Abstract
Acinetobacter baylyi ADP1 is naturally competent and proficient at homologous recombination, so it can be transformed without restriction digests or ligation reactions. Expression vectors for this system, however, are not yet widely available. Here we describe the construction and characterization of inducible expression vectors that replicate as plasmids in A. baylyi or integrate into a nonessential part of its chromosome. These tools will facilitate the engineering of genes and genomes in this promising model organism.
TEXT
Acinetobacter baylyi ADP1 is arguably superior to Escherichia coli for the engineering of genes and genomes (4, 10). Mid-log A. baylyi cells are naturally competent (11) and catalyze the homologous recombination of DNA efficiently. A. baylyi extrudes 1 to 3 μg of chromosomal DNA/ml of liquid culture (16), so sexual recombination occurs under normal growth conditions. The transformation of A. baylyi does not require a refrigerated centrifuge, a −80°C freezer, or an electroporator. The synthesis of primers and genes can be outsourced, so a motivated student could potentially engineer and express genes in an inexpensively equipped laboratory (with an Internet-connected computer, a thermocycler, a gel electrophoresis apparatus, and an incubated shaker). Here we describe, and make available, a series of inducible A. baylyi expression vectors that either replicate as plasmids or integrate efficiently into the chromosome.
Assembly strategy.
We desired a family of A. baylyi expression vectors with different inducible promoters, selectable markers, and replication mechanisms (chromosomal integration or plasmid replication) because no single expression vector is ideal for all applications. We did not know a priori which promoters and ribosome binding sites would work in A. baylyi (particularly in combination with foreign genes and replicons), so we expected some trial and error. We adopted the BioBrick standard because it is well-suited for combinatorial assembly projects, including the construction of vector families (15). A BioBrick is a cloned sequence of DNA (of any length) that is flanked by standard restriction sites (EcoRI-NotI-XbaI-payload-SpeI-NotI-PstI); the compatibility of overhangs created by restriction endonucleases XbaI and SpeI enables the ligation of any two appropriately digested BioBricks to produce a single tandem BioBrick.
BioBrick-accepting plasmid.
A multiple cloning site (MCS; SphI-ribosome binding site-NcoI-EcoRV-HindIII) flanked by the BioBrick restriction sites was created by PCR amplification of an intentional primer dimer; the resulting product was purified, restriction digested, and cloned into the DraIII and AflIII sites of pSL1180 (Pharmacia) to create a custom BioBrick-accepting vector (pIM1154 in Table 1, or pIMBB [2]). This plasmid retained its ColE1 origin, which replicates efficiently in E. coli but not in A. baylyi (7, 8), and beta-lactamase marker. DNA sequences that do not contain internal restriction sites (EcoRI, NotI, XbaI, SpeI, or PstI) become BioBricks when cloned into the MCS of this plasmid. They can subsequently be iteratively combined with other BioBricks for the assembly of the desired vectors.
Table 1.
Plasmids used in this study
| Plasmid | Relevant insert(s) | Description, source, and/or reference |
|---|---|---|
| Source plasmids | Each source plasmid contains regulators, promoters, and markers (not flanked by BioBrick restriction sites) | Internal EcoRI, NotI, XbaI, SpeI, and PstI sites must also be eliminated |
| pET28a+ | Kanr | Novagen |
| pCDF Duet | pCDF-NheI-Spcr | Novagen |
| pACYC184 | Tetr | New England Biolabs |
| lacZ-gfp-pCA24N | lacI-PT5 | 9 |
| pRBL | lacI-Ptac | 14 |
| lacI-pCA24N | lacI (no gfp) | 9 |
| recA-gfp-pCA24N | recA (not a BioBrick) | 9 |
| gusA-gfp-pCA24N | gusA (not a BioBrick) | 9 |
| lacY-gfp-pCA24N | lacY (not a BioBrick) | 9 |
| pBAD myc his a | araC-PBAD (not a BioBrick) | Invitrogen |
| pSL1180 | MCS (not a BioBrick) | Pharmacia |
| pWH1266 | 8 | |
| Non-BioBrick precursors | Plasmids contain internal restriction sites, not BioBrick compatible | All used to create lacI-PT5-gusA BioBrick (pIM1202) |
| pIM1117 | lacI-PT5-pRBL | PCR product of lacI-PT5 from pCA24N + pRBL |
| pIM1119 | lacI-PT5-pCDF Duet | lacI-PT5 from pIM1117 + pCDF Duet |
| pIM1155 | lacI-PT5-lacI-pCDF Duet | lacI from lacI-pCA24N + pIM1119 |
| pIM1159 | lacI-PT5-lacI-pIMBB | lacI-PT5-lacI from pIM1115 + pIM1154 |
| BioBrick precursors | BioBricks used to assemble regulators, promoters, and selectable markers | All conform to BioBrick standard but are not directly useful for vector assembly |
| pIM1154 | MCS (SphI-ribosome binding site-NcoI-EcoRV-HindIII) | BioBrick cloning vector (pIMBB) (2) |
| pIM1171 | lacI-PT5-recA-pIMBB | PCR product of recA from recA-gfp-pCA24N + pIM1179; precursor of pIM1202 |
| pIM1179 | lacI-PT5-lacI-pIMBB | lacI from lacI-pCA24N + pIM1171; precursor of pIM1202 |
| pIM1278 | PT5-lacI-pIMBB-3 | Upstream lacI removed with XhoI and SphI; precursor of pIM1466 |
| pIM1458 | PT5-lacI-pIMBB-4 | pIM1278 without NcoI or HindIII site |
| pIM1180 | lacI-PT5-lacY-pIMBB | lacY from lacY-gfp-pCA24N + pIM1171; precursor of pIM1265 |
| pIM1211 | Tetr-lacY-pIMBB | Tetr from pACYC184 + pIM1180; precursor of pIM1265 |
| BioBrick elements | BioBricks used to assemble expression vectors | |
| pIM1157 | Kanamycin phosphotransferase (Kanr) | Derived from pET28a+ (Novagen) |
| pIM1212 | Spectinomycin adenyltransferase (pCDF-NheI-Spcr) | Derived from pCDF Duet (Novagen) |
| pIM1265 | Tetracycline/H+ antiporter (Tetr) | Derived from pACYC184 (New England Biolabs) |
| pIM1462 | PT5-lacOc-tdk | Derived from ASKA collection |
| pIM1446 | gusA | gusA from pIM1202 + pIM1154 |
| pIM1218 | lacI-PT7-gusA | Derived from pCDF Duet (Novagen) |
| pIM1219 | lacI-Ptac-gusA | Derived from pRBL (14) |
| pIM1221 | araC-PBAD-gusA | Derived from pBAD myc his A (Invitrogen) |
| pIM1217 | pobR-Ppob-gusA | Derived from A. baylyi genome |
| pIM1202 | lacI-PT5-gusA | Derived from pCA24N (9) |
| pIM1460 | PT5-lacOc-gusA | Derived by site-directed mutagenesis in pIM1219 |
| pIM1466 | PT5-lacI-gusA | Derived from pCA24N |
| pIM1215 | ADP1 prophage segment 2.1 | PCR product from A. baylyi genome + pIM1154 |
| pIM1216 | ADP1 prophage segment 2.2 | PCR product from A. baylyi genome + pIM1154 |
| pIM1480 | ADP1 prophage segment 4.1 | PCR product from A. baylyi genome + pIM1154 |
| pIM1502 | ADP1 prophage segment 4.2 | PCR product from A. baylyi genome + pIM1154 |
| Integration vectors | ||
| pIM1356 | pp2.1-pobR-Ppob-gusA-Spcr-pp2.2 | pIM1215 + pIM1217 + pIM1212 + pIM1216 |
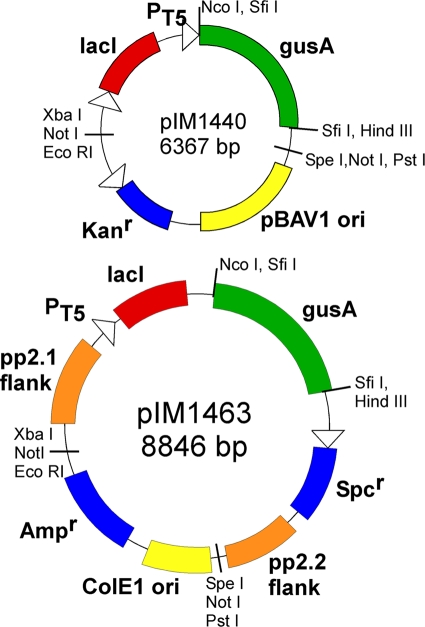
| pIM1463 | pp2.1-PT5-lacI-gusA-Spcr-pp2.2 | pIM1215 + pIM1466 + pIM1212 + pIM1216; available from Addgene |
| pIM1517 | pp4.1-araC-PBAD-gusA-PT5-lacOc-tdk-Spcr-pp4.2 | pIM1480 + pIM1221 + pIM1462 + pIM1212 + pIM1502 |
| pIM1251 | pp2.1-PT5-lacI-gusA-Kanr-pp2.2 | pIM1215 + pIM1466 + pIM1157 + pIM1216 |
| pIM1253 | pp2.1-PT5-lacI-gusA-Tetr-pp2.2 | pIM1215 + pIM1466 + pIM1265 + pIM1216 |
| pBAV1k-based expression vectors | ||
| pIM1522 | PT5-lacO-lacO-gfp | Broad-host-range plasmid (pBAV1k-T5-gfp [GenBank accession no. HQ191434.1]); available from Addgene (3) |
| pIM440 | lacI-PT5-gusA-pBAV1k | pIM1202 + pIM1522 (GenBank accession no. JF828582); available from Addgene |
| pIM1441 | lacI-PT7-gusA-pBAV1k | pIM1218 + pIM1522 (GenBank accession no. JF828583); available from Addgene |
| pIM1445 | lacI-Ptac-gusA-pBAV1k | pIM1219 + pIM1522 (GenBank accession no. JF828584); available from Addgene |
| pIM1442 | araC-PBAD-gusA-pBAV1k | pIM1221 + pIM1522 (GenBank accession no. JF828585); available from Addgene |
| pIM1444 | pobR-Ppob-gusA-pBAV1k | pIM1217 + pIM1522 (see Table S2 in the supplemental material) |
| pWH1266-based plasmid vectors | All Kanr instead of Ampr | Expression from these vectors was constitutive |
| pIM1266 | MCS (SphI-ribosome binding site-NcoI-EcoRV-HindIII) | Kanr from pET28a+ replaces TEM-1 beta-lactamase marker in pIM1154 |
| pIM1272 | pWH1266 origin | PCR product of pWH1266 ori + pIM1266 |
| pIM1311 | pWH1266 origin | pIM1272 without HindIII site |
| pIM1317 | lacI-PT5-gusA-pWH1266 ori | pIM1202 + pIM1311 |
| pIM1318 | pobR-Ppob-gusA-pWH1266 ori | pIM1217 + pIM1311 |
| pIM1319 | lacI-PT7-gusA-pWH1266 ori | pIM1218 + pIM1311 |
| pIM1320 | lacI-Ptac-gusA-pWH1266 ori | pIM1219 + pIM1311 |
BioBrick cloning.
Markers from E. coli plasmids that confer resistance to kanamycin (Kanr), spectinomycin (Spcr), and tetracycline (Tetr) were cloned into pIM1154, thereby converting them into BioBricks (pIM1157, pIM1212, and pIM1265, respectively). A series of expression cassette BioBricks, each containing a regulator gene, a promoter, a ribosome binding site, and the E. coli gusA reporter gene (lacI-PT7-gusA, lacI-Ptac-gusA, araC-PBAD-gusA, pobR-Ppob-gusA, lacI-PT5-gusA, PT5-lacOc-gusA, and PT5-lacI-gusA), was constructed by PCR amplification and cloning (yielding pIM1218, pIM1219, pIM1221, pIM1217, pIM1202, pIM1460, and pIM1466, respectively; see the supplemental material). In each case, the reporter gene was preceded by a ribosome binding site and an NcoI site on the 5′ end and an HindIII site on the 3′ end (Fig. 1). The origin of replication of an endogenous A. baylyi plasmid, pWH1266 (8), was PCR amplified and cloned into pIMBB-Kanr (pIM1266) to create pWH1266-pIMBB-Kanr (pIM1311). Finally, four 1-kb sequences derived from the large prophage region of A. baylyi (arbitrarily designated pp2.1, pp2.2, pp4.1, and pp4.2) were cloned into pIM1154 to create pIM1215, pIM1216, pIM1480, and pIM1502, respectively.
Fig 1.
Examples of plasmid (top) and integration (bottom) vectors. Each contains a single BioBrick flanked by restriction sites EcoRI-NotI-XbaI (prefix) and SpeI-NotI-PstI (suffix). The BioBrick in the integration vector (bottom) was assembled from smaller BioBricks carrying the flanks, expression cassette (regulator-promoter-reporter gene), and selectable marker. The vectors rely upon the function of foreign promoters and ribosome binding sites (each pair is indicated by an open triangle) in A. baylyi ADP1.
Plasmid assembly.
We constructed two sets of A. baylyi ADP1 expression vectors, one based upon pWH1266, the aforementioned endogenous Acinetobacter plasmid (8), and the other upon pBAV1k, a broad-host-range plasmid that is stably maintained at high copy number in Gram-negative and Gram-positive bacteria (3). Several expression cassettes, namely, lacI-PT7-gusA, lacI-Ptac-gusA, pobR-Ppob-gusA, and lacI-PT5-gusA, were cloned in parallel into the BioBrick cloning sites of pWH1266-pIMBB-Kanr (to create pIM1319, pIM1320, pIM1318, and pIM1317, respectively) and pBAV1k (to create pIM1441, pIM1445, pIM1444, and pIM1440, respectively). An arabinose-induced vector, araC-PBAD-gusA-pBAV1k (pIM1442), was also constructed. A. baylyi cells were separately transformed with each of the resulting expression vectors (Table 1) spread onto Luria broth agar plates supplemented with 50 μg/ml kanamycin (LB-kan plates) and incubated overnight at 37°C.
Plasmid function.
Individual transformed colonies were used to inoculate liquid LB-kan cultures. The cultures were propagated to saturation by overnight agitation at 37°C; saturated cultures were diluted 1:30 in fresh medium, propagated to mid-log phase, and induced with 10 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG; for PT7, Ptac, and PT5), 10 mM l-arabinose (for PBAD), 5 mM p-hydroxybenzoic acid (for Ppob), or sterile water (to provide an uninduced negative control for all expression vectors). The cultures were incubated overnight, harvested by centrifugation, and lysed. The cell extracts were reacted with saturating (1 mM) concentrations of 4-methylumbelliferyl-β-d-glucuronide (BMUG); the formation of the 4-methylumbelliferone product was monitored in a microtiter plate spectrofluorimeter (the Molecular Devices M5 reader). We observed efficient (>100-fold) induction for the phage (PT7 and PT5) and E. coli (Ptac and PBAD) promoters on the pBAV1k-based vectors (Fig. 2, top 8 bars). All four of these plasmids should be useful for the heterologous expression and engineering of other genes in A. baylyi.
Fig 2.
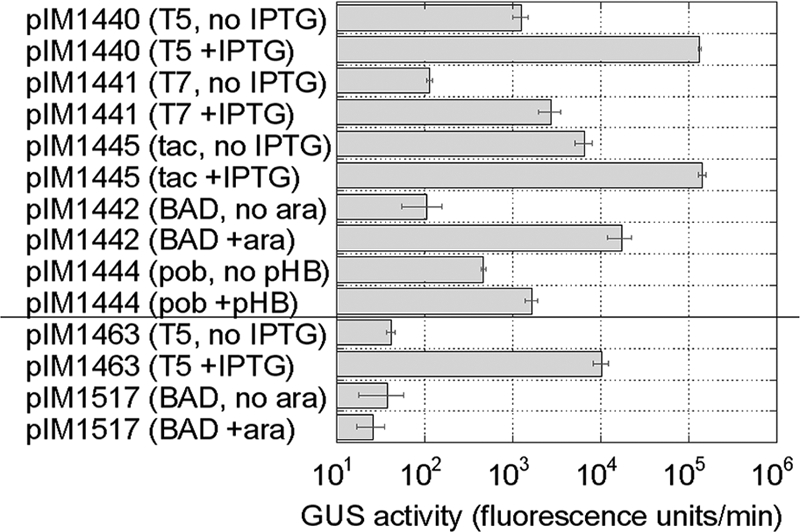
Novel expression vectors function in A. baylyi ADP1. Wild-type A. baylyi ADP1 cells were transformed with plasmid and integration vectors (described in Table 1). Each transformed strain was separately propagated to mid-log phase and challenged with an inducer (IPTG, arabinose, or p-hydroxybenzoate) or water (negative control) for 3 h. The cells were harvested by centrifugation and lysed; the cell extract was reacted with BMUG, and the formation of the fluorescent 4-methylumbelliferone product was monitored in a microtiter plate spectrofluorimeter. The activity was derived from the slope (fluorescence per unit of time) and plotted on the log scale. Error bars represent standard errors from three independent trials.
Native promoter and plasmid origin.
Induction of gusA from the native pob promoter with p-hydroxybenzoic acid, however, was far less efficient (<4-fold). Overall expression levels were relatively modest (Fig. 2, compare 9th and 10th bars with top 8 bars). The pob promoter is well-characterized (5, 6), but its performance might not scale up at high copy number (58 copies/cell [3]). We were also puzzled by the behavior of the pWH1266-pIMBB-Kanr-based vectors. All of them produced modest amounts of β-glucuronidase (GUS), with no detectable difference in the presence or absence of an inducer (data not shown). The pWH1266 origin enables replication within A. baylyi, but unlike the pBAV1k origin, it apparently affects the function of adjacent foreign promoters.
Integration vector assembly.
The BioBricks were also assembled into integration vectors (Fig. 1 and Table 1). Each was constructed in the ColE1-based BioBrick-accepting plasmid pIM1154, which does not replicate efficiently in A. baylyi (7, 8). Each was flanked by 1-kb sequences derived from the large prophage of the A. baylyi chromosome (nucleotides 2116840 to 2169436 of the chromosomal sequence [1]) to integrate the intervening payload into this nonessential region. A. baylyi strains were transformed with integration vectors and spread on LB agar plates supplemented with 50 μg/ml spectinomycin (for pIM1356, pIM1463, and pIM1517) or 50 μg/ml kanamycin (for pIM1251). The tetracycline resistance marker derived from E. coli plasmid pBR322 (pIM1253) did not enable the selection of transformed A. baylyi in our hands.
Integration vector function.
A. baylyi transformed with the PT5-lacI-gusA integration vector (pIM1463) expressed almost 250-fold more GUS activity in the presence of 1 mM IPTG than in its absence (Fig. 2, 11th and 12th bars). In contrast, the induction of an integrated araC-PBAD-gusA expression cassette (pIM1517) with 100 mM l-arabinose did not increase GUS activity relative to that in uninduced controls (Fig. 2, 13th and 14th bars), even though the same expression cassette carried by a multicopy plasmid was efficiently induced. Endogenous A. baylyi sigma factors apparently do not bind the PBAD promoter very tightly, as high copy numbers are necessary for detectable induction. A. baylyi transformed with the pobR-Ppob-gusA integration vector (pIM1356) produces various amounts of GUS activity, either in the presence (1.8 mM) or in the absence of p-hydroxybenzoic acid.
Real-time PCR analysis of the gusA gene showed that PT5-lacI-gusA (pIM1463) A. baylyi strains always had a single copy of the gusA gene (0.7 ± 0.1/cell, relative to the obligate, single-copy relA or rel[ACIAD3326] gene). In contrast, different pobR-Ppob-gusA (pIM1356) A. baylyi isolates contained highly variable gusA copy numbers (23 ± 0.7, 340 ± 35, 114 ± 14, or 531 ± 128 copies/cell). We suspect that the unpredictable copy number variation of pobR-Ppob-gusA (pIM1356) is a function of gene amplification (12, 13), so it is not reliable as an expression vector. We remain confident, however, that the other expression vectors with nonnative promoters, namely, the PT5-lacI-gusA integration vector (pIM1463) and the plasmid expression vectors (pIM1440, pIM1441, pIM1445, and pIM1442), will enable the engineering of genes and genomes in A. baylyi.
Nucleotide sequence accession numbers.
Nucleotide sequences of newly constructed plasmids have been deposited in GenBank and are listed in Table 1 (excluding HQ191434.1).
Supplementary Material
ACKNOWLEDGMENTS
We thank Valérie de Crécy-Lagard (University of Florida) and Laura Cuff (University of Georgia) for their advice and insightful comments.
This work was supported by grants from the NIH (1 R01 GM074264 and 1 R01 GM086824).
Footnotes
Published ahead of print 21 October 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Barbe V, et al. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48:463–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bryksin AV, Matsumura I. 2010. Rational design of a plasmid origin that replicates efficiently in both gram-positive and gram-negative bacteria. PLoS One 5:e13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Berardinis V, Durot M, Weissenbach J, Salanoubat M. 2009. Acinetobacter baylyi ADP1 as a model for metabolic system biology. Curr. Opin. Microbiol. 12:568–576 [DOI] [PubMed] [Google Scholar]
- 5. DiMarco AA, Averhoff B, Ornston LN. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DiMarco AA, Ornston LN. 1994. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J. Bacteriol. 176:4277–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gralton EM, Campbell AL, Neidle EL. 1997. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology 143(Pt. 4):1345–1357 [DOI] [PubMed] [Google Scholar]
- 8. Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51 [DOI] [PubMed] [Google Scholar]
- 9. Kitagawa M, et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 10. Metzgar D, et al. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palmen R, Hellingwerf KJ. 1997. Uptake and processing of DNA by Acinetobacter calcoaceticus—a review. Gene 192:179–190 [DOI] [PubMed] [Google Scholar]
- 12. Reams AB, Neidle EL. 2004. Gene amplification involves site-specific short homology-independent illegitimate recombination in Acinetobacter sp. strain ADP1. J. Mol. Biol. 338:643–656 [DOI] [PubMed] [Google Scholar]
- 13. Reams AB, Neidle EL. 2003. Genome plasticity in Acinetobacter: new degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol. Microbiol. 47:1291–1304 [DOI] [PubMed] [Google Scholar]
- 14. Ren ZJ, Baumann RG, Black LW. 1997. Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: construction of a T4 phage hoc gene display vector. Gene 195:303–311 [DOI] [PubMed] [Google Scholar]
- 15. Shetty RP, Endy D, Knight TF., Jr 2008. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.