Abstract
Salmonella enterica and Escherichia coli O157:H7 are major food-borne pathogens causing serious illness. Phage SFP10, which revealed effective infection of both S. enterica and E. coli O157:H7, was isolated and characterized. SFP10 contains a 158-kb double-stranded DNA genome belonging to the Vi01 phage-like family Myoviridae. In vitro adsorption assays showed that the adsorption constant rates to both Salmonella enterica serovar Typhimurium and E. coli O157:H7 were 2.50 × 10−8 ml/min and 1.91 × 10−8 ml/min, respectively. One-step growth analysis revealed that SFP10 has a shorter latent period (25 min) and a larger burst size (>200 PFU) than ordinary Myoviridae phages, suggesting effective host infection and lytic activity. However, differential development of resistance to SFP10 in S. Typhimurium and E. coli O157:H7 was observed; bacteriophage-insensitive mutant (BIM) frequencies of 1.19 × 10−2 CFU/ml for S. Typhimurium and 4.58 × 10−5 CFU/ml for E. coli O157:H7 were found, indicating that SFP10 should be active and stable for control of E. coli O157:H7 with minimal emergence of SFP10-resistant pathogens but may not be for S. Typhimurium. Specific mutation of rfaL in S. Typhimurium and E. coli O157:H7 revealed the O antigen as an SFP10 receptor for both bacteria. Genome sequence analysis of SFP10 and its comparative analysis with homologous Salmonella Vi01 and Shigella phiSboM-AG3 phages revealed that their tail fiber and tail spike genes share low sequence identity, implying that the genes are major host specificity determinants. This is the first report identifying specific infection and inhibition of Salmonella Typhimurium and E. coli O157:H7 by a single bacteriophage.
INTRODUCTION
Salmonella and Escherichia coli O157:H7 are important food-borne pathogens that cause food poisoning. Salmonellosis is a major illness accompanied by headache, diarrhea, vomiting, and high fever due to Salmonella infection in the epithelial tissue of animals and humans via contaminated foods (3). E. coli O157:H7 is an enterohemorrhagic Shiga toxin producer causing serious food-borne illnesses, such as hemorrhagic colitis, hemolytic uremic syndrome, thrombocytopenia, and kidney failure (33, 45). E. coli O157:H7 present at a very low dose can cause infection, and infection of children and the elderly can be fatal, indicating that it is one of the most serious food-borne pathogens (32, 49). In the United States, more than 1.4 million cases of food-borne salmonellosis have been reported per year, with 17,000 hospitalizations and 600 deaths (46), and food-borne E. coli O157:H7 causes more than 73,000 illnesses, 2,100 hospitalizations, and 60 deaths every year (45, 46). Therefore, even though various food preservatives have been developed to control these pathogens, development of safe and effective new agents to control food-borne pathogens is urgently needed.
Bacteriophages are bacterial viruses that invade specific bacterial cells and utilize the host DNA replication and protein biosynthesis systems for their replication (66). While phage genomes integrate into the host chromosome as prophages in the lysogenic cycle, in the lytic cycle, they disrupt bacterial metabolism and lyse the bacterial host and thus have bactericidal activity (27). In addition, they infect only specific host bacteria without affecting other bacteria in the environment, giving them host specificity (12). Initial human trials of phage therapy by oral administration of phage T4 showed a high safety profile without side effects, suggesting that phage therapy should be safe for human applications (9). Due to their bactericidal properties, host specificity, and safety in humans, bacteriophage treatment has recently been preferred over antibiotic treatment in specific cases, such as for food-borne and antibiotic-resistant pathogens. Bacteriophages have also been considered as biocontrol agents for food safety applications or as therapeutic agents for bacterial infections (16, 50, 53).
While phage therapy has been widely used in the former Soviet Union for decades, the discovery of antibiotics reduced the use of the therapy in Western countries (66). However, recently, this alternative approach to pathogen control and therapeutics has been revisited due to problematic antibiotic treatments and the emergence of antibiotic-resistant pathogens (50). For the application of phages in foods, a Salmonella phage, ΦP7, was tested as a food additive for the control of food-borne Salmonella and produced a significant reduction in Salmonella on a meat surface in a day (7). Furthermore, oral feeding of a Salmonella phage cocktail to broiler chickens as a therapeutic agent showed a rapid reduction in Salmonella colonization in the gut, suggesting that phage therapy may be a good alternative to antibiotic treatment (8). Addition of a phage cocktail containing three different phages to control E. coli O157:H7 in meat slices showed that seven of the nine samples were completely free of E. coli O157:H7 and two samples showed less than 10 CFU/ml, demonstrating significant control of E. coli O157:H7 in contaminated meat by phage treatment (52). In addition, treatment of vegetables, such as cantaloupe and lettuce, with a phage cocktail to control E. coli O157:H7 showed a significant reduction in the bacteria (64). Although broad-host-range phages infecting several different genera of bacteria were previously reported (6, 31, 34) and many phages have been developed and evaluated for control of food-borne Salmonella and E. coli O157:H7, bacteriophages that simultaneously control these two pathogens have been rarely reported (24).
In this study, we isolated and characterized a novel bacteriophage, SFP10, that inhibits both of these food-borne pathogens, suggesting it could be highly effective for their control. Complete genome sequence analysis of SFP10 and comparative genomic analysis with Salmonella phage Vi01 and Shigella phage phiSboM-AG3 were conducted. Information about SFP10 will be useful in the development of broad-host-range phage control of multiple food-borne pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Prophage-cured Salmonella enterica serovar Typhimurium strain LT2 (19) from the Cancer Research Center (Colombia, MO) was used for isolation of Salmonella-infecting phages from the collected slurry samples. All bacteria listed in Table 1 were grown at 37°C for 12 h in Luria-Bertani (LB) broth medium (Difco, Detroit, MI). For agar medium, the broth medium was supplemented with 1.5% agar (Difco).
Table 1.
Host range of phage SFP10
| Bacterial isolate | Plaque formationa | Sourceb or reference |
|---|---|---|
| S. enterica serovar Typhimurium | ||
| SL1344 | +++ | NCTC |
| LT2 | +++ | 44 |
| ATCC 14028s | +++ | ATCC |
| UK1 | +++ | 74 |
| NCTC 12023 | +++ | NCTC |
| KCTC 1425 | +++ | KCTC |
| DT104 | +++ | 58 |
| S. enterica serovar Enteritidis | ||
| ATCC 13076 | +++ | ATCC |
| S. enterica serovar Typhi | ||
| Ty2-b | − | IVI |
| S. enterica serovar Paratyphi | ||
| A IB 211 | + | IVI |
| B IB 231 | + | IVI |
| C IB 216 | − | IVI |
| S. enterica serovar Dublin | ||
| IB 2973 | +++ | IVI |
| E. coli | ||
| K-12 MG1655 | − | 26 |
| ATCC 25922 | − | ATCC |
| O1:K1:H7 KVCC-BA2354 | − | KVCC |
| O112ab:H8 KVCC-BA2396 | − | KVCC |
| O126:H2 KVCC-BA2406 | − | KVCC |
| E. coli O157:H7 | ||
| ATCC 35150 | +++ | ATCC |
| ATCC 43890 | ++ | ATCC |
| O157:NM 3204-92 | +++ | 21 |
| O157:NM H0482 | +++ | 21 |
| Collection of Gram-negative bacteria | ||
| Shigella flexneri 2a strain 2457T | − | IVI |
| Shigella boydii IB 2474 | − | IVI |
| Yersinia enterocolitica ATCC 23715 | − | ATCC |
| Vibrio fischeri ATCC 700601 | − | ATCC |
| Pseudomonas aeruginosa ATCC 27853 | − | ATCC |
| Cronobacter Sakazakii ATCC 29544 | − | ATCC |
| Collection of Gram-positive bacteria | ||
| Enterococcus faecalis ATCC 29212 | − | ATCC |
| Staphylococcus aureus ATCC 29213 | − | ATCC |
| Staphylococcus epidermis ATCC 35983 | − | ATCC |
| Bacillus subtilis ATCC 23857 | − | ATCC |
+++, EOP of 1 to 0.5; ++, EOP of 0.5 to 0.2; +, EOP less than 0.2; −, no susceptibility to SFP10.
NCTC, National Collection of Type Cultures; ATCC, American Type Culture Collection; KCTC, Korean Collection of Type Cultures; IVI, International Vaccine Institute; KVCC, Korean Veterinary Culture Collection.
Bacteriophage isolation and propagation.
Slurry samples were collected from the Seoul Forest in South Korea and used for selection of Salmonella-specific bacteriophages. Procedures for bacteriophage isolation and propagation with the Salmonella host strain, prophage-cured S. Typhimurium LT2, were followed as previously described (35). To isolate the bacteriophages, 25 g of each sample was mixed with 225 ml of sodium chloride–magnesium sulfate (SM) buffer (100 mM NaCl, 10 mM MgSO4 · 7H2O, and 50 mM Tris-HCl, pH 7.5; Sigma, St. Louis, MO) in sterile bags. After homogenization, 25 ml of each diluted sample was mixed with 25 ml of 2× LB broth, and the mixture was incubated with shaking at 37°C for 12 h. After incubation, 0.5 ml of chloroform was added to the mixture and incubated for 5 min at room temperature. The collected samples were centrifuged at 6,000 × g for 10 min, and the supernatants were filtered using 0.22-μm filters (Millipore, Billerica, MA). Ten milliliters of each filtrate was mixed with 50 ml of LB broth medium containing 1% prophage-cured S. Typhimurium LT2 (final concentration) as a propagation strain, and then the mixture was incubated at 37°C for 12 h with shaking. The culture was centrifuged at 6,000 × g for 10 min, and the supernatant containing phage was filtered using a 0.22-μm filter to remove bacterial cells. This supernatant was used for plaque formation in molten 0.4% LB soft agar containing 1% (final concentration) prophage-cured S. Typhimurium LT2. Individual plaques were picked, and phage was eluted with SM buffer, replated, and repicked more than five times for isolation of pure individual phage. When the optical density (OD) of the culture of prophage-cured S. Typhimurium LT2 reached 1.0 at a 600-nm wavelength, the bacteria were infected with SFP10 at a multiplicity of infection (MOI) of 1 and incubated at 37°C for 4 h. For purification of isolated phage, cell debris was removed by subsequent centrifugation at 6,000 × g for 10 min, the supernatant was filtered with 0.22-μm filters, and phage particles were precipitated by treatment with polyethylene glycol (PEG) 6000 (Junsei, Japan). Finally, CsCl density gradient ultracentrifugation (Himac CP 100β; Hitachi, Japan) with stepped CsCl (step densities = 1.3, 1.45, 1.5, and 1.7 g/ml) at 78,500 × g for 2 h was performed at 4°C. The band containing viral particles was recovered by puncturing the centrifuge tube with a sterilized needle, followed by dialysis using 1 liter of standard dialysis buffer (10 mM NaCl, 10 mM MgSO4, and 1 M Tris-HCl at pH 8.0) for 1 h. Phage was stored at 4°C for further experiments.
Electron microscopy.
Purified SFP10 phage was used for transmission electron microscope (TEM) analysis. TEM analysis was conducted following the procedure of Kim and Ryu (35). Based on the morphology of the SFP10 phage, identification and classification were done according to the guidelines of the International Committee on Taxonomy of Viruses (20).
Host range.
One hundred microliters of each test bacterial culture was added to 5 ml of molten 0.4% LB agar, and the mixture was overlaid on 1.5% LB agar plates. Ten microliters of each serially diluted SFP10 phage suspension from 102 to 1011 PFU/ml was spotted on the overlaid plates and incubated at 37°C. After incubation, the sensitivity of test bacteria to SFP10 phage was determined by the degrees of clarity in the spots. Efficiency of plating (EOP) was determined by comparison of the phage titer of the SFP10-sensitive strain with that of the reference strain, S. Typhimurium SL1344. This test was performed in triplicate.
One-step growth curve.
When the OD of the culture of the reference strain at a 600-nm wavelength reached 1.0, 50 ml of the culture was harvested. SFP10 phage was added at an MOI of 0.01 and adsorbed for 5 min at room temperature. To remove the excess phage, the mixture was centrifuged, and the supernatant was discarded. The cell pellets were resuspended in the same volume of fresh LB broth and incubated aerobically at 37°C. Two sets of samples were collected every 5 min. These two sets of samples were immediately 10-fold serially diluted and plated for phage titration. Before the titration, the second set of samples was treated with 1% chloroform (final concentration) to release intracellular phage to determine the eclipse period. Based on the number of PFU per ml, the latent period and burst size were determined.
In vitro adsorption assays.
The in vitro adsorption assay with reference strains, S. Typhimurium SL1344 and E. coli O157:H7 ATCC 43890, was performed as previously described (35). The phage adsorption constant rate was calculated as previously described (36). When the ODs of the reference strains, S. Typhimurium SL1344 and E. coli O157:H7 ATCC 43890, reached 1.0 at a 600-nm wavelength, 50 ml of each culture was transferred and diluted 10-fold using fresh LB broth medium. SFP10 phage was added to each diluted culture at an MOI of 0.01 and incubated at 37°C for 15 min. Each minute, samples were collected, and the bacterial cells were removed by centrifugation at 13,000 × g for 1 min and filtered using 0.22-μm filters. Finally, The numbers of PFU in the collected supernatant samples were determined by serial dilution and standard plaque assay using each reference strain. Based on the ratio between the initial titer and test titers, the adsorption of SFP10 phage to each reference strain was determined.
Transposon mutagenesis and selection of SFP10-resistant mutants.
Random gene mutation of SFP10-sensitive S. Typhimurium SL1344 was performed using the EZ-Tn5 <R6Kγori/KAN-2>Tnp Transposome Kit according to the manufacturer's procedure (Epicentre, Madison, WI). For electroporation of the EZ-Tn5 transposome into the SL1344 strain, electrocompetent cells were freshly prepared and used as follows: a 2% overnight seed culture was subinoculated into 8 ml of fresh LB broth and incubated with shaking at 37°C for 1.5 h. After incubation, cells were harvested by centrifugation at 5,071 × g for 10 min, and the cell pellet was resuspended with 1 ml of molecular-grade water. The pellets were washed three times with the same volume of molecular-grade water and resuspended with 100 μl of molecular-grade water. For electroporation, 1 μl of Tn5 transposome (33 ng/μl) was added to the SL1344 competent cells and mixed briefly. Electroporation was conducted with the mixture in an ice-cold 2-mm electroporation cuvette at 2.45 kV, 200 Ω, and 25 μF using a Gene-Pulser Xcell system (Bio-Rad, Hercules, CA). After electroporation, 1 ml of SOC medium (Super Optimal broth with catabolite repression) was added immediately, and the culture was incubated with shaking at 37°C for 1 h. A total of 2,000 independent random mutants were selected on LB agar containing 50 μg/ml kanamycin sulfate (Sigma). With these selected mutants, the random mutant library was constructed and stored at −80°C in 15% sterilized glycerol (final concentration). For selection of SFP10-resistant mutants, duplicate inoculation of each selected mutant was done in two 96-well plates containing LB broth medium with 50 μg/ml kanamycin sulfate, and the plates were incubated at 37°C for 1.5 h. After incubation, one of the plates was infected with SFP10 phage (MOI = 1), and both plates were incubated at 37°C for an additional 3 h. To identify SFP10-resistant mutants, the ODs of the two plates were measured at 600-nm wavelength using an iMark microplate absorbance reader (Bio-Rad). Rescue cloning of transposed genome DNA and partial sequencing were performed for confirmation of transposon insertion sites, according to the protocol of the EZ-Tn5 <R6Kγori/KAN-2>Tnp Transposome Kit (Epicentre).
Construction of the rfaL deletion mutants and complementation.
An S. Typhimurium SL1344 strain with deletion of the rfaL gene encoding O-antigen ligase was constructed using the one-step gene inactivation method (17). The kanamycin resistance (Kmr) cassette from plasmid pKD13 was amplified using primers rfaL-lamb-F, containing the sequence upstream of the start codon of the rfaL gene following the priming site 1 sequence of pKD13 (5′-CTGGTTTTTCTTTTTGTTGCCACGTATTTTCTGGATGGTATGTAGGCTGGAGCTGCTTCG-3′), and rfaL-lamb-R, containing the sequence downstream of the stop codon of the rfaL gene linked to the priming site 4 sequence of pKD13 (5′-TGGATAATCGACAACGCGTTTATTATAAACACCATCATACATTCCGGGGATCCGTCGACC-3′). The resulting PCR product was used to transform the wild-type strain containing pKD46 (17), and the PCR product was integrated into the rfaL gene in the chromosome. Finally, the Kmr cassette was removed using the pCP20 plasmid (15). For complementation of the rfaL deletion mutant and the rfbG-Tn5 insertion mutant, the rfaL or rfbG region of S. Typhimurium SL1344 was amplified using the rfaL-complementing primers rfaL-pUHE-F(EcoRI) (5′-GCCACAAGCGAATTCGGAAGATT-3′) and rfaL-pUHE-R(BamHI) (5′-TACCGTAATAAGGATCCGCGCGTT-3′) or the rfbG-complementing primers rfbG-pUHE-F (BamHI) (5′-CTGTCATTACTTTGGATCCTTAAACTTA-3′) and rfbG-pUHE-R (EcoRI) (5′-AATGGCTTTTGAATTCCCAGGTTTC-3′), respectively. The PCR products were digested with BamHI and EcoRI and ligated into the BamHI/EcoRI-digested pUHE21-lacIq expression vector harboring the ampicillin-resistance gene (65). After confirmation of the rfaL and rfbG region sequence in the vectors by partial sequencing, pUHE21-lacIq::rfaL and pUHE21-lacIq::rfbG were transformed into the rfaL deletion mutant and the rfbG-Tn5 insertion mutant, respectively. The complementation of the corresponding genes was confirmed by colony PCR and bacteriophage SFP10 susceptibility. The rfaL gene in E. coli O157:H7 ATCC 43890 was inactivated using a TargeTron Kit, according to the manufacturer's procedure (Sigma). To mutate the RNA portion of the intron, PCR was performed using rfaL-TargeTron primers, rfaL-IBS (5′-AAAAAAGCTTATAATTATCCTTATATAACAGGTAAGTGCGCCCAGATAGGGTG-3′), rfaL-EBS1d (5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCAGGTAATCTAACTTACCTTTCTTTGT-3′), and rfaL-EBS2 (5′-TGAACGCAAGTTTCTAATTTCGGTTTTATATCGATAGAGGAAAGTGTCT-3′). The PCR product was cut with HindIII and BsrGI and ligated into a linearized pACD4K-C vector included in the kit. Electroporation into E. coli O157:H7 ATCC 43890 was performed. The transformant was selected on LB agar containing 25 μg/ml chloramphenicol, and the retargeted intron was expressed and integrated into the rfaL gene by induction with 100 mM (final concentration) IPTG (isopropyl-β-d-thiogalactopyranoside). The ΔrfaL mutant was selected on LB agar containing 50 μg/ml kanamycin sulfate. The selected mutant was confirmed by PCR using rfaL inactivation-confirming primers, rfaL-F(O157) (5′-CTTCTCATTTATTAGTGCGTTGGGC-3′) and rfaL-R(O157) (5′-CATCGAGTCAGAAATGCTACGGTGT-3′).
Bacterial challenge test.
Two reference strains, S. Typhimurium SL1344 and E. coli O157:H7 ATCC 43890, were inoculated into LB broth medium and grown at 37°C for 12 h with shaking, and then 1% of each culture was subinoculated into 100 ml of fresh LB broth medium and incubated with shaking at 37°C. Samples were collected every hour, the OD at 600 nm was taken, and the samples were serially diluted and plated in triplicate. When the OD at 600 nm reached 1.0, the culture was divided into two 50-ml samples, and SFP10 phage was added to one of the two samples at an MOI of 100. Both samples were grown further and collected every hour, serially diluted, and plated in triplicate. The numbers of CFU in the plates were determined by serial dilution and standard viable-cell counting. For statistical analysis, Microsoft Excel was used for a Student's t test with a P value threshold of ≤0.05. The bacteriophage-insensitive mutant (BIM) frequency was determined as previously described by O'Flynn et al. (52).
Stability test under various temperatures and pHs.
For evaluation of phage stability under various temperature conditions, SFP10 phage (final concentration, 108 PFU/ml) were added to fresh LB broth, and the phage suspensions were incubated at 20, 30, 40, 50, 60, 70, and 75°C for 1 h. After incubation, phage titers were enumerated using a standard plaque assay with a reference strain, S. Typhimurium SL1344. The phage suspensions were held at 4°C as controls. For pH stability of SFP10 phage, phage (final concentration, 108 PFU/ml) was added to LB broth that was pH adjusted with HCl or NaOH to a pH range of 1 to 12, and the phage suspensions were incubated at 37°C for 24 h. After incubation, the phage titer of the cultures was enumerated using a standard plaque assay with the same reference strain. Phage suspensions were also held at pH 7 as controls.
Genome sequencing and bioinformatics analysis.
Before isolation of SFP10 genomic DNA, contaminating bacterial DNA was removed by DNase I (20 units/ml, final concentration; New England BioLabs, Ipswich, MA) treatment at 37°C for 30 min. SFP10 genomic DNA was isolated as previously described by Wilcox et al. (71). For construction of the genomic-DNA library for the Genome Sequencer FLX (GS-FLX) Titanium (Roche, Mannheim, Germany), 1 μg of purified genomic SFP10 phage DNA was physically sheared using a HydroShear DNA-shearing machine (Digilab, Holliston, MA), and the ends of each fragment were blunted. Two adapters were added to the blunt ends for PCR amplification and sequencing of the library fragments, and DNA was denatured to generate single-stranded template DNA fragments (sstDNA library). This library was quantified using an Agilent Bioanalyzer 2100 and a DNA1000 kit (Applied Biosystems, Foster City, CA). For separation and individual sequencing of library DNAs, DNA was hybridized to DNA capture beads, and sequencing primer was annealed to the immobilized DNA template. Each DNA template was sequenced with the GS-FLX instrument, with sequencing reagents and nucleotide sequences determined by the onboard computer. Prediction of all open reading frames (ORFs) was carried out using the Glimmer by GAMOLA automatic annotation program (1) and confirmed using GeneMark (5) and FgenesV software (Softberry, Inc., Mount Kisco, NY). Annotation of predicted ORFs was performed, and GenBank files were generated using GAMOLA with BLAST databases before manual checking. Compilation and editing of genome-sequencing and annotation data were conducted using Artemis12 (13). Ribosomal binding sites (RBS) were predicted using RBSfinder (J. Craig' Venter Institute, San Diego, CA) for confirmation of predicted ORFs. To predict gene functions, the GAMOLA and InterProScan programs with conserved protein domain databases were used (73). Cluster of orthologous groups (COG) functional categories were used for functional classification of all genes in the SFP10 genome (68). For phylogenetic analysis of bacteriophages, including the SFP10 genome, by the neighbor-joining method, using P distance values, MEGA4 was used (37). Comparative genomic analysis of SFP10 with homologous phages was done by MUMmer3 (38), BLAST 2 Sequence (NCBI, Bethesda, MD), and ClustalX (40) programs, and visualization of the comparative-analysis results was performed using ACT9 (13).
Nucleotide sequence accession number.
The GenBank accession number for the complete genome sequence and annotation information for broad-host-range bacteriophage SFP10 is HQ259103.
RESULTS
Isolation and host range of phage SFP10.
The purpose of this study was the isolation of Salmonella-specific bacteriophages from environmental samples for the development of biocontrol agents. Using prophage-cured S. Typhimurium LT2 as an indicator strain, a total of 11 bacteriophages were isolated from 21 slurry samples collected from the Seoul Forest in Seoul, South Korea. Interestingly, the host range test of isolated Salmonella-specific phages revealed that one of the phages, designated SFP10, showed specific inhibition against S. enterica serovar Typhimurium, serovar Enteritidis, and serovar Dublin and E. coli O157:H7 (Table 1). However, it did not inhibit, or very poorly inhibited, other Salmonella species and E. coli, as well as various other Gram-positive and Gram-negative bacteria (Table 1). Furthermore, comparative EOP analysis using S. Typhimurium SL1344 as a standard strain also showed the susceptibility of test bacteria to the SFP10 phage, supporting the broad host specificity of the phage (Table 1). This suggested that the SFP10 phage might be a good candidate for development of a new biocontrol agent to inhibit S. enterica and E. coli O157:H7 strains at the same time.
Morphology of phage SFP10.
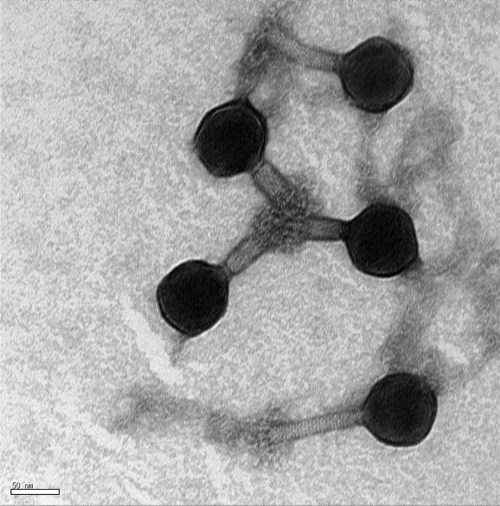
For morphological characterization of phage SFP10, TEM analysis showed an icosahedral head and contractile, nonflexible tails, suggesting that SFP10 belongs to morphotype A1 in the family Myoviridae (Fig. 1). The diameters of the isomeric head and tail were 68.75 nm and 12.90 nm, respectively, and the noncontracted and contracted tail lengths were 131.25 nm and 41.67 nm, respectively. Based on the morphology of SFP10, it is a Vi01-like phage, and its tail fiber structure is very similar to that of phage phiSboM-AG3 (2).
Fig 1.
Electron microscopic image of phage SFP10 negatively stained with 0.2% uranyl acetate. Scale bar, 50 nm.
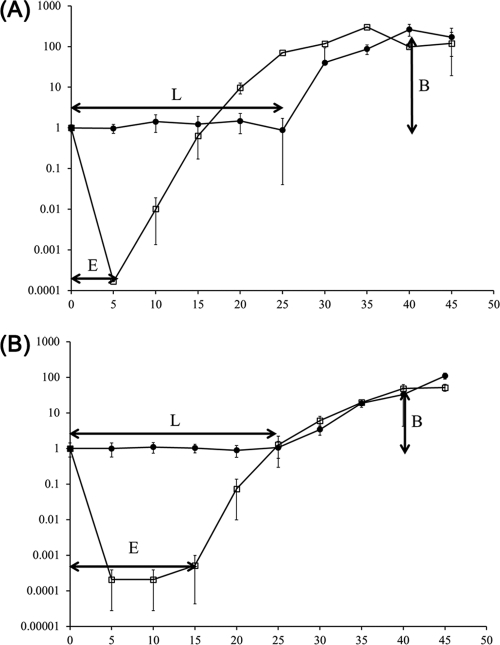
Latent period, burst size, and adsorption constant rate.
To elucidate the ability of SFP10 phage to lyse S. Typhimurium SL1334 and E. coli O157:H7 ATCC 43890, the eclipse and latent periods and burst size of the phage were determined using a one-step growth curve analysis (Fig. 2). The eclipse and latent periods of SFP10 phage were 5 and 25 min in S. Typhimurium SL1344 and 15 min and 25 min in E. coli O157:H7 ATCC 43890. After lysis of the host cell, the burst size was more than 200 PFU per cell in S. Typhimurium SL1344 and 100 PFU per cell in E. coli O157:H7 ATCC 43890. In addition, the adsorption constant rates of SFP10 phage to S. Typhimurium SL1344 and E. coli O157:H7 ATCC 43890 were 2.50 × 10−8 ml/min and 1.91 × 10−8 ml/min, respectively, which are higher than those of T4 and M13 infections of their host strains (36).
Fig 2.
One-step growth curve analysis of S. Typhimurium SL1344 (A) and E. coli O157:H7 ATCC 43890 (B) infected by SFP10 phage. E, eclipse period; L, latent period; B, burst size. Closed circles, non-chloroform-treated sample; open squares, chloroform-treated sample. The error bars indicate standard deviations.
Identification of the phage SFP10 receptor in S. Typhimurium and E. coli O157:H7.
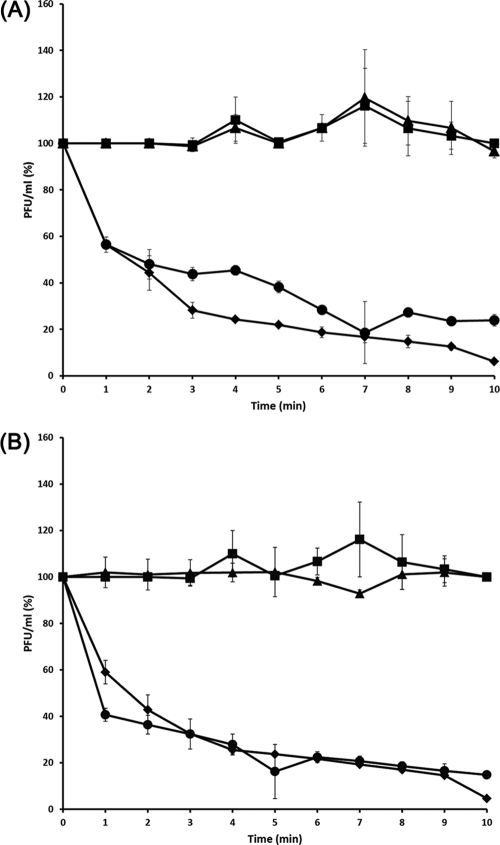
To identify the SFP10 receptor in SFP10-sensitive S. Typhimurium SL1344, a random mutant Tn5 transposon library was constructed and screened for phage resistance. Two phage-resistant mutants were obtained, one with a Tn5 insertion in the rfaL gene and the other with an insertion in the rfbG gene. rfaL encodes O-antigen ligase, and rfbG encodes CDP glucose 4,6-dehydratase. Both genes are involved in lipopolysaccharide (LPS) biosynthesis, indicating that O antigen may be the receptor of SFP10 phage. Comparison of the adsorption ability of SFP10 to host bacteria, including two SFP10-sensitive strains, S. Typhimurium SL1344 and E. coli O157:H7 ATCC 43890; the ΔrfaL and the Tn5 insertion rfbG (ΔrfbG/Tn5) mutants; and a non-SFP10-sensitive strain, E. coli MG1655, revealed that the ΔrfaL and ΔrfbG/Tn5 mutants, as well as E. coli MG1655, did not adsorb SFP10. However, more than 60% of the phage were adsorbed to the SFP10-sensitive strains in 5 min and at least 90% in 10 min (Fig. 3). Complementation of these mutants with the rfaL or rfbG gene expression vector (pUHE21-lacIq::rfaL or pUHE21-lacIq::rfbG) restored the SFP10-sensitive phenotype, demonstrating that the O antigen in the SL1344 strain is a receptor for SFP10 phage (Fig. 3). In addition, the rfaL gene in E. coli O157:H7 was deleted by the TargeTron intron system (Sigma), and the ΔrfaL mutant showed resistance to SFP10 phage, proving that the O antigen in E. coli O157:H7 is also a receptor for SFP10 phage, as in Salmonella (data not shown).
Fig 3.
Confirmation of a phage SFP10 receptor by deletion and complementation of rfaL (A) and Tn5 mutation and complementation of rfbG (B) involved in LPS biosynthesis in S. Typhimurium SL1344. The phage sensitivities of wild-type and mutant strains were tested using an adsorption assay with SFP10 phage. Diamonds, wild-type strain (SFP10 sensitive); squares, E. coli MG1655 (SFP10 resistant). (A) Triangles, ΔrfaL deletion mutant; circles, ΔrfaL deletion mutant complemented with the pUHE21-lacIq::rfaL expression vector. (B) Triangles, ΔrfbG/Tn5 mutant; circles, ΔrfbG/Tn5 mutant complemented with the pUHE21-lacIq::rfbG expression vector. The error bars indicate the standard deviations in triplicate experiments.
Bacterial challenge tests.
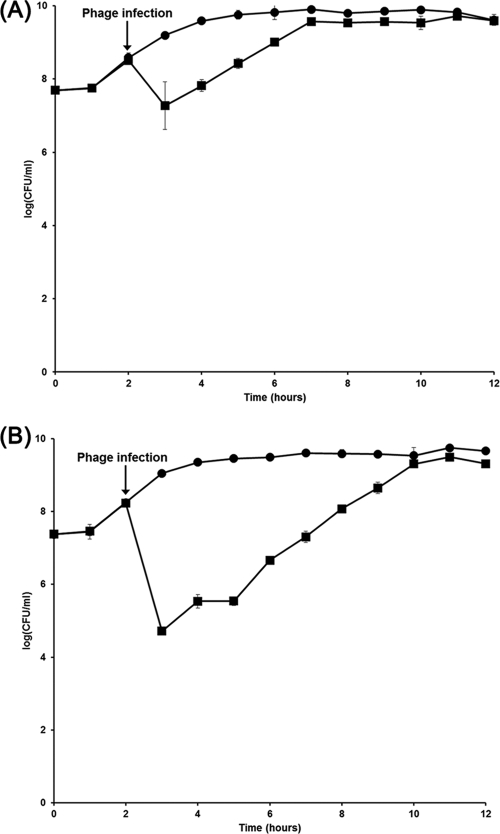
BIMs of SFP10-sensitive S. Typhimurium SL1344 or E. coli O157:H7 ATCC 43890 were determined by growth curve analysis and viable-cell counting after SFP10 infection. One hour after phage addition, the bacterial cell numbers were reduced by 1.77 log(CFU/ml) for strain SL1344 and by 4.34 log(CFU/ml) for E. coli O157:H7 (Fig. 4). The statistical analyses of reduction of these pathogens by SFP10 phage showed that the P values of these reductions are <0.05, suggesting that their reductions by SFP10 phage are significant. However, bacterial counts recovered to control levels, without phage infection, after additional incubation of 5 h for strain SL1344 or 8 h for E. coli O157:H7, indicating the generation of BIMs. Strikingly, the BIM frequency of S. Typhimurium SL1344 (1.19 × 10−2 CFU/ml) was approximately 260-fold higher than that of E. coli O157:H7 ATCC 43890 (4.58 × 10−5 CFU/ml), suggesting different mechanisms of BIM development against SFP10 infection in S. Typhimurium and E. coli O157:H7 (35, 52).
Fig 4.
Bacterial challenge test of phage SFP10 with S. Typhimurium SL1344 (A) and E. coli O157:H7 ATCC 43890 (B). The graphs show viable-cell counts of samples collected every hour. Each strain was infected with phage SFP10 when the OD at 600 nm was 1.0. Circles, non-SFP10-infected sample; squares, SFP10-infected sample. The error bars indicate standard deviations.
Stability under conditions of varying temperature and pH.
For application of SFP10 as a biocontrol agent to inhibit food-borne pathogenic bacteria, such as Salmonella and E. coli O157:H7, its viable stability needed to be confirmed under various stress conditions, such as temperature and pH. There was no significant loss of SFP10 phage count between 20 and 60°C, with 37°C the optimum temperature for phage activity. However, the phage count was reduced by 54% at 70°C and was completely inactivated at 75°C, indicating that phage SFP10 has moderate heat resistance (data not shown). A pH stability test of the phage also showed that it was highly stable between pH 4 and 10. However, SFP10 phage was completely abolished under strong acid (pH <2) or strong alkali (pH >12) conditions (data not shown). These resistances to stress of SFP10 would be useful for various applications as a biocontrol agent against pathogens.
Bacteriophage genome analysis.
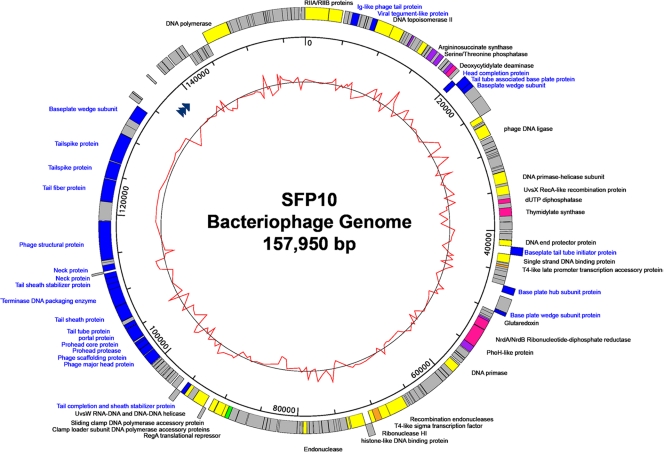
Phage SFP10 genome sequencing was performed using the 454 pyrosequencing approach. The general characteristics of the genome include a total of 157,950 bp with an overall G+C content of 44.53%, 201 predicted ORFs, and 4 tRNAs (Fig. 5 and Table 2). However, only 63 ORFs (31.34%) were predicted and determined to be functional, based on gene prediction and annotation of the genome. The origins of replication regions were not detected by the OriLoc program (23).
Fig 5.
Genome map of phage SFP10. The outer circle indicates the gene coding regions by strand. The color of each gene refers to the functional category: phage structure (blue), replication/recombination/repair (yellow), nucleotide metabolism (pink), transcription (orange), translation (green), or additional function (purple). The arrowheads in the first inner circle indicate the locations of tRNAs. The inner circle with a red line indicates the GC content. The legends for phage structural proteins are blue. The scale units are base pairs.
Table 2.
General characteristics of three homologous bacteriophage genomes
| Characteristic | Value for bacteriophage: |
||
|---|---|---|---|
| SFP10 | Salmonella Vi01 | Shigella phiSboM-AG | |
| Length (bp) | 157,950 | 157,061 | 158,006 |
| Overall G+C content (%) | 44.53 | 45.22 | 50.40 |
| No. of annotated genes | 201 | 208 | 216 |
| Avg gene length (bp) | 721 | 698 | 678 |
| Gene density (no. of genes/kb) | 1.272 | 1.324 | 1.367 |
| Gene coding content (%) | 91.8 | 92.5 | 92.7 |
| Gene GC content (%) | 44.75 | 45.38 | 50.81 |
| No. of tRNAs | 4 | 6 | 4 |
| No. of putative tail fiber proteins | 1 | 2 | 1 |
| No. of putative tailspike proteins | 2 | 3 | 2 |
This phage genome contains complete genes for phage structure and genes for replication/recombination/repair, nucleotide metabolism, transcription, translation, and additional functions. The phage structural genes encode head structure proteins (major capsid protein, scaffolding protein, prohead protease, head completion protein, and prohead core protein), tail/neck structure proteins (tail tube protein, tail sheath protein, tail sheath stabilizer proteins, tail completion protein, neck proteins, tail spike proteins, tail fiber protein, baseplate wedge subunits, tail tube-associated base plate protein, and baseplate hub subunit), and accessory structural proteins (Ig-like virion protein and baseplate tail tube initiator protein). Therefore, this module appears to contain all required genes for complete recovery of phage head and tail structures. The replication/recombination/repair gene module encodes replication proteins (rIIA and rIIB proteins, DNA topoisomerase II proteins, T4-like loader of DNA helicase, DNA helicases, DNA primase, phage clamp loader subunits, sliding clamp protein, DNA polymerase, DNA end protector protein, DexA exonuclease, and T4-like endonuclease) and recombination/repair proteins (recombinases, UvsW helicase, UvsY DNA recombination/repair protein, and single-stranded DNA binding protein), suggesting that SFP10 phage has its own replication/recombination/repair systems. The module of nucleotide metabolism genes encodes deoxycytidylate deaminase, dUTP diphosphatase, thymidylate synthase, and NrdA/NrdB ribonucleotide-diphosphate reductase. The module of transcription/translation genes encodes T4-like sigma transcription factor, its accessory protein, and the RegA translational repressor protein. Interestingly, this phage genome encodes several additional proteins, such as argininosuccinate synthase and serine/threonine phosphatase for amino acid biosynthesis, glutaredoxin for redox function, and PhoH-like protein for phosphate starvation, suggesting additional functions available to the host.
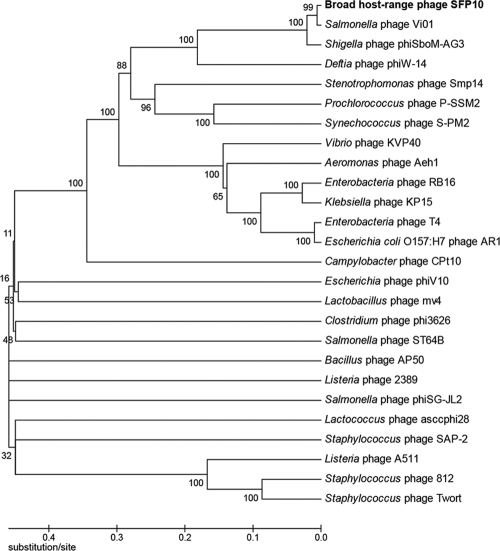
The phylogenetic analysis of conserved major capsid proteins (MCPs) from various bacteriophage genomes, including SFP10 phage, showed that SFP10 phage is located near Salmonella phage Vi01 (57) and Shigella phage phiSboM-AG3 (Fig. 6). Although DNA identity among these phage genomes is more than 90% at the DNA level, they have different host specificities (Vi01 for Salmonella, phiSboM-AG3 for Shigella, and SFP10 for Salmonella and E. coli O157:H7), indicating that they may have genes for different host-interacting proteins in their genomes.
Fig 6.
Phylogenetic analysis of MCPs from various bacteriophages. The MCPs were compared by ClustalW multiple alignments, and the phylogenetic tree was generated by the MEGA4 program using the neighbor-joining method with P distance values.
Comparative genomic analysis of SFP10 with other, related bacteriophages.
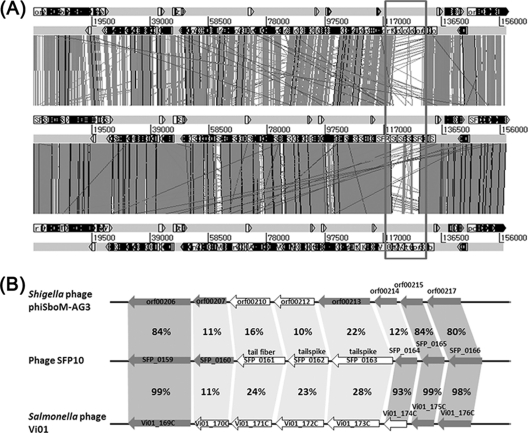
To elucidate the mechanisms of the different host specificities of the three phylogenetically close bacteriophages, comparative genomic analysis was conducted using the MUMmer3 and BLAST genome alignment programs (Fig. 7A and Table 2). While most genes are highly similar among the three genomes, gene clusters encoding a phage tail fiber protein and two tail spike proteins are quite different (Fig. 7A). Comparative analysis of these genes at the amino acid level using the BLASTP program revealed that they share less than 20% amino acid identity, suggesting that the genes may determine the host specificities (Fig. 7B). Interestingly, while one of the tail fiber proteins of SFP10 (SFP_0161) is homologous to that of E. coli O157:H7 phage phiV10 (54), two tail spike proteins (SFP_0162 and SFP_0163) in the SFP10 gene cluster are homologous to those of Salmonella phages (57, 70). This may be the main reason for the dual infectivities of Salmonella and E. coli O157:H7 by SFP10 (Table 3). Although the SFP10 and Vi01 phages can infect Salmonella, they have different host specificities. Phage SFP10 infects Salmonella O antigen type 1, and phage Vi01 is specific for the Salmonella Vi capsular antigen, likely due to a difference in infection receptors determined by the SFP_0162 and Vi01_171C genes (57) (Table 3). Even though three genes (orf00210, orf00213, and orf00214) in the gene cluster of phiSboM-AG3 are similar to those of the Salmonella Vi01 phage in the narrow regions, their amino acid sequence identities were too low to deduce the receptor of phiSboM-AG3 (Table 3), suggesting the different host specificity of phiSboM-AG3 (2).
Fig 7.
Comparative analysis of three phage genomes (A) and gene clusters involved in host specificity for infection from three phage genomes (B). (A) Phage SFP10 (middle), Shigella phiSboM-AG3 phage (top), and Salmonella phage Vi01 (bottom). The variable regions in the three phage genomes involved in host specificity for infection are boxed. (B) The white arrows indicate host specificity genes, and the gray arrows indicate hypothetical proteins. The identities of amino acids between homologous genes are indicated as percentages.
Table 3.
Comparative analysis of ORFs in host specificity-related regions of three similar bacteriophage genomes
| ORF | Predicted function | Lengtha | % Identityb | BLASTP best matches | Receptorc | Reference |
|---|---|---|---|---|---|---|
| Broad-host-range phage SFP10 | ||||||
| SFP_0160 | Conserved hypothetical protein | 770 | 71 over 162 aa | Tailspike protein from Salmonella phage Det7 | LPS | 70 |
| SFP_0161 | Probable phage tail fiber protein | 926 | 50 over 688 aa | Putative tail fiber from E. coli O157:H7 phage phiV10 | LPS (O157 antigen) | 54 |
| 48 over 437 aa | Hypothetical tail fiber from E. coli O157:H7 phage JK06 | ND | ||||
| SFP_0162 | Phage tailspike protein | 698 | 51 over 709 aa | Tailspike protein from Salmonella phage Det7 | LPS | 70 |
| 36 over 620 aa | Tailspike protein from Salmonella phage SETP3 | LPS (O12 antigen) | 18 | |||
| SFP_0163 | Conserved hypothetical protein partially similar to phage tailspike protein | 1,167 | 83 over 400 aa | Hemolysin-type calcium-binding protein from Salmonella phage Vi01 | Vi capsular antigen | 57 |
| SFP_0164 | Hypothetical protein | 396 | 93 over 396 aa | Putative tail fiber protein from Salmonella phage Vi01 | Vi capsular antigen | 57 |
| Salmonella phage Vi01 | ||||||
| Vi01_170C | Tail spike protein 1 | 596 | 58 over 163 aa | Hypothetical protein from Shigella phage phiSboM-AG3 | ND | 70 |
| 55 over 76 aa | Tailspike protein from Salmonella phage Det7 | LPS | ||||
| Vi01_171C | Vi maturation-adhesion tailspike | 732 | 61 over 293 aa | Predicted tail fiber protein from Salmonella phage Vi06 | Vi capsular antigen | 57 |
| 63 over 264 aa | Maturation/adhesion protein from Salmonella phage E1 | Vi capsular antigen | 56 | |||
| Vi01_172C | Tailspike 3 | 846 | 52 over 695 aa | Head-binding domain of phage tailspike protein from Escherichia fergusonii ATCC 35469 | ND | |
| Vi01_173C | Probable tail fiber | 1,057 | 64 over 281 aa | Hypothetical protein from Shigella phage phiSboM-AG3 | ND | |
| Vi01_174C | Putative tail fiber | 396 | 65 over 406 aa | Hypothetical protein from Shigella phage phiSboM-AG3 | ND | |
| Shigella phage phiSboM-AG3 | ||||||
| orf00207 | Conserved hypothetical phage protein | 594 | 47 over 182 aa | Tailspike protein from Salmonella phage Det7 | LPS | 70 |
| orf00210 | Tailspike protein | 753 | 66 over 198 aa | Maturation/adhesion protein from Salmonella phage Vi01 | Vi capsular antigen | 57 |
| orf00212 | Tailspike protein | 673 | 57 over 96 aa | Conserved phage protein from Enterobacteria phage Eco1230-10 | ND | 62 |
| orf00213 | Conserved hypothetical phage protein | 1,019 | 64 over 442 aa | Hemolysin-type calcium-binding protein from Salmonella phage Vi01 | Vi capsular antigen | 57 |
| orf00214 | Hypothetical phage protein | 403 | 65 over 406 aa | Putative uncharacterized protein from Salmonella phage Vi01 | Vi capsular antigen | 57 |
Number of amino acids.
Amino acid (aa) sequence identity.
Predicted phage absorption site. ND, not determined.
DISCUSSION
Outbreaks of food-borne pathogens, such as Salmonella and E. coli O157:H7, have been problematic in the food safety industry. While the control of these pathogens using various antibiotics has resulted in antibiotic-resistant bacteria, bacteriophages have attracted much interest and have been studied for their use in biocontrol and as therapeutic agents (16, 50, 51, 53). Typically, bacteriophages have a very narrow spectrum for bacterial inhibition and high host specificity for infection (30). Therefore, to increase the phage infection range and inhibition efficiency, phage cocktails containing different phages have been developed and used for inhibition of pathogens such as Salmonella and E. coli O157:H7 (52, 67, 69). Although a few broad-host-range phages have been reported (6, 31, 34), the bacteriophage AR1 is the only phage reported to date that can inhibit both Salmonella and E. coli O157:H7 (24, 42). AR1 is reported to infect many serotypes of E. coli and S. enterica serovar Enteritidis and serovar Choleraesuis, but not serovar Typhimurium. The aim of this study was to isolate and characterize a bacteriophage specifically inhibiting E. coli O157:H7 and Salmonella Typhimurium, the most critical and the most frequently reported food-borne pathogens, respectively. To accomplish this aim of the study, we isolated phage SFP10 from natural slurry samples, which inhibited both Salmonella Typhimurium and E. coli O157:H7. While the phage showed broad infection ability against S. enterica serovar Enteritidis and serovar Dublin and some serovar Paratyphi isolates, it showed a very specific host range against E. coli O157:H7 (Table 1). This is the first report identifying specific infection and inhibition of major food-borne pathogens, Salmonella Typhimurium and E. coli O157:H7, by a single bacteriophage.
Various components of the bacterial outermost cell layer, such as flagella (61, 63), O antigen of LPS (4, 39, 47), OmpC (28, 67, 72), and BtuB (29, 35), are used as bacteriophage receptors. Identification of the phage receptor is essential for the application of phages in the biocontrol of pathogens (25). We found that the SFP10 receptor in Salmonella is the O antigen, similar to phage P22. All SFP10-sensitive S. enterica strains, such as serovars Typhimurium, Enteritidis, Paratyphi A and B, and Dublin, share O-antigen serotype 1 (O1 antigen) (48), while other, non-SFP10-sensitive Salmonella strains, such as serovars Typhi and Paratyphi C, do not have the O1 antigen, suggesting the O1 antigen may be a specific receptor for SFP10 phage infection of Salmonella.
The high host specificity of bacteriophages has been useful for the inhibition of specific bacterial hosts (30). For the protection of food from pathogens, phages represent an ideal approach to control specific pathogens while preserving beneficial bacteria in foods, such as fermented foods and foods containing live probiotics (16). Phage SFP10 has a specific bacterial inhibition spectrum for pathogenic Salmonella and E. coli O157:H7; EOP analysis supports high host specificity and stable inhibition for all SFP10-sensitive bacteria, suggesting that the phage may be useful as a biocontrol agent. In 2006, the Food and Drug Administration (FDA) approved the use of a commercial phage cocktail (ListShield; Intralytix, Inc.) as a biocontrol agent for direct use on foods to prevent Listeria monocytogenes contamination (10, 50), substantiating the safety of phage treatment for control of food-borne pathogens. Interestingly, one-step growth analysis of SFP10 phage showed a much shorter latent period and a larger burst size than another broad-host-range phage, AR1, showing a 40-min latent period and a 38-PFU burst size against E. coli O157:H7 (24), suggesting that SFP10 phage might be a better biocontrol agent against E. coli O157:H7. Generally, most Myoviridae bacteriophages showed latent periods of between 21 and 120 min and burst sizes between 50 and 100 PFU per infected host cell (14, 22, 55, 59, 60). The relatively short latent period and large burst size of SFP10 indicated high lytic activity and robust propagation of the phage.
Stability tests of phage SFP10 under various conditions of temperature and pH showed high stability, suggesting that it should be active and stable under various conditions of food processing and storage. Furthermore, BIM analysis of SFP10 phage showed that its frequencies of BIM are very low for E. coli O157:H7, suggesting the phage is active and stable with low emergence of SFP10-resistant mutants. However, we do not understand clearly why Salmonella showed high frequencies of BIM. The SFP10 phage resistance in Salmonella was also transient, similar to that of SPC35 phage (35). The underlying mechanism for high frequencies of BIM observed in Salmonella should be elucidated for efficient control of Salmonella by SFP10 phage.
Genome sequence analysis of phage SFP10 revealed novel insights into its genomic characteristics and potential functions for infection and propagation. This genome analysis showed that it contains the complete set of genes involved in DNA replication. However, the replication genes are not located in a gene cluster but scattered throughout the genome. For complete replication of the genome, the SFP10 genome encodes DNA polymerase with sliding clamp and sliding clamp loader complexes, DNA helicase with loader, DNA primase, DNA end protector, and the DNA topoisomerase II complex (Fig. 5). In addition, endonucleases, RNase HI, the NrdAB ribonucleotide reductase complex, and DexA exonuclease A are also encoded in the genome, possibly for obtaining nucleotides from host DNA or RNA, similar to phage T4 (11). Interestingly, the genome also has a complete recombination system consisting of a putative exonuclease complex encoded by gp46 and gp47 (SFP_0041 and 0042) for preparation of recombination, UvsX recombinase, UvsY recombination mediator protein, UvsW helicase, and single-stranded DNA binding protein for subsequent recombination. However, phage SPF10 does not form lysogens in Salmonella hosts. Furthermore, previous experiments revealed that this recombination system may be involved in DNA repair rather than phage DNA integration (41), supporting a lack of SFP10 lysogenization in Salmonella.
Overall, phylogenetic analysis of various bacteriophage major capsid proteins revealed that broadly host-specific phage SFP10 is closely related to Shigella phiSboM-AG3 phage and Salmonella Vi01 phage. However, comparative genomic analysis showed that even though they share most functional genes for phage reconstruction, host specificity-related genes are quite different, which may affect host infection. A recent phage-engineering study involving tail fiber protein replacement showed changes in host specificity, substantiating this hypothesis (43). It is intriguing that the tail fiber protein in phage SFP10 may target the LPS receptor of host E. coli O157:H7, similar to phage phiV10, but the tail spike protein in the same gene cluster may target the LPS receptor of host S. enterica, similar to phage Det7, suggesting how phage SFP10 can infect both bacteria (Table 3). The receptor of phage Vi01 was reported to be a Vi capsular antigen of Salmonella, and the tail spike protein encoded by Vi01_171C gene is a receptor binding protein (57). However, three host specificity-related genes (orf00210, orf00213, and orf00214) of Shigella phage phiSboM-AG3 showed low homology with a phage Vi01 gene encoding receptor binding protein targeting Vi capsular antigen (Table 3). Recent genome sequence analysis of Shigella boydii revealed no gene for biosynthesis of Vi capsular antigen, suggesting that the receptor of phiSboM-AG3 may not be Vi capsular antigen (2). In addition, no Shigella-specific phage gene related to host specificity was detected in BLASTP analysis, probably due to lack of information on other known Shigella phage receptor binding proteins in the GenBank database. However, the high similarity of these genomes suggests that they have evolved from a common ancestor but that the different host specificities were likely obtained after their evolutionary divergence.
Recently, the complete genome sequence of the AR1 phage infecting both E. coli O157:H7 and S. enterica was reported (24, 42). However, comparative genomic analysis of the SFP10 and AR1 phages revealed no homology between the two genomic DNAs (data not shown). In addition, phylogenetic analysis of the two phages revealed that they are not evolutionarily related (Fig. 6). AR1 phage cannot infect S. Typhimurium and uses OmpC as a receptor (42), suggesting AR1 may have a Salmonella infection mechanism different from that of SFP10 phage. Although these phages infect both E. coli O157:H7 and Salmonella, this comparative result shows why they do not share the host specificity-related genes in their genomes.
In this study, a bacteriophage capable of inhibiting two different major food-borne pathogens, Salmonella Typhimurium and E. coli O157:H7, was isolated and characterized. Our results underscore the potential usefulness, stability, and convenience of phage SFP10 for food safety and protection. This study also provides novel insights into bacteriophage targeting of multiple food-borne pathogens and describes the potential for new biocontrol agents. In addition, further genomic and mutational studies of phage SFP10 may provide insight into single-phage control of multiple other food-borne pathogens.
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (no. 20090078983). M.P. was the recipient of a graduate fellowship provided by the MEST through the Brain Korea 21 Project.
Footnotes
Published ahead of print 21 October 2011
REFERENCES
- 1. Altermann E, Klaenhammer TR. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7:161–169. [DOI] [PubMed] [Google Scholar]
- 2. Anany H, et al. 2011. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol. J. 8:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbara G, et al. 2000. Role of antibiotic therapy on long-term germ excretion in faeces and digestive symptoms after Salmonella infection. Aliment. Pharmacol. Ther. 14:1127–1131 [DOI] [PubMed] [Google Scholar]
- 4. Baxa U, et al. 1996. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 71:2040–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. 2007. Salmonella host range of bacteriophages that infect multiple genera. Poult. Sci. 86:2536–2540 [DOI] [PubMed] [Google Scholar]
- 7. Bigwood T, Hudson JA, Billington C, Carey-Smith GV, Heinemann JA. 2008. Phage inactivation of foodborne pathogens on cooked and raw meat. Food Microbiol. 25:400–406 [DOI] [PubMed] [Google Scholar]
- 8. Borie C, et al. 2008. Bacteriophage treatment reduces Salmonella colonization of infected chickens. Avian Dis. 52:64–67 [DOI] [PubMed] [Google Scholar]
- 9. Bruttin A, Brussow H. 2005. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob. Agents Chemother. 49:2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cairns BJ, Payne RJH. 2008. Bacteriophage therapy and the mutant selection window. Antimicrob. Agents Chemother. 52:4344–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calendar R. 2006. The bacteriophages, 2nd ed. Oxford University Press, New York, NY [Google Scholar]
- 12. Campbell A. 2003. The future of bacteriophage biology. Nat. Rev. Genet. 4:471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carver T, et al. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang H-C, et al. 2005. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage ΦSMA5. Appl. Environ. Microbiol. 71:1387–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 16. Coffey B, Mills S, Coffey A, McAuliffe O, Ross RP. 2010. Phage and their lysins as biocontrol agents for food safety applications. Annu. Rev. Food Sci. Technol. 1:449–468 [DOI] [PubMed] [Google Scholar]
- 17. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Lappe N, Doran G, O'Connor J, O'Hare C, Cormican M. 2009. Characterization of bacteriophages used in the Salmonella enterica serovar Enteritidis phage-typing scheme. J. Med. Microbiol. 58:86–93 [DOI] [PubMed] [Google Scholar]
- 19. Erickson M, et al. 2009. Competition among isolates of Salmonella enterica ssp. enterica serovar Typhimurium: role of prophage/phage in archived cultures. FEMS Microbiol. Lett. 294:37–44 [DOI] [PubMed] [Google Scholar]
- 20. Fauquet C, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed). 2005. Virus taxonomy. Eighth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 21. Feng P, Fields PI, Swaminathan B, Whittam TS. 1996. Characterization of nonmotile variants of Escherichia coli O157 and other serotypes by using an antiflagellin monoclonal antibody. J. Clin. Microbiol. 34:2856–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foschino R, Perrone F, Galli A. 1995. Characterization of two virulent Lactobacillus fermentum bacteriophages isolated from sour dough. J. Appl. Microbiol. 79:677–683 [Google Scholar]
- 23. Frank AC, Lobry JR. 2000. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 16:560–561 [DOI] [PubMed] [Google Scholar]
- 24. Goodridge L, Gallaccio A, Griffiths MW. 2003. Morphological, host range, and genetic characterization of two coliphages. Appl. Environ. Microbiol. 69:5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodridge LD. 2010. Designing phage therapeutics. Curr. Pharm. Biotechnol. 11:15–27 [DOI] [PubMed] [Google Scholar]
- 26. Hayashi K, et al. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hendrix RW. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506–511 [DOI] [PubMed] [Google Scholar]
- 28. Ho TD, Slauch JM. 2001. OmpC is the receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella. J. Bacteriol. 183:1495–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong J, et al. 2008. Identification of host receptor and receptor-binding module of a newly sequenced T5-like phage EPS7. FEMS Microbiol. Lett. 289:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70:217–248 [DOI] [PubMed] [Google Scholar]
- 31. Jensen EC, et al. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 33. Karmali MA, Steele BT, Petric M, Lim C. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 321:619–620 [DOI] [PubMed] [Google Scholar]
- 34. Khan MA, Satoh H, Katayama H, Kurisu F, Mino T. 2002. Bacteriophages isolated from activated sludge processes and their polyvalency. Water Res. 36:3364–3370 [DOI] [PubMed] [Google Scholar]
- 35. Kim M, Ryu S. 2011. Characterization of a T5-like coliphage SPC35 and differential development of resistance to SPC35 in Salmonella Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77:2042–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kropinski AM. 2009. Measurement of the rate of attachment of bacteriophage to cells, p. 151–155 In Clokie MRJ, Kropinski AM. (ed), Bacteriophages: methods and protocols, vol. 1 Isolation, characterization, and interactions Humana Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 37. Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Landstrom J, et al. 2008. Interaction of a Salmonella enteritidis O-antigen octasaccharide with the phage P22 tailspike protein by NMR spectroscopy and docking studies. Glycoconj. J. 25:137–143 [DOI] [PubMed] [Google Scholar]
- 40. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 41. Li X, Heyer W-D. 2008. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18:99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liao WC, et al. 2011. T4-like genome organization of the Escherichia coli O157:H7 lytic phage AR1. J. Virol. 85:6567–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mahichi F, Synnott AJ, Yamamichi K, Osada T, Tanji Y. 2009. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol. Lett. 295:211–217 [DOI] [PubMed] [Google Scholar]
- 44. McClelland M, et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 45. Mead PS, Griffin PM. 1998. Escherichia coli O157:H7. Lancet 352:1207–1212 [DOI] [PubMed] [Google Scholar]
- 46. Mead PS, et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mizoguchi K, et al. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 69:170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murray PR, Baron EJ. 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC. [Google Scholar]
- 49. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Flaherty S, Ross RP, Coffey A. 2009. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33:801–819 [DOI] [PubMed] [Google Scholar]
- 51. O'Flaherty S, et al. 2005. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl. Environ. Microbiol. 71:1836–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Flynn G, Ross RP, Fitzgerald GF, Coffey A. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ. 2008. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 104:1–13 [DOI] [PubMed] [Google Scholar]
- 54. Perry LL, et al. 2009. Sequence analysis of Escherichia coli O157:H7 bacteriophage ΦV10 and identification of a phage-encoded immunity protein that modifies the O157 antigen. FEMS Microbiol. Lett. 292:182–186 [DOI] [PubMed] [Google Scholar]
- 55. Petty NK, Foulds IJ, Pradel E, Ewbank JJ, Salmond GPC. 2006. A generalized transducing phage ΦIF3 for the genomically sequenced Serratia marcescens strain Db11: a tool for functional genomics of an opportunistic human pathogen. Microbiology 152:1701–1708 [DOI] [PubMed] [Google Scholar]
- 56. Pickard D, et al. 2008. Molecular characterization of the Salmonella enterica serovar Typhi Vi-typing bacteriophage E1. J. Bacteriol. 190:2580–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pickard D, et al. 2010. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J. Bacteriol. 192:5746–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Poppe C, et al. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559–565 [PMC free article] [PubMed] [Google Scholar]
- 59. Raya RR, et al. 2011. Naturally resident and exogenously applied T4-like and T5-like bacteriophages can reduce Escherichia coli O157:H7 levels in sheep guts. Bacteriophage 1:15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raya RR, et al. 2006. Isolation and characterization of a new T-even bacteriophage, CEV1, and determination of its potential to reduce Escherichia coli O157:H7 levels in sheep. Appl. Environ. Microbiol. 72:6405–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Samuel ADT, et al. 1999. Flagellar determinants of bacterial sensitivity to χ-phage. Proc. Natl. Acad. Sci. U. S. A. 96:9863–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Santos TMA, Bicalho RC. 2011. Complete genome sequence of vB_EcoM_ECO1230-10: a coliphage with therapeutic potential for bovine metritis. Vet. Microbiol. 148:267–275 [DOI] [PubMed] [Google Scholar]
- 63. Schade SZ, Adler J, Ris H. 1967. How bacteriophage χ attacks motile bacteria. J. Virol. 1:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sharma M, Patel JR, Conway WS, Ferguson S, Sulakvelidze A. 2009. Effectiveness of bacteriophages in reducing Escherichia coli O157:H7 on fresh-cut cantaloupes and lettuces. J. Food Prot. 72:1481–1485 [DOI] [PubMed] [Google Scholar]
- 65. Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sulakvelidze A, Alavidze Z, Morris JG., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tanji Y, et al. 2004. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 64:270–274 [DOI] [PubMed] [Google Scholar]
- 68. Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wall SK, Zhang J, Rostagno MH, Ebner PD. 2010. Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Appl. Environ. Microbiol. 76:48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walter M, et al. 2008. Structure of the receptor-binding protein of bacteriophage Det7: a podoviral tail spike in a myovirus. J. Virol. 82:2265–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wilcox SA, Toder R, Foster JW. 1996. Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res. 4:397–398 [DOI] [PubMed] [Google Scholar]
- 72. Yu SL, Ko KL, Chen CS, Chang YC, Syu WJ. 2000. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. J. Bacteriol. 182:5962–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 74. Zhang X, Kelly SM, Bollen WS, Curtiss R, III 1997. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 Δcrp and Δcdt deletion mutants. Infect. Immun. 65:5381–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]