Abstract
Lactobacillus rhamnosus GG, a probiotic with good survival capacity in the human gut, has well-documented adhesion properties and health effects. Recently, spaCBA-encoded pili that bind to human intestinal mucus were identified on its cell surface. Here, we report on the phenotypic analysis of a spaCBA pilus knockout mutant in comparison with the wild type and other adhesin mutants. The SpaCBA pilus of L. rhamnosus GG showed to be key for efficient adherence to the Caco-2 intestinal epithelial cell (IEC) line and biofilm formation. Moreover, the spaCBA mutant induces an elevated level of interleukin-8 (IL-8) mRNA in Caco-2 cells compared to the wild type, possibly involving an interaction of lipoteichoic acid with Toll-like receptor 2. In contrast, an L. rhamnosus GG mutant without exopolysaccharides but with an increased exposure of pili leads to the reduced expression of IL-8. Using Transwells to partition bacteria from Caco-2 cells, IL-8 induction is blocked completely regardless of whether wild-type or mutant L. rhamnosus GG cells are used. Taken together, our data suggest that L. rhamnosus GG SpaCBA pili, while promoting strong adhesive interactions with IECs, have a functional role in balancing IL-8 mRNA expression induced by surface molecules such as lipoteichoic acid.
INTRODUCTION
The human gastrointestinal (GI) tract lives in close harmony with a complex microbiota that contributes to digestion, pathogen exclusion, and optimal functioning of the epithelial barrier and immune system (36). Interest in the beneficial attributes of the human GI microbiota has led to the identification of various bacterial strains that are used frequently as probiotics. The main modes of action by which probiotics can promote human health are classified into three categories (23). These include (i) inhibition of pathogens, (ii) improvement of the epithelial barrier function, and (iii) modulation of host immune responses. The identity of the various probiotic molecules that exert these beneficial properties remains largely unknown. Moreover, whether cell-mediated adhesion to host surfaces is important for any of these three probiotic mechanisms is still a topic of debate (23, 24).
Lactobacillus rhamnosus GG is a well-documented probiotic strain (8). Examples of its proven clinical benefits include preventing and relieving certain types of diarrhea (11), reducing the incidence of respiratory infections in children (15) and impeding atopic disease (18). Up to now, only a few probiotic effector molecules, including lactic acid as an antimicrobial agent against Salmonella enterica serovar Typhimurium (7), secreted proteins that mediate homeostasis of intestinal epithelial cells (IECs) (43), and genomic DNA with anti-inflammatory effects in IECs (9), have been identified in L. rhamnosus GG. We have chosen this strain as a model for genetic studies to identify other probiotic molecules. Phenotypic comparison between the wild type and knockout mutants lacking a putative probiotic molecule offers the advantage that the functional role of these targeted molecules can be studied in situ with live bacteria.
Recently, comparative genomics of L. rhamnosus GG has revealed the presence of a gene cluster that encodes SpaCBA polymeric pili containing an SpaC mucus binding adhesin at the tip, which is missing in the dairy strain L. rhamnosus LC705 (19). Until then, Gram-positive pili were described only for pathogenic strains. These pili seem to function mainly in colonization and biofilm formation (28). The display of an adhesin at the tip of the extended pilus fiber has been suggested to facilitate the initial stages of bacterial adherence to host cells. Interestingly, the piliated pathogens form additional contacts with host cells through the binding of cell wall-anchored auxiliary pilin proteins and a variety of nonpilus adhesins. This ensuing “intimate zone of adhesion” with the host is suggested to permit the efficient delivery of virulence factors by these pathogens (28).
In a study analogous to those characterizing the role of pili in pathogenesis, we aimed to investigate whether the pili of the probiotic L. rhamnosus GG are key adhesion and immunomodulatory factors for IECs. Herein, we first performed a functional analysis of a mutant with knockout of the SpaCBA pilus-related genes and compared its phenotype with those of knockout mutants of other putative adhesins. In addition, the adhesive role of the spaCBA-encoded pili for mediating some immunomodulatory interactions with IECs was explored.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Wild-type L. rhamnosus GG and its mutant derivatives (Table 1) were grown at 37°C without agitation in MRS or lactobacilli AOAC medium (Difco). Escherichia coli and S. Typhimurium SL1344 cells (14) were grown with shaking at 37°C in Luria-Bertani (LB) medium (34). When required, the following antibiotics were used at the indicated final concentrations: tetracycline, 10 μg/ml; ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and erythromycin, 10 μg/ml for L. rhamnosus GG and 100 μg/ml for E. coli.
Table 1.
L. rhamnosus GG-derived strains used in this studya
| Strain | Genotype | Target locus | Inactivated molecule | Adhesion domain | Reference |
|---|---|---|---|---|---|
| ATCC53103 | Wild type | 8 | |||
| CMPG5230 | ΔmabA::Tcr | LGG_01865 | MabA = large modulatory protein of adhesion and biofilm formation | PF07564 (fibronectin) | 40 |
| CMPG5351 | ΔwelE::Tcr | LGG_02043 | Long galactose-rich EPS molecules | 26 | |
| CMPG5355 | ΔadhA::Tcr | LGG_02923 | Putative adhesion exoprotein | PF07523 (bacterial Ig-like domain) | This study |
| CMPG5356 | Δmbf::Tcr | LGG_02337 | Mbf = mucus binding factor | PF06458 (mucus) | This study; 41 |
| CMPG5357 | ΔspaCBA::Tcr | LGG_00442–LGG_00444 | spaCBA-encoded pili; SpaA = putative major pilin subunit, SpaB = putative minor pilin subunit at the base, SpaC = putative large-sized tip adhesin | SpaC: PF00092 (Von Willebrand factor type A domain) and PF05738 (collagen) | This study |
| CMPG5358 | ΔspaFED::Tcr | LGG_02370 | SpaD is the putative major backbone protein of cryptic SpaFED-type pili in L. rhamnosus GG | SpaD: PF05737 (collagen) | This study |
| CMPG5365 | ΔwelE::Tcr ΩspaC::Eryr | LGG_02043 LGG_00442 | Galactose-rich EPS and spaCBA-encoded pili (double mutant) | This study |
For more details on the adhesion domains, we refer to reference 19.
DNA manipulations.
Standard protocols were used for all DNA manipulations, including restriction digests, ligations, and transformation of E. coli (34). Plasmid DNA was isolated using the QIAprep spin kit according to the manufacturer's instructions (Qiagen). DNA amplification by PCR was performed using a Taq DNA polymerase (Roche) according to the manufacturer's recommended procedure. PCR primers (Table 2) were synthesized by Integrated DNA Technologies (Coralville, IA) and, when required, also included a restriction site at the 5′ ends to facilitate DNA cloning. Purified DNA fragments were recovered from 1.0% agarose gels by using the Qiagen gel extraction kit. L. rhamnosus GG cells were made electrocompetent by using the protocol described previously (6).
Table 2.
Primer sequences used in this study for the construction and verification of correct allelic replacement in the L. rhamnosus GG knockout mutants
| Primer name | Sequence (5′–3′)a | Restriction site | Target region |
|---|---|---|---|
| Pro-1058 | ATGGATCCCCTATCGCAAGCGCCGCAATGG | BamHI | 5′ HR1 spaCBA |
| Pro-1059 | ATCCCGGGCGTTTCACCACGACTTGTGTGGC | SmaI | 3′ HR1 spaCBA |
| Pro-1389 | ATAAGCTTGCCACTTTTCGCTGCACCGTGACG | HindIII | 5′ HR2 spaCBA |
| Pro-1390 | ATGAATTCCAGATGGGGCACGAATCGAGCTGG | EcoRI | 3′ HR2 spaCBA |
| Pro-1050 | ATACTAGTGGTTCGACACTGCTGTTTCGTAAG | SpeI | 5′ HR1 mbf |
| Pro-1051 | ATCCCGGGCGGGACAAAGATTGCCATCATGCC | XmaI | 3′ HR1 mbf |
| Pro-1048 | ATGTCGACCTTTCCAGCGACGCCCGAATG | SalI | 5′ HR2 mbf |
| Pro-1049 | ATGCGGCCGCGCTACCAGCCATAGCTTTCCTG | NotI | 3′ HR2 mbf |
| Pro-1052 | ATCCCGGGGTGGCCGCGGCTGCTAAACTGCTG | SmaI | 5′ HR1 adhA |
| Pro-1053 | ATACTAGTGCCAGCGCTTCTTGGCCTTGTAC | SpeI | 3′ HR1 adhA |
| Pro-1054 | ATGCGGCCGCGTTACGAGTATTGGCGCCAAGGC | NotI | 5′ HR2 adhA |
| Pro-1055 | ATTCTAGACTGACTGGCTGTTGTTTTGCCGTC | XbaI | 3′ HR2 adhA |
| Pro-1060 | ATCCCGGGGGGAACAAAGGTATCGGGCTG | SmaI | 5′ HR1 spaD |
| Pro-1061 | ATACTAGTGCTGCCAAGAGACTGTGCCCG | SpeI | 3′ HR1 spaD |
| Pro-1062 | ATGCGGCCGCGACAGGCGGAATCGGACTCTTCG | NotI | 5′ HR2 spaD |
| Pro-1063 | ATTCTAGAGCTCTGCCAACCTGATACATACCCC | XbaI | 3′ HR2 spaD |
| Pro-1733 | CGACGCTCTTGCTTGACTCAT_ | 5′ spaCBA::TcR | |
| Pro-1732 | CAGTTAAGCAATCAATCGTTACC_ | 3′ spaCBA::TcR | |
| Pro-1727 | CACGAAAGAGGGGCAAGAC_ | 5′ inlJ::TcR | |
| Pro-1728 | TAGATTTACTTTCATCCACTACTG_ | 3′ inlJ::TcR | |
| Pro-1730 | TACAGTCCATTCAACAAGCAGA_ | 5′ adhA::TcR | |
| Pro-1731 | CAACTTTAGGAATCAAATTAGCC_ | 3′ adhA::TcR | |
| Pro-1735 | CGTAACAGCCGACTTGTCC_ | 5′ spaD::TcR | |
| Pro-1734 | TTAGGGAGTCTGTTACTACTAC_ | 3′ spaD::TcR |
Underlining indicates restriction recognition sites.
Construction of knockout mutants by double homologous recombination.
Knockout mutants of L. rhamnosus GG were constructed according to strategies described previously (22). Briefly, open reading frames (ORFs) encoding putative adhesins targeted for inactivation were identified in the L. rhamnosus GG genome sequence (19). PCR primers were designed to amplify two ca.-1,000-bp-long homologous regions (HRs) that flank the 5′ and 3′ ends of the target gene(s) (Table 2). Each of the HR-containing PCR fragments were subsequently ligated into the respective multiple cloning sites (MCSs) upstream and downstream of the tetracycline resistance gene in the pCMPG10205 plasmid. The pCMPG10205 plasmid is derived from pFAJ5301 (22) by ligation of a tetracycline resistance gene from L. plantarum MD5057 (5) in the EcoRI site. Following heat shock transformation of chemically competent E. coli DH5α cells, the resulting suicide plasmids of each targeted adhesin gene were isolated and then electroporated into highly competent cultures of wild-type L. rhamnosus GG by using a method described previously (22). Positive knockout mutants, wherein successful double homologous recombination and allelic replacement have taken place, were selected according to tetracycline resistance and erythromycin sensitivity. Complete allelic replacement was confirmed by PCR using the Pro-249, Pro-250, and HR 5′-end and 3′-end primers (Table 2) as described before (22). The ΔwelE ΩspaC double mutant CMPG5365 is derived from the welE mutant CMPG5351 by integration of plasmid pCMPG10102, which insertionally inactivates the spaC gene (19). The spaCBA mutant CMPG5357 was complemented by introduction of a pLAB1301-derived plasmid (17), containing the spaCBA operon with its native promoter after PCR amplification with primers Pro-6188 (5′-ATGAATTCGACGCTCTTGCTTGACTCAT-3′) and Pro-3884 (5′-ATGAATTCTGAATTGTAGCACGGTC-3′) and ligation in the EcoRI cloning site.
Caco-2 cells.
The Caco-2 cell line, purchased from the American Type Culture Collection (ATCC) (Rockville, MD), was routinely maintained at 37°C with 5% CO2 and 90% relative humidity in 75-cm2 tissue culture flasks containing Dulbecco modified Eagle medium (DMEM)–F-12 (Invitrogen) (1:1) supplemented with 10% fetal bovine serum (FBS; HyClone). Every 3 days, when Caco-2 cell monolayers reached 70 to 80% confluence, cells were reseeded with a 1:7 split in fresh culture medium. For adhesion and immunomodulation experiments, Caco-2 cells were grown in 12-well culture plates (Cellstar) at a density of 4 × 104 cells/cm2. Confluence was reached within 3 or 4 days after seeding, and fully differentiated monolayers were obtained 15 days after seeding. Differentiation was confirmed by Western blot analysis testing for saccharase-isomaltase as described previously (38).
Adhesion assay to Caco-2 cells.
Experiments to assess the adhesion of various L. rhamnosus GG strains to epithelial Caco-2 cells were carried out by both the adhesion assay method described previously (26) and microscopic techniques. For the adhesion assay, a 1.5-ml volume of L. rhamnosus GG cells (1 × 107 CFU/ml or 108 CFU/ml) was added to tissue culture plate wells containing fully differentiated Caco-2 cells, which were allowed to incubate at 37°C for 1.5 h to mediate adherence. The cells were then rinsed twice with phosphate-buffered saline (PBS) prewarmed to 37°C and, after the addition of a 0.1-ml volume of 1× trypsin-EDTA (Invitrogen), incubated for 10 min at 37°C. Next, a 0.9-ml volume of PBS was added to each well, the cell suspension was mixed, and a set of serial dilutions was prepared and plated out on solid MRS medium. Following incubation at 37°C for 72 h, L. rhamnosus GG colonies were enumerated. The adhesion ratio, expressed as a percentage, was calculated by comparing the total number of bacterial colonies counted after adhesion to the number of cells in the bacterial suspension added originally to the tissue culture plate wells. Three independent experiments were performed in which all strains were tested in triplicate. The adhesion pattern of the L. rhamnosus GG strains to IECs was also visualized by epifluorescence microscopy. Hereto, L. rhamnosus GG strains were fluorescently labeled with fluorescein isothiocyanate (FITC) (0.5 mg/ml) (Sigma) and incubated with differentiated Caco-2 cells for 1 h. The samples were then rinsed twice with PBS and examined by epifluorescence microscopy using a Zeiss Axio Imager Z1 microscope equipped with an AxioCam MRm Rev.3 monochrome digital camera. Antibody-mediated inhibition experiments using anti-SpaC rabbit serum, similar to those described previously (19), were also included. Briefly, L. rhamnosus GG cells were preincubated for 10 min with anti-SpaC serum (diluted 1:100), after which the protocol as outlined above was continued.
EM and immunogold labeling.
Immunoelectron microscopy (immuno-EM) of wild-type L. rhamnosus GG and the EPS mutant (CMPG5351) was performed essentially as described previously (19). In brief, cells grown to stationary phase were bound to copper grids and treated with anti-SpaC antibodies, which were then labeled with protein A-conjugated gold particles (10 nm). Grids were negatively stained and examined with a JEOL 1200-EXII transmission electron microscope.
Biofilm formation experiments.
Biofilms of L. rhamnosus GG strains were grown for 72 h in AOAC medium and then evaluated by crystal violet staining as previously described (27). Biofilm formations of wild-type L. rhamnosus GG (positive control) and sterile growth medium (negative control) were included. Each of the analyses was performed eight times in experiments that were repeated in triplicate. The results were normalized against the positive control. When indicated, anti-SpaC antibody (diluted 1:100) was added to the biofilm medium.
Induction of cytokine gene expression in Caco-2 cells.
The Caco-2 cells growing in 12-well tissue culture plates (Costar) were deprived of FBS 1 day before the mRNA induction experiments. Wild-type and mutant L. rhamnosus GG cells were grown overnight in AOAC medium and then centrifuged at 4,000 × g at 4°C for 10 min. After rinsing with PBS, cells were resuspended in DMEM without FBS and adjusted to a final concentration of 1 × 108 CFU/ml. A 1.5-ml volume of the L. rhamnosus GG cell suspension was then added to the wells containing Caco-2 cells and incubated at 37°C (5% CO2 and 90% humidity) for 1.5 h. Included as controls were an identical number of S. Typhimurium SL1344 cells in the same medium (positive control) and DMEM unsupplemented with FBS (negative control). Afterwards, the cells were rinsed twice with prewarmed PBS and a 0.2-ml volume of PBS was added to each of the wells. RNA was extracted from the Caco-2 cells by using the high pure RNA isolation kit (Roche) following the manufacturer's protocol. Cytokine gene expression measurement by quantitative reverse transcription-PCR (qRT-PCR) is outlined below. To compare the effects of soluble factors released from the various L. rhamnosus GG strains, epithelial cells were incubated with the L. rhamnosus GG strains in a Transwell culture system (Nunc). Herein, L. rhamnosus GG cells in an upper chamber and Caco-2 cells in a lower chamber are separated by a 0.2-μm-pore-size permeable filter membrane support (Nunc), thereby minimizing any direct contact between bacterial cells and IECs. In addition, Caco-2 cells were coincubated for 1.5 h with recombinant L. rhamnosus GG SpaC pilin (2 μg/ml), exopolysaccharide (EPS) (2 μg/ml), and lipoteichoic acid (LTA) (1 μg/ml) molecules from L. rhamnosus GG and flagellin proteins from S. Typhimurium (3 μg/ml). Recombinantly produced SpaC pilin (19), L. rhamnosus GG-derived EPS (26), L. rhamnosus GG-derived LTA (30), and S. Typhimurium flagellin (16) were obtained as described previously. To investigate the involvement of TLR2, Caco-2 cells were pretreated for 1 h with anti-hTLR2-IgA monoclonal antibody (InvivoGen) (2 μg/ml), after which the Caco-2 cells were incubated with the L. rhamnosus GG cells for 1.5 h as outlined above.
Analysis of cytokine mRNA levels by qRT-PCR.
To determine cytokine mRNA expression by real-time quantitative PCR (RT-qPCR), commercially available methodologies for performing total cellular RNA extraction (high pure RNA isolation kit; Roche), reverse transcription (SuperScript III first-strand synthesis system; Invitrogen), and real-time DNA amplification (TaqMan universal PCR master mix; Applied Biosystems) were used. For RT-qPCR amplification, the StepOnePlus Real Time PCR system (Applied Biosystems, Lennik, Belgium) was used. All primers and probes were designed based on published sequences and chemically synthesized by Eurogentec (Seraing, Belgium) (Table 3). At least one primer or probe was designed to span an intron region of the matching cytokine gene. Purified plasmid DNA specific for each targeted cytokine gene served as cDNA plasmid standards and was used to quantify the respective cytokine in the test samples. The relative abundance of each mRNA species was measured by qPCR using 40 amplification cycles, with each cycle consisting of a denaturation step (94°C for 15 s) and a 1-min combined annealing-extension step (60°C). qPCR data are presented as a ratio of the mRNA level for cytokine genes over that for a housekeeping gene. Peptidylprolyl cis-trans isomerase A (PPIA), one of the most stably expressed genes in Caco-2 cells (13), served as the housekeeping gene. Nontemplate controls were included for each run.
Table 3.
Primers and probes used in this study for mRNA measurementsa
| Target mRNA | Primer or probe | Sequence (5′-3′) | Reference |
|---|---|---|---|
| PPIA | FW | CGCGTCTCCTTTGAGCTGTT_ | 13 |
| RV | CTGACACATAAACCCTGGAAT AATTC_ | ||
| TP | CAGACAAGGTCCCAAAGACAGCAGAAAATTT_ | ||
| IL-8 | FW | TGGCAGCCTTCCTGATTTCT_ | 2 |
| RV | TTAGCACTCCTTGGCAAAACTG_ | ||
| TP | CAGCTCTGTGTGAAGGT_ | ||
| TNF | FW | TCTTCTCGAACCCCGAGTGA_ | 10 |
| RV | CCTCTGATGGCACCACCAG_ | ||
| TP | TAGCCCATGTTGTAGCAAACCCTCAAGCT_ | ||
| IL-10 | FW | GTGATGCCCCAAGCTGAGA | 10 |
| RV | CACGGCCTTGCTCTTGTTTT | ||
| TP | CCAAGACCCAGACATCAAGGCGCA |
FW, forward primer; RV, reverse primer; TP, TaqMan probe, dually labeled with 5′ 6-carboxyfluorescein and 3′ 6-carboxytetramethylrhodamine; PPIA, peptidyl-prolyl cis-trans isomerase A; IL-8, interleukin-8; IL-10, interleukin-10; TNF, tumor necrosis factor.
Statistical analysis.
Significant differences in the data between wild-type L. rhamnosus GG and its mutant derivatives were determined using the unequal variance t test, in which P values of <0.05 were considered statistically significant.
RESULTS
SpaCBA pili play a key role in L. rhamnosus GG adhesion to IECs.
In previous studies, wild-type L. rhamnosus GG has been shown to display a high adhesion capacity to IECs and mucus (reviewed in reference 8). Recently, the SpaC pilin tip adhesin and mucus binding factor Mbf of L. rhamnosus GG were found to be involved in adhesion to human mucus (19, 41). In this study, we aimed to identify key adhesins of L. rhamnosus GG for binding to Caco-2 IECs based on a comparative knockout mutant analysis. Several LGG genes were selected for constructing knockouts based on the presence of putative adhesion domains in the encoding proteins (19) (Table 1). Subsequent adhesion assays revealed that the adhesion of L. rhamnosus GG to Caco-2 IECs is drastically reduced only for the pilus spaCBA knockout mutant CMPG5357 as compared to the wild type (Fig. 1A). Complementation of this mutant could restore adhesion to near-wild-type levels (data not shown). In comparison, the adhesion capacity of the mabA mutant CMPG5230 showed a moderate reduction (Fig. 1A), whereas the other putative adhesin mutants did not differ significantly from wild-type L. rhamnosus GG. The EPS-deficient welE mutant CMPG5351 showed a markedly increased adhesion capacity (Fig. 1A). Previously (25), we correlated the phenotype of CMPG5351 with the removal of the EPS layer and the subsequent increased surface accessibility for adhesion proteins. Here, we performed several experiments to characterize the augmented adhesion capacity of the EPS mutant in relation to the SpaCBA pilus fibers. First, the adhesion capacity of a double knockout of the spaCBA operon and the welE gene (CMPG5365) was shown to be significantly impaired (Fig. 1A). Next, by performing competition adhesion experiments with anti-SpaC serum, which blocks the accessibility to SpaCBA pilus fibers, we showed that the adhesion capacity of wild-type L. rhamnosus GG and the EPS mutant was reduced appreciably in both strains (Fig. 1A). Moreover, we were able to visualize an increased exposure of the SpaC-containing pili in the EPS mutant CMPG5351 by immunogold TEM (Fig. 1B). Only fully elongated pilus fibers were found to extend past the EPS layer in the wild-type strain. In contrast, SpaCBA pili and possibly SpaC monomeric proteins were observed throughout the exposed cell surface of the EPS-lacking mutant CMPG5351. Using epifluorescence microcopy, either with or without anti-SpaC serum, we further characterized the different adhesion patterns of the wild type and the pilus (CMPG5357) and EPS (CMPG5351) mutants. The adhesion of the pilus mutant appeared less specific and clearly attenuated, whereas that of the EPS mutant was tight and clustered but easily disrupted upon treatment with SpaC antiserum (Fig. 1C).
Fig 1.
The adhesion capacity of wild-type L. rhamnosus GG and its knockout mutant derivatives to IECs. (A) Overnight-grown L. rhamnosus GG cells were coincubated with the Caco-2 cells for 1.5 h, and thereafter the proportion of adherent bacteria, expressed as a percentage, was determined. Wild-type L. rhamnosus GG is indicated as LGG WT, and further details about the mutant strains are described in Table 1. The results shown are for 107 CFU/ml added, but similar trends were observed with 108 CFU/ml. Inclusion of anti-SpaC serum for antibody-mediated inhibition experiments is denoted by SpaC As. Each experiment was done in triplicate, and corresponding standard deviations are indicated by error bars. Data set comparisons (mutant strains versus the wild type) considered significant (P < 0.05) are indicated with an asterisk. (B) Immuno-EM analysis of L. rhamnosus GG and its EPS-deficient welE mutant (CMPG5351). SpaC-containing pili are identified by anti-SpaC antibodies conjugated to gold particles, which are seen as small dark dots (arrows). (C) Visualization of the adhesion pattern between various L. rhamnosus GG strains and epithelial Caco-2 cells by epifluorescence microscopy. Cells of wild-type L. rhamnosus GG (LGG WT) and the spaCBA (CMPG5357) and welE (CMPG5351) mutant strains are indicated. Inclusion of anti-SpaC antibody is denoted by SpaC As.
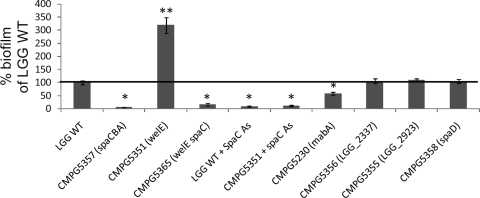
SpaCBA pili play a key role in L. rhamnosus GG biofilm formation.
Previously, we reported that L. rhamnosus GG can form biofilms on polystyrene and glass substrates more efficiently than related Lactobacillus strains (27). Here, we demonstrate that the spaCBA pilus mutant CMPG5357 can no longer form biofilms on polystyrene (Fig. 2) and glass (data not shown). In contrast, the EPS mutant CMPG5351 exhibits a substantial increase in biofilm formation (Fig. 2). As we suggested previously, this might be linked to the improved cell surface accessibility of adhesins such as pilus-like protein appendages (26). Moreover, we also observed that the capacity to form biofilms by the EPS- and SpaCBA pilus-deficient double mutant and by anti-SpaC-treated wild-type L. rhamnosus GG or EPS mutant is considerably reduced (Fig. 2), further highlighting the essential role of surface piliation for effective biofilm formation by L. rhamnosus GG. On the other hand, as we reported recently (40), the biofilm formation capacity of the mabA mutant CMPG5230 was reduced by only 2-fold (Fig. 2). In addition, the other putative adhesin mutants that were tested for their ability to produce biofilms showed little difference from the wild-type strain (Fig. 2).
Fig 2.
Comparison of biofilm formation of wild-type L. rhamnosus GG (LGG WT) and its knockout mutant derivatives. Formation of L. rhamnosus GG biofilms on polystyrene pegs occurring after 72 h of incubation in lactobacilli AOAC medium was quantified by crystal violet staining. Biofilm formation of the mutant strains was normalized to that of wild-type L. rhamnosus GG (set to 100%). Data are expressed as means ± standard deviations, and the data set comparisons (mutant strains versus the wild type) considered significant (P < 0.05) are indicated with an asterisk (double asterisks indicate increased biofilm formation compared to LGG WT).
SpaCBA pili modulate cytokine induction by L. rhamnosus GG in IECs.
Having established that the SpaCBA pili play a key role in adhesion to IECs, we subsequently aimed to investigate whether SpaCBA-mediated adhesion impacts on cytokine induction in the Caco-2 epithelial cell line. We first used a pathway-focused macroarray to obtain a general view on possible differences in the expression profiles of Caco-2 cells cocultured with the L. rhamnosus GG wild type or the spaCBA pilus mutant CMPG5357. These data suggest that the cytokine expression in Caco-2 cells is indeed modulated differently by the wild type and the CMPG5357 pilus mutant. For example, in pilus mutant-treated cells, gene expression for the proinflammatory markers interleukin-8 (IL-8) and tumor necrosis factor (TNF) is upregulated by more than 2.5-fold, whereas gene expression for the anti-inflammatory marker interleukin-10 (IL-10) is downregulated by a similar magnitude (data not shown). Confirmation of these data was obtained by quantifying cytokine mRNA expression using TaqMan qRT-PCR. Given that mRNA expression in Caco-2 cells is significantly higher for IL-8 than for either TNF or IL-10, we used IL-8 as the immunological marker for further characterizing the different responses induced by the L. rhamnosus GG strains. In comparison with wild-type L. rhamnosus GG, the pilus mutant (CMPG5357) accounted for an ∼2-fold increase in IL-8 mRNA levels (Fig. 3). Importantly, however, IL-8 mRNA induction by the SpaCBA pilus mutant remained much lower than that observed for the gastrointestinal pathogen Salmonella Typhimurium (Fig. 3). Unlike the wild type and pilus mutant strain CMPG5357, the L. rhamnosus GG EPS-deficient mutant CMPG5351, whose surface piliation is more exposed and accessible (Fig. 1B), causes a decrease in IL-8 mRNA expression (Fig. 3).
Fig 3.
Induction of IL-8 mRNA expression in IECs. A comparative analysis of mRNA induction of IL-8 in epithelial Caco-2 cells by various L. rhamnosus GG strains (108 CFU/ml) or purified recombinant SpaC pilin protein (2 μg/ml) and isolated L. rhamnosus GG EPS molecules (2 μg/ml) or LTA (1 μg/ml) was performed by TaqMan qRT-PCR. Included as positive controls were S. Typhimurium SL1344 cells (108 CFU/ml) and purified S. Typhimurium flagellin (3 μg/ml). DMEM served as the negative control. The presented data are the average of four independent experiments, and standard deviations are indicated. Data are expressed as means ± standard deviations, and the data set comparisons (mutant strains and isolated compounds versus the LGG wild type) considered significant (P < 0.05) (see Materials and Methods) are indicated with an asterisk (double asterisks indicate increased induction compared to LGG WT).
Involvement of SpaCBA pilus-mediated adhesion in immunomodulatory interactions. Based on the aforementioned results, it is apparent that the IL-8 mRNA induction by wild-type L. rhamnosus GG and the SpaCBA pilus and EPS mutants (Fig. 3) is inversely related to their adhesion capacity (Fig. 1A). This suggests that pili do not stimulate IL-8 production in Caco-2 cells. It can even be postulated that the presence of pili on the surface of L. rhamnosus GG cells dampens IL-8 mRNA induction in Caco-2 cells. Subsequently, we performed a series of experiments to gain better insight into how wild-type L. rhamnosus GG and the different mutant strains (CMPG5357 and CMPG5351) can affect induction of IL-8 in Caco-2 cells as well as on the role played by the SpaCBA pilus-mediated adhesion process. In the first experiment, we used a Transwell system, in which the bacteria and host cells are partitioned by a 0.22-μm membrane, to prevent all direct cell-cell contacts between the various L. rhamnosus GG strains and the Caco-2 cells. This way, none of the strains could induce IL-8 mRNA expression above the background level of DMEM (Fig. 4). These findings indicate that direct cell-cell contact is required for cell surface components of L. rhamnosus GG to trigger IL-8 mRNA induction. In a second experiment, we investigated whether purified recombinant SpaC pilin subunit and isolated EPS have a direct effect on IL-8 mRNA expression. In sharp contrast to the response elicited with purified S. Typhimurium flagellin (3 μg/ml) (Fig. 3), coincubation of Caco-2 cells with SpaC protein monomer (2 μg/ml) or EPS molecules (2 μg/ml) does not lead to substantive changes in IL-8 mRNA expression relative to the DMEM background control (Fig. 3). However, highly pure isolated LTA molecules from LGG (1 μg/ml), which have been previously suggested to be proinflammatory (4), can markedly increase IL-8 mRNA expression (Fig. 3).
Fig 4.
Role of direct cell-cell contact and TLR2-dependent modulation in L. rhamnosus GG-induced expression of IL-8 mRNA in IECs. The pattern of IL-8 mRNA induction in Caco-2 cells in response to various L. rhamnosus GG strains (108 CFU/ml) was determined by qRT-PCR. DMEM was included as the negative control. Direct contact between L. rhamnosus GG and Caco-2 cells was prevented by using the Transwell culture system equipped with a 0.22-μm-pore-size permeable membrane. In addition, the involvement of TLR2-mediated signaling was demonstrated by the 1-h pretreatment of Caco-2 cells with anti-human TLR2-IgA monoclonal antibody (2 μg/ml). The presented data are the average of three independent experiments. Data are expressed as means ± standard deviations. The data set comparisons (impact of Transwell or TLR2 antibody versus original situation) considered significant (P < 0.05) are indicated with an asterisk.
Finally, we also investigated the involvement of Toll-like receptor 2 (TLR2), an innate immune receptor that can detect a variety of microbe-associated molecular patterns (MAMPs) such as LTA, lipoproteins, and glycans (24). To test whether TLR2 contributes to the immunomodulatory capacity of the L. rhamnosus GG strains, we examined the effect of pretreatment of the Caco-2 cells with human TLR2-specific antibody. As shown in Fig. 4, antibody blockage of TLR2 abolished the increase in IL-8 mRNA expression that was associated with the SpaCBA pilus mutant and the LGG wild type but did not have a significant impact on the effect of the EPS mutant. Taken together, results from the Transwell experiment indicate that contact of L. rhamnosus GG with the Caco-2 cells is required for IL-8 mRNA induction by surface molecules such as LTA via TLR2 interaction, while the phenotype of the spaCBA mutant (Fig. 1A) indicates that “loose contact” (less than 1% adhesion) is sufficient. On the other hand, a strong adhesion of L. rhamnosus GG cells mediated by the SpaCBA pili appears to confer an attenuating effect on IL-8 mRNA induction.
DISCUSSION
Pili (or fimbriae) are elongated filamentous protein structures that protrude from the bacterial cell walls of both Gram-negative and Gram-positive bacteria (20, 32, 37). Unlike the pilus-like appendages in Gram-negative bacteria, Gram-positive pili represent a polymerized assemblage of different protein subunits (called pilins), which are covalently linked by the transpeptidase action of sortase enzymes. There is only a limited understanding about the biological role of the Gram-positive pilus, although reports in pathogens are steadily increasing (28, 32, 35, 37). Recently, we documented for the first time that surface piliation is associated with the probiotic L. rhamnosus GG (19, 26). Here, we investigated the functional significance of these multisubunit pili (called SpaCBA) in relation to biofilm formation, adhesion to IECs, and cytokine induction in IECs. In addition to the function attributed previously to the SpaC pilin subunit for binding to human intestinal mucus (19), our results point to an important role for the SpaCBA pilus fibers in the adherence of L. rhamnosus GG to human IECs. Furthermore, by incorporating knockout mutants of other putative adhesin genes in our study, we could confirm that the spaCBA-encoded pilus acts as a key mediator of the strong adhesive interactions that exist between L. rhamnosus GG and Caco-2 cells.
Given that the outer surface of L. rhamnosus GG cells, depending on the growth conditions, consists of a thick layer of EPS molecules (26), adhesin subunits (SpaC) affixed to the lengthy SpaCBA pilus fibers that extend through and beyond the surrounding EPS layer will likely facilitate the establishment of initial distal contacts between this probiotic strain and its targeted host cells. We have previously observed that the large-sized surface protein MabA can also modulate binding to Caco-2 cells (40) and that the recombinant Mbf protein can also bind to human mucus (41), but the mutant analysis in this study showed that their overall role in L. rhamnosus GG adhesion to IECs is a minor one compared to that of the SpaCBA pilus. However, a combination of different surface-localized proteins for host cell binding in L. rhamnosus GG agrees with the proposed “zipper model” for pilus-mediated adhesion among Gram-positive pathogens, in which initial bacterial contact with host cells is made with the long and adhesive pilus fibers, followed by other surface adhesins helping the formation of an intimate zone of contact between the bacterial and host cells (37). In light of the peristaltically driven high shear flow throughout the intestine, intestinal colonization by L. rhamnosus GG by use of this type of adhesion strategy might explain why in a comparative human intervention study this probiotic strain persisted for a longer length of time in the GI tract than the nonpiliated L. rhamnosus LC705 strain (19).
In many mucosal pathogens, such as Enterococcus faecalis (29) and group B streptococci (21), pili are recognized as important contributors in biofilm formation and pathogenesis. Although the importance of in situ biofilm formation by commensal or probiotic lactobacilli in the human intestine is not yet well understood (25), it can be envisaged that surface-attached microcolonies and biofilms of bacteria will likely increase their persistence and residence time in the GI tract (23). In our present study, we found that the SpaCBA pilus is a crucial element in the mechanism for in vitro biofilm formation by the probiotic L. rhamnosus GG, which seems to explain its unique in vitro high biofilm-forming ability (27), which is in agreement with the fact that pilus-mediated biofilm formation is rare in lactobacilli. However, as the in vivo biofilm-forming capacity of L. rhamnosus GG is rare in adult GI tracts (25), other factors such as nutritional flexibility and capacity to compete with the endogenous microbiota are also important in determining biofilm formation in situ.
As pili can mediate pathogenic adherence to the host epithelium, it is plausible that pili also influence mucosal immune responses (28, 37). However, even though in one study it was demonstrated that a piliated pneumococcal strain, relative to its nonpiliated mutant, can trigger a TNF-elicited inflammatory response in a mouse model (1), and aside from a few reports that describe Gram-negative pili as mediators of intestinal inflammation (3, 39), a role for pilus-like structures in stimulating pro- or anti-inflammatory signaling pathways in IECs remains largely undefined. In contrast, flagellins are well-documented mediators of immunomodulatory activity for pathogenic intestinal bacteria such as enteropathogenic Salmonella (12). Here, we aimed to investigate whether the pili from a probiotic Gram-positive strain are involved in mediating immunomodulatory signaling molecules in human IECs. Because of its relatively high and stimulus-dependent dynamic expression in the Caco-2 cells, we used IL-8 mRNA induction as an immunomodulatory marker. IL-8 is a chemokine produced by a variety of cell types, including epithelial cells and leukocytes, and it plays central roles in localized inflammation. IL-8 is regulated primarily at the level of gene transcription and is known as an early response gene (33). Our results indicate that the nonpiliated L. rhamnosus GG spaCBA mutant strain causes increased induction of this proinflammatory marker compared to the wild type in Caco-2 cells. In contrast, the EPS mutant, likely through its increased exposure of SpaCBA pili, causes a reduction in the IL-8 mRNA response, which suggests an inverse relationship between the stronger pilus-mediated adherence of this L. rhamnosus GG derivative strain and the triggering of IL-8 mRNA. Transwell experiments highlight the importance of direct cell contact for the induction of IL-8 mRNA by cell surface components of L. rhamnosus GG. Experiments with isolated molecules suggest that surface molecules such as LTA, but not EPS or SpaC pilins under the tested conditions, are principal inducers of IL-8. Thus, our mutant analysis shows that IL-8 mRNA induction in IECs after contact with L. rhamnosus GG differs remarkably depending on the presence of pili (strong adhesive contacts such as in the EPS mutant and the LGG wild type versus the loose association of the spaCBA mutant). Future studies are aimed at unraveling the molecular details of this pilus-mediated immunomodulatory activity. The SpaCBA pili could directly induce anti-inflammatory pathways or indirectly lead to anti-inflammatory signaling by promoting the release of anti-inflammatory factors like the p75 and p40 soluble proteins characterized previously (42, 43). The latter possibility is conceivable and analogous to the model proposed by Mandlik et al. (28), in which Gram-positive pathogens can deliver their virulence factors more efficiently after first establishing a pilus-mediated intimate interaction with host cells. As our current results also indicate, detailed unraveling of the signaling interactions between L. rhamnosus GG and IECs involves TLR2 and various bacterial ligands, such as pili, LTA, and EPS, but also other host receptors and bacterial factors (24).
To conclude, we have studied the functionality of SpaCBA piliation in L. rhamnosus GG by analyzing different phenotypic traits of mutants affected in surface properties. SpaCBA pili were shown to be a key factor involved in adherence to human IECs, biofilm formation, and dampening of IL-8 mRNA expression in IECs provoked by other cell surface components of L. rhamnosus GG such as LTA. To our knowledge, this is the first report that demonstrates such a role for pili in a beneficial Gram-positive bacterium. Given the widespread genomic potential of pilus-encoding genes in commensal, probiotic, and pathogenic strains (31), forthcoming studies on the different types of pili should focus on the identification of host cell receptors and the determination of their function in immunological signaling and host defense mechanisms, both in vitro and in vivo.
ACKNOWLEDGMENTS
At the time of the experiments, Sarah Lebeer was a postdoctoral researcher funded by the Fund for Scientific Research (FWO) of Flanders, Belgium. Ingmar Claes holds a Ph.D. grant from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT–Vlaanderen). Funding of this project at K.U.Leuven was in part through the Fund for Scientific Research, Flanders (FWO) (grant G.0236.07) and in part through TEKES (Finnish Funding Agency for Technology and Innovation) (grant 201/08). Work performed at the University of Helsinki was funded by an Academy of Finland research grant (118165) and was part of the Center of Excellence in Microbial Food Safety Research (MiFoSa) and the Research Program on Nutrition, Foods, and Health (ELVIRA).
We acknowledge Mariya Petrova, Raf Mols, and Patrick Augustijns for their invaluable help with the culture of the Caco-2 cells. Anneleen Demuynck is gratefully acknowledged for LTA isolation, purification, and structural analysis. We also thank Ellen Dilissen at K.U.Leuven for her technical assistance with the setup of the qRT-PCR experiments as well as Esa Pohjolainen, Katariina Kojo, and Marko Sutinen at the University of Helsinki for their excellent technical expertise in purifying recombinant SpaC protein. Docent Ilkka Palva (University of Helsinki) is thanked for helpful comments. Soile Tynkkynen, Riitta Korpela, and Tuomas Salusjärvi (Valio Ltd., Research and Development, Helsinki, Finland) are thanked for their support throughout this project.
Footnotes
Published ahead of print 21 October 2011
REFERENCES
- 1. Barocchi MA, et al. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103:2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bullens DM, et al. 2006. IL-17 mRNA in sputum of asthmatic patients: linking T cell driven inflammation and granulocytic influx? Resp. Res. 3:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carvalho FA, et al. 2009. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206:2179–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Claes IJJ, et al. 2010. Impact of lipoteichoic acid modification on the performance of the probiotic Lactobacillus rhamnosus GG in experimental colitis. Clin. Exp. Immunol. 162:306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Danielsen M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98–103 [DOI] [PubMed] [Google Scholar]
- 6. De Keersmaecker SCJ, et al. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl. Environ. Microbiol. 72:4923–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Keersmaecker SCJ, et al. 2006. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol. Lett. 259:89–96 [DOI] [PubMed] [Google Scholar]
- 8. Doron S, Snydman DR, Gorbach SL. 2005. Lactobacillus GG: bacteriology and clinical applications. Gastroenterol. Clin. North Am. 34:483–498 [DOI] [PubMed] [Google Scholar]
- 9. Ghadimi D, Vrese MD, Heller KJ, Schrezenmeir J. 2010. Effect of natural commensal-origin DNA on Toll-like receptor 9 (TLR9) signaling cascade, chemokine IL-8 expression, and barrier integrity of polarized intestinal epithelial cells. Inflamm. Bowel Dis. 16:410–427 [DOI] [PubMed] [Google Scholar]
- 10. Giulietti A, et al. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386–401 [DOI] [PubMed] [Google Scholar]
- 11. Guandalini S, et al. 2000. Lactobacillus GG administered in oral rehydration solution to children with acute diarrhea: a multicenter European trial. J. Pediatr. Gastroenterol. Nutr. 30:54–60 [DOI] [PubMed] [Google Scholar]
- 12. Hayashi F, et al. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103 [DOI] [PubMed] [Google Scholar]
- 13. Hayeshi R, et al. 2008. Comparison of drug transporter gene expression and functionality in Caco-2 cells from 10 different laboratories. Eur. J. Pharm. Sci. 35:383–396 [DOI] [PubMed] [Google Scholar]
- 14. Hoiseth SK, Stocker BAD. 1981. Aromatic dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 15. Hojsak I, et al. 2010. Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: a randomized, double-blind, placebo-controlled trial. Clin. Nutr. 29:312–316 [DOI] [PubMed] [Google Scholar]
- 16. Ikeda JS, et al. 2001. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 69:3021–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Josson K, et al. 1989. Characterization of a Gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9–20 [DOI] [PubMed] [Google Scholar]
- 18. Kalliomäki M, et al. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076–1079 [DOI] [PubMed] [Google Scholar]
- 19. Kankainen M, et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kline KA, Dodson KW, Caparon MG, Hultgren SJ. 2010. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 18:224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Konto-Ghiorghi Y, et al. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lebeer S, et al. 2007. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189:860–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lebeer S, Vanderleyden J, De Keersmaecker S. 2008. Genes and molecules of Lactobacillus supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lebeer S, Vanderleyden J, De Keersmaecker S. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 25. Lebeer S, et al. 2011. FISH analysis of Lactobacillus biofilms in the gastrointestinal tract of different hosts. Lett. Appl. Microbiol. 52:220–226 [DOI] [PubMed] [Google Scholar]
- 26. Lebeer S, et al. 2009. Identification of a gene cluster for the biosynthesis of a long galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 75:3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lebeer S, Verhoeven TLA, Perea Vélez M, Vanderleyden J, De Keersmaecker SCJ. 2007. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73:6768–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nallapareddy SR, et al. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116:2799–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perea Vélez M, et al. 2007. Functional analysis of d-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73:3595–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plyusnin I, Holm L, Kankainen M. 2009. LOCP–locating pilus operons in Gram-positive bacteria. Bioinformatics 25:1187–1188 [DOI] [PubMed] [Google Scholar]
- 32. Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell. Mol. Life Sci. 66:613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roebuck KA. 1999. Regulation of interleukin-8 gene expression. J. Interferon Cytokine Res. 19:429–438 [DOI] [PubMed] [Google Scholar]
- 34. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Scott JR, Zahner D. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol. Microbiol. 62:320–330 [DOI] [PubMed] [Google Scholar]
- 36. Sonnenburg JL, Angenent LT, Gordon JI. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569–573 [DOI] [PubMed] [Google Scholar]
- 37. Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. 2006. Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4:509–519 [DOI] [PubMed] [Google Scholar]
- 38. Trugnan G, Rousset M, Chantret I, Barbat A, Zweibaum A. 1987. The posttranslational processing of sucrase-isomaltase in HT-29 cells is a function of their state of enterocytic differentiation. J. Biol. Chem. 104:1199–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tükel C, et al. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58:289–304 [DOI] [PubMed] [Google Scholar]
- 40. Vélez PM, et al. 2010. Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation. FEMS Immunol. Med. Microbiol. 59:386–398 [DOI] [PubMed] [Google Scholar]
- 41. von Ossowski I, et al. 2011. Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 77:4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan F, et al. 2011. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Invest. 121:2242–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan F, et al. 2007. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132:562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]