to phages from Corynebacterineae. Virology 378:226–232 [DOI] [PubMed] [Google Scholar]
- 9. Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676 [DOI] [PubMed] [Google Scholar]
- 10. Davies MR, McMillan DJ, Van Domselaar GH, Jones MK, Sriprakash KS. 2007. Phage 3396 from a Streptococcus dysgalactiae subsp. equisimilis pathovar may have its origins in Streptococcus pyogenes. J. Bacteriol. 189:2646–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de los Reyes FL. 2010. Foaming, p 215–259 In Seviour RJ, Nielsen PH. (ed), Microbial ecology of activated sludge. IWA Publishing, London, United Kingdom [Google Scholar]
- 14. Dirix G, et al. 2004. Peptide signal molecules and bacteriocins in Gram-negative bacteria: a genome wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides 25:1425–1440 [DOI] [PubMed] [Google Scholar]
- 15. Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27–33 [DOI] [PubMed] [Google Scholar]
- 16. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujisawa H, Morita M. 1997. Phage DNA packaging. Genes Cells 2:537–545 [DOI] [PubMed] [Google Scholar]
- 18. Goodfellow M, Maldonado LA, 2006. The families Dietziaceae, Gordoniaceae, Nocardiaceae and Tsukamurellaceae, p 843–888 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes 3. Archaea Bacteria: Firmicutes, Actinomycetes Springer, New York, NY [Google Scholar]
- 19. Haft DH, Selengut D, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hatfull GF, Cresawn SG, Hendrix RW. 2008. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol. 159:332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hatfull GF, et al. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemmerich C, Buechlein A, Podicheti R, Revanna KV, Dong Q. 2010. An Ergatis-based prokaryotic genome annotation web server. Bioinformatics 26:1122–1124 [DOI] [PubMed] [Google Scholar]
- 23. Kragelund C, et al. 2007. Ecophysiology of mycolic acid-containing Actinobacteria (Mycolata) in activated sludge foams. FEMS Microbiol. Ecol. 61:174–184 [DOI] [PubMed] [Google Scholar]
- 24. Kutter E, et al. 2010. Phage therapy in clinical practice: treatment of human infections. Curr. Pharm. Biotechnol. 11:69–86 [DOI] [PubMed] [Google Scholar]
- 25. Lagesen K, et al. 2007. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lechevalier MP, Lechevalier HA. 1974. Nocardia amarae sp. nov., an actinomycete common in foaming activated sludge. Int. J. Syst. Bacteriol. 24:278–288 [Google Scholar]
- 27. Lemmer H, Kroppenstedt RM. 1984. Chemotaxonomy and physiology of some actinomycetes isolated from scumming activated sludge. Syst. Appl. Microbiol. 5:124–135 [Google Scholar]
- 28. Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. 2004. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 11:157–162 [DOI] [PubMed] [Google Scholar]
- 29. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahony J, McAuliffe O, Ross RP, van Sinderen D. 2011. Bacteriophages as biocontrol agents of food pathogens. Curr. Opin. Biotechnol. 22:157–163. [DOI] [PubMed] [Google Scholar]
- 31. Orvis J, et al. 2010. Ergatis: a web interface and scalable software system for bioinformatics workflows. Bioinformatics 26:1488–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Payne K, Sun QJ, Sacchettini Hatfull GF. 2009. Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 73:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petrovski S, et al. 2011. An examination of the mechanisms for stable foam formation in activated sludge systems. Water Res. 45:2146–2154 [DOI] [PubMed] [Google Scholar]
- 34. Petrovski S, Seviour RJ, Tillett D. 2011. Genome sequence and characterization of the Tsukamurella phage TPA2. Appl. Environ. Microbiol. 77:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petrovski S, Seviour RJ, Tillett D. 2011. Characterization of the genome of the polyvalent lytic bacteriophage GTE2, which has potential for biocontrol of Gordonia-, Rhodococcus-, and Nocardia-stabilized foams in activated sludge plants. Appl. Environ. Microbiol. 77:3923–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pinto AV, et al. 2005. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17:113–120 [DOI] [PubMed] [Google Scholar]
- 37. Piuri M, Hatfull GFA. 2006. Peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62:1569–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rao VB, Feiss M. 2008. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42:647–681 [DOI] [PubMed] [Google Scholar]
- 39. Santos MA. 1991. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 19:5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seviour RJ, et al. 2008. Ecophysiology of the Actinobacteria in activated sludge systems. Antonie van Leeuwenhoek 94:21–33 [DOI] [PubMed] [Google Scholar]
- 42. Shevchenko A, et al. 1996. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24:893–896 [DOI] [PubMed] [Google Scholar]
- 43. Soddell JA. 1999. Foaming, p 161–202 In Seviour RJ, Blackall LL. (ed), Microbiology of activated sludge. Kluwer, Dordrecht, The Netherlands [Google Scholar]
- 44. Soddell JA, Seviour RJ. 1990. Microbiology of foaming in activated sludge plants—a review. J. Appl. Bacteriol. 69:145–176 [Google Scholar]
- 45. Stratton HM, Brooks PR, Griffiths PC, Seviour RJ. 2002. Cell surface hydrophobicity and mycolic acid composition of Rhodococcus strains isolated from activated sludge foams. J. Ind. Microbiol. Biotechnol. 28:264–267 [DOI] [PubMed] [Google Scholar]
- 46. Thomas JA. 2005. Actinophages in activated sludge. PhD thesis. La Trobe University, Melbourne, Australia [Google Scholar]
- 47. Thomas JA, Soddell JA, Kurtböke DI. 2002. Fighting foam with phages. Water Sci. Technol. 46:511–664 [PubMed] [Google Scholar]
- 48. Weaver LH, Matthews BW. 1987. Structure of bacteriophage T4 lysozyme refined at 1.7 A resolution. J. Mol. Biol. 193:189–199 [DOI] [PubMed] [Google Scholar]
- 49. Withey SE, Cartmell E, Avery LM, Stephenson T. 2005. Bacteriophages potential for application in wastewater treatment processes. Sci. Total Environ. 339:1–18 [DOI] [PubMed] [Google Scholar]
- 50. Xu J, Hendrix RW, Duda RL. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11–21 [DOI] [PubMed] [Google Scholar]
- 51. Yeo HJ, et al. 2002. Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins. Mol. Microbiol. 43:855–867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.