Abstract
The two-component regulatory system, GraRS, appears to be involved in staphylococcal responses to cationic antimicrobial peptides (CAPs). However, the mechanism(s) by which GraRS is induced, regulated, and modulated remain undefined. In this study, we used two well-characterized MRSA strains (Mu50 and COL) and their respective mutants of graR and vraG (encoding the ABC transporter-dependent efflux pump immediately downstream of graRS), and show that (i) the expression of two key determinants of net positive surface charge (mprF and dlt) is dependent on the cotranscription of both graR and vraG, (ii) reduced expression of mprF and dlt in graR mutants was phenotypically associated with reduced surface-positive charge, (iii) this net reduction in surface-positive charge in graR and vraG mutants, in turn, correlated with enhanced killing by a range of CAPs of diverse structure and origin, including those from mammalian platelets (tPMPs) and neutrophils (hNP-1) and from bacteria (polymyxin B), and (iv) the synthesis and translocation of membrane lysyl-phosphatidylglycerol (an mprF-dependent function) was substantially lower in graR and vraG mutants than in parental strains. Importantly, the inducibility of mprF and dlt transcription via the graRS-vraFG pathway was selective, with induction by sublethal exposure to the CAPs, RP-1 (platelets), and polymyxin B, but not by other cationic molecules (hNP-1, vancomycin, gentamicin, or calcium-daptomycin). Although graR regulates expression of vraG, the expression of graR was codependent on an intact downstream vraG locus. Collectively, these data support an important role of the graRS and vraFG loci in the sensing of and response to specific CAPs involved in innate host defenses.
INTRODUCTION
Two-component regulatory systems (TCRS) are prototypical signal transduction mechanisms utilized by most bacteria to monitor and respond to environmental stimuli. These systems typically use a membrane protein sensor and a response regulator activated via a phosphorelay to control target gene transcription (48). It has been recently shown that GraRS, a TCRS in Staphylococcus aureus, plays a functional role in expression of the heterotypic versus homotypic vancomycin-intermediate resistance phenotypes (VISA) (9, 18, 26). In addition, mutations in graRS or its adjacent ABC transporter genes vraFG (encoding an ATPase and a permease) render strains hypersusceptible to vancomycin, as well as to polymyxin B (PMB; a cyclic cationic bacterium-derived peptide) (26). Extending this observation, Li et al. demonstrated that graRS (also called aps, for antibiotic peptide sensor) and vraFG are coinvolved in promoting resistance to distinct cationic antimicrobial peptides (CAPs) in S. aureus (23). In several strain backgrounds, GraRS has been shown to regulate expression of the immediate downstream locus, vraFG, as well as mprF, and dltABCD (17, 23, 26). MprF is a lysyl-phosphatidylglycerol (L-PG) synthase which adds positively charged lysine molecules to phosphatidylglycerol within the S. aureus cell membrane and also functions as an outer membrane translocase for L-PG (33, 44, 46). Besides MprF, the dltABCD operon also contributes to the net positive surface charge by covalently attaching d-alanine to cell wall teichoic acids(46). Since both the mprF and dlt operons participate in maintaining overall staphylococcal surface positive charge (33, 36, 44), we proposed that mutations in graRS could impact susceptibility to CAPs, potentially via a surface charge-dependent mechanism. Indeed, downregulation of these graRS-regulated genes in graRS mutants has been linked to increased susceptibility to selected CAPs (23, 26). However, the exact molecular mechanisms by which GraRS regulates expression of mprF, dlt, and vraFG genes in mediating CAP resistance are not well understood.
In the present study, we utilized isogenic graR and vraG parent-mutant strain pairs in two distinct methicillin-resistant S. aureus (MRSA) genetic backgrounds, Mu50 and COL, to characterize the contribution of these two linked loci to (i) the induction of mprF and dlt expression by sublethal concentrations of a range of CAPs, (ii) the modulation of cell surface charge, and (iii) in vitro resistance to a cadre of CAPs of distinct structures and origins.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in the present study are listed in Table 1. S. aureus Mu50, a prototypical clinical VISA isolate, has been well characterized phenotypically (e.g., homotypic VISA) and is virulent in vivo in animal models (7, 8, 22, 45). Similarly, S. aureus COL, a prototypical MRSA laboratory strain with a known genome, has been studied extensively in vitro and is virulent in a number of animal models (10, 15). All mutant strains were generated by allelic replacement using the plasmid pMAD, resulting in deletion of the coding sequence, as described previously (2, 26). For selected studies, we utilized the MU50 parental strain, its vraG deletion mutant, and a complemented vraG mutant containing a plasmid expressing vraFG in trans (Table 1).
Table 1.
Strains and plasmid used in this study
| S. aureus strain or plasmid | Description | Reference |
|---|---|---|
| S. aureus strains | ||
| Mu50 | MRSA, VISA, wild-type strain | 22 |
| Mu50 ΔgraR | graR in-frame deletion mutant of Mu50 | 26 |
| Mu50 ΔvraG | vraG in-frame deletion mutant of Mu50 | 26 |
| COL | MRSA, wild-type strain | 15 |
| COL ΔgraR | graR in-frame deletion mutant of COL | 26 |
| COL ΔvraG | vraG in-frame deletion mutant of COL | 26 |
| Plasmid | ||
| pEPSA5::vraFG | pEPSA5 expressing vraFG genes from Mu50 | 26 |
All S. aureus strains were grown in either tryptic soy broth (TSB; Difco Laboratories, Detroit, MI) or Mueller-Hinton broth (MH; Difco Laboratories, Detroit, MI) for individual experiments. Liquid cultures were grown in Erlenmeyer flasks at 37°C with shaking (250 rpm) in a volume that was no greater than 10% of the flask volume. All strains were maintained at −70°C until thawed before each experimental run.
CAPs.
PMB was purchased from Sigma Chemicals Co. (St. Louis, MO). Human neutrophil peptide-1 (hNP-1), a prototypical α-defensin, was purchased from Peptide International (Louisville, KY). RP-1 (a synthetic 18-amino-acid congener modeled in part upon α-helical microbicidal domains of platelet factor-4 family PMPs), was prepared and authenticated as detailed before (49, 56). Of note, the antistaphylococcal mechanisms of RP-1 recapitulate that of native PMP-1 (49). Because of the large amounts of peptide required for both susceptibility and gene induction studies, the RP-1 peptide was used instead of thrombin-induced platelet microbicidal proteins isolated and purified from fresh mammalian platelets (49, 56). Peptides hNP-1 and RP-1 were used for both in vitro killing assays (see below) and gene induction studies for the study strains; PMB was used in selected gene induction experiments, The CAPs described above differ in primary and secondary structures, mechanisms of action, and cationicity (ranging from +4 to +6 at neutral pH) (49, 50). For gene induction experiments, these three CAPs were utilized at the following exposure concentrations: hNP-1, 50 μg/ml; RP-1, 50 μg/ml; and PMB, 60 μg/ml. These concentrations did not exert substantial killing of the bacterial inoculum over a 30-min exposure period (data not shown).
In addition to the cationic peptides above, we utilized the following cationic molecules to examine for target gene inducibility: vancomycin, calcium-daptomycin, and gentamicin. Each agent was purchased from their respective pharmaceutical sources, reconstituted according to manufacturer's instructions and utilized at the following concentrations for gene induction studies: vancomycin, 8 μg/ml; calcium-daptomycin, 2 μg/ml; and gentamicin, 512 μg/ml. As for the CAPs described above, these peptide concentrations did not exert substantial killing of the bacterial inoculum over the 30-min gene induction period (data not shown).
CAP susceptibility testing.
Standard MIC testing in nutrient broth may underestimate most CAP activities (50, 57). Accordingly, in vitro bactericidal assays were carried out with RP-1 and hNP-1 as described previously using a 2-h microdilution method in Eagle minimal essential medium (20, 50). We used an inoculum of 103 CFU of exponential-growth-phase cells and CAP concentrations of RP-1 at 3 μg/ml and hNP-1 at 20 μg/ml. These CAP concentrations were selected based on extensive pilot studies showing their inability to substantially reduce starting inocula of either parental strain over the 2-h exposure period. The data were calculated and expressed as the relative percent surviving CFU (± the standard deviation [SD]) of CAP-exposed versus CAP-unexposed cells, with the survival of each parental strain set at 100%. A minimum of three independent runs was performed for each CAP.
Isolation of RNA.
For RNA isolation, fresh overnight cultures of S. aureus strains were used to inoculate TSB to an optical density at 600 nm (OD600) of 0.1. Cells were harvested during the exponential growth phase (at 2 h), the early stationary phase (at 6 h), and the late stationary phase (at 12 h). Total RNA was isolated from the cell pellets using an RNeasy kit (Qiagen, Valencia, CA) and a FastPrep FP120 instrument (Bio 101, Vista, CA), according to the manufacturer's recommended protocols.
Transcription analyses.
Quantitative real-time PCR (qRT-PCR) analyses were performed as described previously (5). Briefly, 1 μg of DNase-treated RNA was reverse transcribed using a SuperScript III first-strand synthesis kit (Invitrogen) according to the manufacturer's protocols. Quantification of the cDNA levels was performed according to the instructions of the Power SYBR Green Master Mix kit (Applied Biosystems) on an ABI Prism 7000 sequence detection system (Applied Biosystems). The primers used to amplify mprF were qRT-mprF-F (5′-TTGTAGGTTTCGGTGGCTTT-3′) and qRT-mprF-R (5′-GATGCATCGAAAACATGGAA-3′). The dltA and gyrB genes were similarly detected using respective specific primers as described before (5).
RT-PCR was performed as described previously (52). Briefly, mprF, dltA, and vraF cDNAs were generated using Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Beverly, MA) and gene-specific reverse primers (26). The RT-PCR primers used for the detection of the gyrA transcripts have been described previously (39). Amplification was performed with initial denaturation at 95°C for 1 min, followed by 18 to 20 cycles of denaturation at 95°C for 30s, annealing at 52°C for 30s, and extension at 72°C for 30s, followed in turn by a final extension at 72°C for 5 min. For all quantitation studies, gene expression of the parental strain was normalized to “1”, and those of all mutants were quantified relative to the parental strain accordingly.
To assess the induction of mprF and dlt by CAPs and other cationic molecules, RT-PCR analyses were performed on RNA samples from cultures of the strain sets exposed to hNP-1, RP-1, PMB, vancomycin, calcium-daptomycin, or gentamicin. Briefly, overnight cultures of the strain sets were used to inoculate 20 ml of TSB to an OD600 of 0.1 and allowed to grow for 2 h (∼108 CFU/ml) before the addition of hNP-1 (50 μg/ml), RP-1 (50 μg/ml), PMB (60 μg/ml), vancomycin (8 μg/ml), calcium-daptomycin (2 μg/ml), or gentamicin (512 μg/ml). The cultures were incubated for an additional 30 min before the RNA was harvested. The sublethality of these CAP concentrations over 30 min was confirmed by quantitative culture (data not shown).
Net cell surface charge.
To quantify relative cell surface charge in our parental and mutant constructs, we used one of two assays with polycations: (i) poly-l-lysine (PLL) binding or (ii) cytochrome c binding (Sigma). The binding of fluorescein isothiocyanate (FITC)-labeled PLL to the S. aureus isolates was performed with a well-described flow cytometric assay (20, 30). In this analysis, the extent of FITC-labeled PLL inversely reflects the relative surface positive charge. The data are expressed as mean relative fluorescence units (± the SD). In selected assays requiring large amounts of reagent, (e.g., CAP induction studies), we also determined binding to cytochrome c as a complementary surface charge assay (26, 37). Previous studies have documented a close correlation between these two surface charge assays in S. aureus (20, 46, 51, 53). The binding of cytochrome c was measured with a spectrophotometric assay which quantifies the amount of the polycation remaining within reaction mixture supernatants following exposure to the study strains, with higher amounts of residual cytochrome c in the supernatants correlating with a more positive surface charge (20, 26, 30, 51). The data are calculated and expressed as the percentage of cytochrome c bound to the cell. The data shown for both surface charge assays are the means (± the SD) of three independent experiments.
Membrane PL contents.
MprF is an enzyme involved in the synthesis and outer membrane translocation of lysyl-phosphatidylglycerol (L-PG), one of the three major S. aureus membrane phospholipids (PLs; L-PG, PG, and cardiolipin [CL]) (12, 33, 36). To quantify the relative proportions of these three PLs in our strain sets, membrane PLs were extracted from S. aureus cell pellets as described previously (30). The target PLs were separated by two-dimensional thin-layer chromatography, removed from the plates, and then quantified spectrophotometrically by a previously described chemical assay (20, 30). The proportion of synthesized l-PG that was translocated to the outer cell membrane leaflet was quantified spectrophotometrically, as detailed before using the l-PG outer-membrane-impermeable UV probe, fluorescamine (29, 30). The data were expressed as the proportionalities (± the SD) of the three PLs. At least three independent experiments were performed to analyze the PL contents.
Cell membrane fluidity.
The relative membrane order characteristics (i.e., fluidity/rigidity) of S. aureus can independently modify interactions with CAPs (29). To assure that any differences seen among the study strains in CAP susceptibility profiles were not attributable to membrane order perturbations, membrane fluidity was determined by fluorescence polarization spectroscopy using the fluorescent probe 1,6-diphenyl-1,3,5-hexatriene (DPH) as described previously (4, 29). The data were quantified by determination of the polarization index (± the SD) (20, 50). These assays were performed at least six times for each strain on separate days.
Statistics.
Data were analyzed by using Kruskal-Wallis analysis of variance, with a P value of <0.05 considered significant.
RESULTS
CAP susceptibility.
To assess the role of graRS and vraFG in resistance to structurally distinct host defense CAPs, we examined the in vitro susceptibility profiles of graR and vraG knockout mutants of Mu50 and COL against two prototypical antistaphylococcal CAPs: RP-1 (a synthetic congener of platelet factor-4 family of microbicidal molecules) (55, 56) and hNP-1 (a human neutrophil CAP). As shown in Table 2, deletion of graR or vraG resulted in significantly increased susceptibilities to both RP-1 and hNP-1 killing of the mutants compared to the respective parental strains (P < 0.05). Meehl et al. have also shown that both knockout mutants above were significantly more susceptible to vancomycin and polymyxin B (PMB) in these two strain backgrounds (26), but exhibited only slighly enhanced susceptibility to, calcium-daptomycin and gentamicin, both cationic antimicrobials (26).
Table 2.
Effect of graR and vraG mutations on CAP susceptibilities and PL profiles
| Strain | Mean ± SDa |
|||
|---|---|---|---|---|
| % Survival after 2-h exposure to: |
% L-PG among overall PL content |
|||
| 3 μg of RP-1/ml | 20 μg of hNP-1/ml | Total L-PG (inner + outer) | Outer L-PG only | |
| S. aureus Mu50 (parental) | 100† | 100 | 12.89 ± 0.16 | 4.42 ± 1.13 |
| Mu50 ΔvraG | 65.23 ± 2.03* | 79.47 ± 7.63* | 9.46 ± 2.81* | 1.57 ± 0.4* |
| Mu50 ΔgraR | 33.91 ± 0.81* | 59.78 ± 4.01* | 9.69 ± 0.49* | 1.65 ± 0.24* |
| S. aureus COL (parental) | 100† | 100 | 15.63 ± 0.33 | 5.28 ± 0.05 |
| COL ΔvraG | 49.17 ± 12.96* | 32.19 ± 2.77* | 12.65 ± 1.09* | 4.29 ± 1.83 |
| COL ΔgraR | 53.33 ± 18.86* | 53.23 ± 10.09* | 12.04 ± 1.12* | 4.31 ± 0.86 |
*, P < 0.05 versus the parental strains. †, Parental strains were normalized to 100%.
Regulation of mprF and dltABCD expression by GraRS and VraFG.
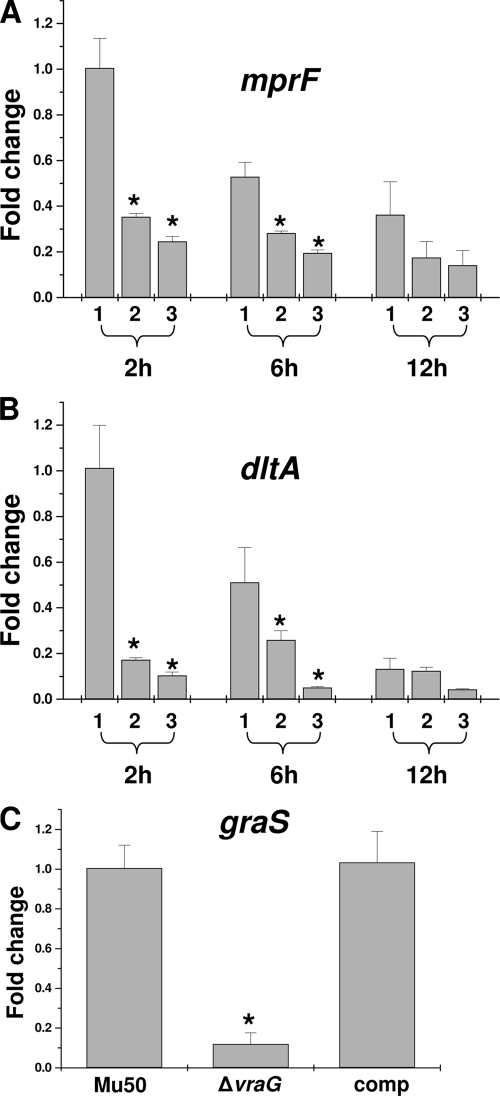
To determine the effect of graR and vraG mutations on expression of mprF and dlt genes, qRT-PCR analyses were performed on RNA samples isolated from cultures of Mu50 strain set. As shown in Fig. 1A and B, in agreement with previous findings with MRSA strain MW2 (23), transcription of the mprF and dlt genes in VISA strain Mu50 was significantly decreased in the isogenic graR mutant ∼4-fold and ∼8-fold, respectively, during the early exponential growth phase (2 h of growth; P < 0.01). Similar to the graR mutant, the vraG mutant strain also displayed significantly decreased early-exponential-phase expression of mprF and dlt compared to the parental Mu50 strain (∼2.5- and ∼5-fold, respectively). Similar, statistically relevant expression outcomes were noted at the late exponential phase (6 h growth) for both mprF and dlt expression. By the stationary phase (12 h growth), the expression differences between parental and graR or vraG mutants were minimal. These data demonstrate that graRS regulates expression of mprF and dlt, at least in part, via a vraFG-dependent mechanism(s), in a growth-phase-dependent manner.
Fig 1.
Transcriptional regulation of mprF and dlt expression by GraRS and VraFG. (A) Relative transcription level of mprF in parental Mu50 (1), its ΔvraG mutant (2), and its ΔgraR mutant (3) strains. (B) Relative transcription of the dlt in Mu50 (bar 1), ΔvraG (bar 2), and ΔgraR (bar 3) strains. (C) Effect of vraG mutation on graRS transcription. RNA samples from parental Mu50, its ΔvraG mutant, and its complemented ΔvraG mutant containing pEPSA5::vraFG were isolated at 2 h postinoculation (exponential growth phase) and subjected to a qRT-PCR analysis. *, P < 0.01.
Furthermore, the effect of vraG on the expression of graRS was also assessed by qRT-PCR. As shown in Fig. 1C, the vraG mutant exhibited ∼9-fold-lower graS expression than the parent during exponential growth, whereas the complemented vraG mutant (containing the plasmid pEPSA5 expressing vraFG) exhibited restored parental-level graS expression. These data demonstrated the interdependency of graRS and vraFG genes and suggest the presence of a positive-feedback loop between graRS and vraFG.
Effect of the graRS and vraFG mutations on cell surface charge.
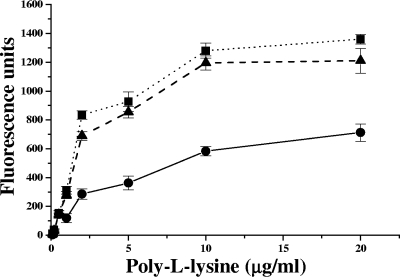
Since GraRS and VraFG positively coregulate expression of mprF and dltABCD, each of which, in turn, is critical for maintaining overall positive surface charge, we assessed the relative surface charges of our strain sets. As shown in Fig. 2, the net positive surface charges of both the graR and vraG mutants were significantly lower than that of the parental strain Mu50, as determined by enhanced fluorescent-PLL binding (P < 0.01). Similar reductions in the net positive surface charge were also observed in the graR and vraG mutant strains compared to the parental COL strain (P < 0.01; data not shown).
Fig 2.
Binding of FITC-labeled PLL to whole S. aureus cells. The graph shows the relative fluorescence units (± the SD) of FITC-labeled PLL bound to Mu50 (●), ΔvraG (■), and ΔgraR (▲) whole cells: the lower the number of fluorescence units, the greater the PLL repulsion and the more positively charged the S. aureus cell envelope (30).
Effect of the graR and vraG mutations on synthesis and translocation of l-PG.
The synthesis and translocation of the positively charged l-PG molecule (which confers increased net positive charge to the cell surface) is dependent on expression of mprF (12, 20, 31, 44, 46, 54). Since graRS regulates the expression of mprF and graRS expression appears to be dependent on an intact vraFG, the impact of graR and vraG on the synthesis and “flipping” of l-PG to the outer surface of the cell membrane was investigated. As shown in Table 2, the total proportion of l-PG synthesized within the overall PL content was significantly reduced in the graR and vraG mutant strains compared to both respective parental strains (P < 0.05). The amount of l-PG translocated to the outer cell membrane was also reduced, especially for the Mu50 strain set. The impact of vraG and graR mutations on translocation of l-PG failed to reach statistical significance in the COL strain set, indicating that the net impact of the GraRS-VraFG regulatory system on global MprF protein function (i.e., l-PG synthesis and translocation) may be strain specific. Of note, these phenotypic data on l-PG production and flipping roughly parallel the impact of the graR and vraG mutations on relative positive surface charge noted above.
Membrane fluidity.
Cell membrane fluidity analyses revealed no substantive differences among the parental, graR, and vraG mutant isolates in either strain background (data not shown).
Induction of mprF and dlt expression by specific CAPs via GraRS-VraFG network.
Previous studies have shown that certain CAPs (e.g., hBD-3; indolicidin) can induce expression of graRS and its downstream regulated genes, including mprF and dlt (23). To assess whether expression of mprF is inducible by our study CAPs (hNP-1, RP-1, or PMB), RT-PCR analyses were performed in parental strain Mu50. As shown in Fig. 3, both RP-1 (50 μg/ml) and PMB (60 μg/ml), but not hNP-1 (50 μg/ml), were associated with increased transcription of mprF. Similarly, expression of dlt was induced by RP-1 and PMB, but not by hNP-1. The expression of both mprF and dlt in MRSA strain COL was also inducible with PMB at the 30- and 60-μg/ml concentrations (data not shown).
Fig 3.
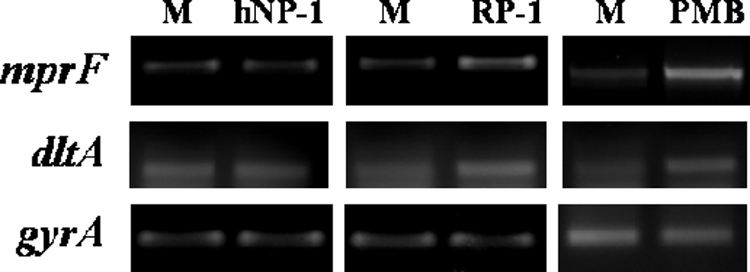
Induction of mprF and dlt transcription by specific CAPs. RT-PCR analyses were performed on RNA samples from cultures of Mu50 strain exposed to hNP-1 (50 μg/ml), RP-1 (50 μg/ml), or PMB (60 μg/ml) for 30 min during exponential growth. M, CAP-free medium alone.
Next, using PMB as a prototypical inducer of mprF and dlt expression in Mu50, we hypothesized that this induction event is dependent on an intact graRS-vraFG network. Thus, RT-PCR analyses were also performed on RNA samples from the parental Mu50 strain and its isogenic graR and vraG mutant strains, in the presence or absence of PMB. As predicted, induction of mprF and dlt expression by PMB was dependent on an intact graRS locus (Fig. 4). Similar to the graR mutant, the vraG mutant strain also failed to induce expression of mprF and dltA in the presence of PMB. In addition, the vraG mutant strain exhibited a more pronounced effect on dlt expression than on mprF expression. Collectively, these data indicate that induction of mprF and dlt gene expression is dependent on the TCRS GraRS, as well as the ABC transporter system, VraFG.
Fig 4.
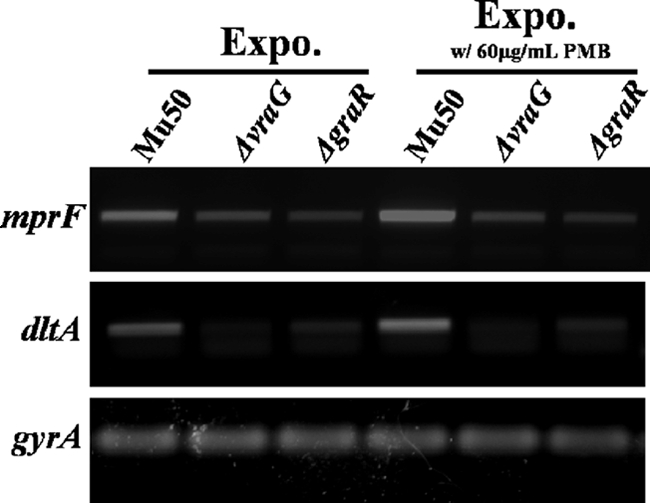
Effect of vraFG and graRS on PMB-induced expression of mprF and dlt genes. RNA samples were isolated from exponential-phase cultures of the strain set in the presence or absence of PMB (60 μg/ml) and subjected to RT-PCR to detect the transcription of mprF, dltA, and gyrA.
Induction of positive cell surface charge by PMB.
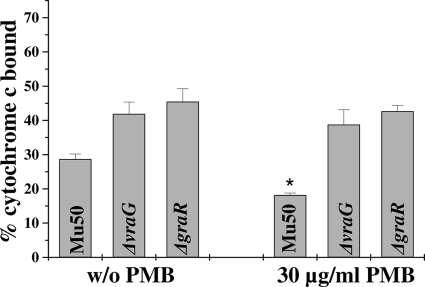
To determine whether the inducibility of dlt and mprF by PMB documented above translated into a detectable phenotype in terms of enhanced surface positive charge, we utilized the cytochrome c binding assay. As seen in Fig. 5, the relative positive surface charge of the parental strain Mu50 increased significantly with PMB induction compared to the uninduced control (i.e., reduced cytochrome c binding; P < 0.05). In contrast, there was no substantial change in surface charge in the presence of PMB for the vraG and graR mutant strains. Of interest, the association of PMB-induced dlt and mprF gene expression with enhancement of the positive surface charge also correlated with increased synthesis and outer membrane translocation of l-PG (1.52- and 1.62-fold increases in total l-PG and outer l-PG, respectively, in the presence of 30 μg of PMB/ml versus the uninduced control; P < 0.05). Thus, PMB-triggered induction of dlt and mprF gene expression translated well into the predicted phenotypic outcomes.
Fig 5.
Effect of PMB induction of the graRS network on relative positive surface charge. The graph shows the percentage of cytochrome c bound after 10 min of incubation in parental strain Mu50 compared to its ΔvraG and ΔgraR deletion mutants. The data represent the means and SDs from three independent experiments. *, P < 0.05. Mu50 parent, PMB-induced versus uninduced.
DISCUSSION
Cationic antimicrobial peptides (CAPs) are crucial components of the innate immune system. Their production is evolutionarily conserved in virtually all groups of organisms, including vertebrates, invertebrates, and plants (3, 16, 34, 47, 55). Such molecules are typically amphipathic, with a net positive charge at physiological pH, features believed to be important in targeting these molecules to the relatively negatively charged bacterial cell membranes (3, 55, 57). In addition to damaging target bacterial membranes by a variety of mechanisms, CAPs may also affect vital intracellular processes including biosynthesis of nucleic acids, proteins, and cell wall components (6, 35, 50). Accordingly, S. aureus has developed a variety of resistance strategies to prevent peptide-induced lethality for a wide range of CAPs from epithelial cells (e.g., LL-37 and hBD-3), phagocytic cells (e.g., hNP-1), and platelets (e.g., tPMPs) (3, 23, 38, 46, 55, 57). These putative “CAP-evasive” mechanisms in S. aureus may include proteolytic degradation of CAPs (42), trapping of CAPs within the cell wall to inhibit access to target cell membranes (19) and increase the net surface positive charge to reduce interactions with CAPs at the bacterial surface (46), efflux of the CAPs by transport pumps (26), and enhanced production of carotenoid pigments to alter membrane fluidity properties (28).
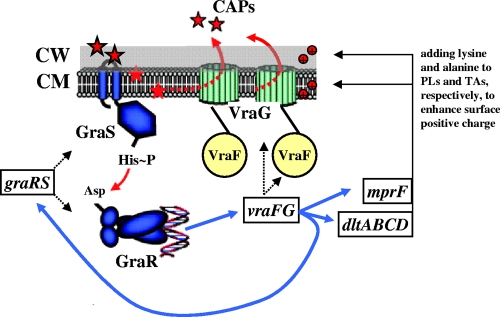
Recently, it has been shown that the TCRS, GraRS, may well play a pivotal role in resistance of S. aureus to CAPs by acting as a CAP “sensor” (23, 26) (Fig. 6). In these studies, it was apparent that there was a relatively selective range of CAPs that could activate the graRS system in S. aureus (e.g., the human cathelicidin, LL-37, or indolicidin, but not hBD-3 or histadin). Li et al. also showed that GraRS appeared to regulate expression of dltABCD and mprF (two important positive surface charge maintenance genes in S. aureus). Furthermore, deletion of the graRS locus led to significantly increased in vitro susceptibility to the positively charged molecule, vancomycin, and the bacterial-derived CAP, PMB as we previously reported (26). In addition to the graR and graS, the first gene of graRS (apsRS) operon encodes GraX (ApsX), a protein of unknown function that may also play a role in CAP resistance (23, 24, 26). Our studies, as well as that of Li et al., demonstrated graRS-mediated regulation of the downstream vraFG, encoding an ABC-transporter-dependent efflux pump (23, 26). This pattern of downstream regulation of TCRS following CAP sensing has also been described in other Gram-positive bacteria. For example, upon exposure to bacitracin, the BceRS TCRS in B. subtilis induces expression of the bacitracin transporter, BceAB, by upregulating bceRS transcription and subsequent expression of the bceAB operon (32).
Fig 6.
Putative model of the GraRS-VraFG network. In this model, GraS senses CAPs selectively, resulting in its autophosphorylation. GraS then phosphorylates GraR, driving mprF and dlt expression in a vraFG-dependent manner. VraFG can also affect expression of mprF and dlt via a positive feedback mechanism. MprF and DltABCD encode proteins which add positively charged lysine and d-alanine to membrane phospholipids and cell wall teichoic acids, respectively, to increase the surface positive charge (44, 46, 53, 54). VraFG may function to efflux CAPs (26). PLs, phospholipids; TAs, teichoic acids. A blue arrow indicates transcriptional upregulation.
Despite these findings, the specific contribution of GraRS to mprF and dltABCD expression and how this regulation system might, in turn, impact susceptibility to specific host defense CAPs remained undefined. In the present study, we used two well-known and well-characterized S. aureus strains (VISA Mu50 and MRSA COL) and their isogenic graR and vraG mutants to explore these issues. A number of interesting findings emerged from these investigations. First, both vraG and graR mutant strains displayed significantly decreased intrinsic transcription of mprF and dltA compared to their respective parental strains. Of interest, graRS has been known to influence mprF and/or dlt expression in some, but not all, previously studied strains (17, 23), Importantly, the present data show that vraFG could also alter the expression of mprF and dlt via graRS, a phenomenon not well appreciated previously (i.e., graRS expression was downregulated in vraG mutant strains).
Recently, it has been shown that the agr operon (via agrC-agrA), a quorum-sensing system, is involved in the regulation of graRS expression (13, 25). In addition, the graRS operon also appears to interact with the WalKR (YycFG) TCRS, which is involved in cell wall lipid metabolism (11). Thus, it is possible that the mutation in vraG may have affected regulation of overlapping TCRS, such as Agr, resulting in indirect downregulation of graRS expression. Taken together, these data provide strong support for our notion of a graRS-vraFG regulatory “loop” with a positive-feedback limb (Fig. 6).
Second, although previous studies have shown regulation of mprF and/or dlt by graRS (23, 40), these studies did not assess the phenotypic consequence of these expression pathways in detail. In the current study, we documented changes in L-PG synthesis and outer membrane translocation that paralleled perturbations in the mprF expression profiles in graR or vraG mutant strains. It should be emphasized that, although the absolute quantitative changes in L-PG synthesis and flipping between parental and mutant strains was relatively small, such modest alterations in membrane lipids can have profound physiologic impacts (1, 41). In addition, lowered expression of both mprF and dlt in the above mutants correlated well with both their reduced positive surface charge phenotype and their increased susceptibility to killing by two mammalian host defense CAPs of platelet and polymorphonuclear leukocyte origins. Recent data from our laboratory and others suggest an alternative to a strict surface-mediated charge-repulsion mechanism in circumstances of enhanced mprF-mediated L-PG synthesis and/or translocation. Kilelee et al. (21) demonstrated the likelihood of formation of “lipid domains” consisting of CAPs plus anionic membrane PLs (e.g., PG or cardiolipin). Thus, in the presence of excess L-PG (e.g., during graRS induction), the relative proportion of membrane PG and/or cardiolipin is reduced, providing fewer “docking sites” for initial membrane localization of CAPs. Whether the vraFG ABC transporter system can also impact CAP susceptibilities in S. aureus via a putative CAP-efflux mechanism remains to be defined.
Third, as noted above, graRS in S. aureus induces expression of mprF, dltABCD, and vraFG in a relatively CAP-selective manner (23). In the present study, we found that the expression of the graRS-mediated effector genes, mprF and dlt, was induced upon exposure to the platelet CAP congener, RP-1, and the bacterial CAP, PMB, but not by the neutrophil CAP, hNP-1. In addition, three other positively charged compounds—vancomycin, gentamicin, and calcium-daptomycin—also failed to induce transcription of mprF (data not shown). Collectively, these findings are in line with those of Li et al. (23) and suggest a relative selectivity in the induction of the sensor GraS by specific CAPs, thus suggesting that GraS may be a putative CAP membrane sensor protein in S. aureus. Ongoing investigations are focused on defining specific structural features of individual CAPs that may trigger GraRS sense and response functions.
Fourth, it is noteworthy that there was a relative disconnect between the ability to induce the graRS TCRS and phenotypic resistance to the same cadre of potential inducer molecules. Thus, for example, both RP-1 and PMB each induced the graRS network, and strains with an intact graRS pathway were substantially more resistant to killing in vitro by these same peptides. In contrast, neither hNP-1 nor vancomycin could induce the graRS network, and yet strains with an intact graRS pathway were substantially more resistant to killing in vitro by these same peptides versus corresponding graRS mutants (26). Furthermore, certain cationic antibiotics (gentamicin and daptomycin) were not sensed by the graRS system, and in vitro susceptibility profiles to these compounds were not impacted by genetic perturbations within this network. Collectively, these observations (i) confirm relatively selective cationic peptide sensing by the graRS system and (ii) phenotypic changes induced by sense response to one peptide may provide in vitro cross-resistance to killing by other structurally unrelated cationic molecules (e.g., induction by RP-1 with resultant enhanced resistance to the noninducer, hNP-1). This suggests that there may be common mechanisms of resistance among such diverse CAPs.
We recognize that there are several limitations to the CAP susceptibility and induction studies above: (i) these investigations were performed in relatively austere artificial media, not closely reflective of the host milieu (e.g., absence of plasma proteins); (ii) moreover, invading bacteria are likely exposed to a cadre of CAPs at sites of infection; and (iii) physiologic CAP concentrations are undoubtedly orders of magnitude above those used in our in vitro analyses (14, 27, 43). Future studies will be designed to adjudicate these limitations.
In contrast to S. aureus, the GraRS homolog in S. epidermidis induced mprF and dltABCD expression upon exposure to CAPs nonselectively (24). Although there is a significant amino acid sequence homology (70%) between the GraS (sensor kinase) of S. aureus and S. epidermidis, sequence divergence occurs in the 9-amino-acid extracellular loop (33%) flanked by two transmembrane domains at the N terminus of the S. epidermidis GraS protein (23, 26). Of interest, when the GraS extracellular loop of S. aureus is replaced with that from S. epidermidis, expression of dlt was induced promiscuously upon exposure to a number of CAPs (e.g., hBD3), similar to what has been found with the parental S. epidermidis strain (23). In addition, we have recently shown that deletion of this 9-amino-acid extracellular loop in S. aureus could render the resultant mutant strain incapable of responding to CAPs that normally induce mprF and dltABCD expression in the parent (e.g., RP-1 and PMB [unpublished data]). At present, we are attempting to define the critical residues within the GraS extracellular loop of S. aureus that are involved in induction of GraRS-regulated genes upon exposure to host defense CAPs.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (AI-39108 to A.S.B., AI-39001 to M.R.Y., and AI-56114 to A.L.C.) and American Heart Association grants SDG0630219N and AID09GRNT2180065 to Y.Q.X.
We thank Andres Taboada for excellent technical assistance with the CAP susceptibility testing.
Footnotes
Published ahead of print 10 October 2011
REFERENCES
- 1. Aricha B, et al. 2004. Differences in membrane fluidity and fatty acid composition between phenotypic variants of Streptococcus pneumoniae. J. Bacteriol. 186: 4638–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 70: 6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bader MW, et al. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122: 461–472 [DOI] [PubMed] [Google Scholar]
- 4. Bayer AS, et al. 2000. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect. Immun. 68: 3548–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertsche U, et al. 2011. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and d-alanylation. Antimicrob. Agents Chemother. 55: 3922–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3: 238–250 [DOI] [PubMed] [Google Scholar]
- 7. Climo MW, Patron RL, Archer GL. 1999. Combinations of vancomycin and β-lactams are synergistic against staphylococci with reduced susceptibilities to vancomycin. Antimicrob. Agents Chemother. 43: 1747–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44: 2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui L, Tominaga E, Neoh H-M, Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50: 1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Lencastre H, et al. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5: 163–175 [DOI] [PubMed] [Google Scholar]
- 11. Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70: 1307–1322 [DOI] [PubMed] [Google Scholar]
- 12. Ernst C, et al. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5: e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falord M, Mader U, Hiron A, Debarbouille M, Msadek T. 2011. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6: e21323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganz T, Selsted ME, Lehrer RI. 1990. Defensins. Eur. J. Haematol. 44: 1–8 [DOI] [PubMed] [Google Scholar]
- 15. Gill SR, et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187: 2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hancock REW, Diamond G. 2000. The role of cationic antimicrobial peptides in innate host defenses. Trends Microbiol. 8: 402–410 [DOI] [PubMed] [Google Scholar]
- 17. Herbert S, et al. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 3: e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howden BP, et al. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52: 3755–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin T, et al. 2004. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172: 1169–1176 [DOI] [PubMed] [Google Scholar]
- 20. Jones T, et al. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kilelee E, Pokorny A, Yeaman MR, Bayer AS. 2010. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: implications for daptomycin resistance. Antimicrob. Agents Chemother. 54: 4476–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuroda M, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357: 1225–1240 [DOI] [PubMed] [Google Scholar]
- 23. Li M, et al. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66: 1136–1147 [DOI] [PubMed] [Google Scholar]
- 24. Li M, et al. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104: 9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuo M, Oogai Y, Kato F, Sugai M, Komatsuzawa H. 2011. Growth-phase dependence of susceptibility to antimicrobial peptides in Staphylococcus aureus. Microbiology 157: 1786–1797 [DOI] [PubMed] [Google Scholar]
- 26. Meehl M, Herbert S, Gotz F, Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51: 2679–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mezzano S, et al. 1992. Glomerular localization of platelet factor 4 in streptococcal nephritis. Nephron 61: 58–63 [DOI] [PubMed] [Google Scholar]
- 28. Mishra NN, et al. 2011. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55: 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mishra NN, et al. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus (MRSA). Antimicrob. Agents Chemother. 53: 2312–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mukhopadhyay K, et al. 2007. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153: 1187–1197 [DOI] [PubMed] [Google Scholar]
- 31. Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48: 4800–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohki R, et al. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49: 1135–1144 [DOI] [PubMed] [Google Scholar]
- 33. Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. 2004. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150: 45–51 [DOI] [PubMed] [Google Scholar]
- 34. Pelegrini P, et al. 2009. A novel antimicrobial peptide from Crotalaria pallida seeds with activity against human and phytopathogens. Curr. Microbiol. 59: 400–404 [DOI] [PubMed] [Google Scholar]
- 35. Peschel A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10: 179–186 [DOI] [PubMed] [Google Scholar]
- 36. Peschel A, et al. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peschel A, et al. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274: 8405–8410 [DOI] [PubMed] [Google Scholar]
- 38. Peschel A, Sahl H-G. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4: 529–536 [DOI] [PubMed] [Google Scholar]
- 39. Rice KC, et al. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185: 2635–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sass P, Bierbaum G. 2009. Native graS mutation supports the susceptibility of Staphylococcus aureus strain SG511 to antimicrobial peptides. Int. J. Med. Microbiol. 299: 313–322 [DOI] [PubMed] [Google Scholar]
- 41. Shinitzky M, Barenholz Y. 1978. Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim. Biophys. Acta 515: 367–394 [DOI] [PubMed] [Google Scholar]
- 42. Sieprawska-Lupa M, et al. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48: 4673–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soong LB, Ganz T, Ellison A, Caughey GH. 1997. Purification and characterization of defensins from cystic fibrosis sputum. Inflamm. Res. 46: 98–102 [DOI] [PubMed] [Google Scholar]
- 44. Staubitz P, Neumann H, Schneider T, Wiedemann I, Peschel A. 2004. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 231: 67. [DOI] [PubMed] [Google Scholar]
- 45. Utaida S, Pfeltz RF, Jayaswal RK, Wilkinson BJ. 2006. Autolytic properties of glycopeptide-intermediate Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 50: 1541–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weidenmaier C, et al. 2005. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect. Immun. 73: 8033–8038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Welling MM, et al. 1998. Antibacterial activity of human neutrophil defensins in experimental infections in mice is accompanied by increased leukocyte accumulation. J. Clin. Invest. 102: 1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26: 369. [DOI] [PubMed] [Google Scholar]
- 49. Xiong YQ, Bayer AS, Elazegui L, Yeaman MR. 2006. A synthetic congener modeled on a microbicidal domain of thrombin-induced platelet microbicidal protein-1 recapitulates staphylocidal mechanisms of the native molecule. Antimicrob. Agents Chemother. 50: 3786–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiong YQ, Mukhopadhyay K, Yeaman MR, Adler-Moore J, Bayer AS. 2005. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob. Agents Chemother. 49: 3114–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang S-J, et al. 2010. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 54: 3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang S-J, et al. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 187: 5893–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang SJ, et al. 2009. Enhanced expression of dltABCD is associated with development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200: 1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang SJ, et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 53: 2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yeaman MR, Bayer AS, Koo S-P, Foss W, Sullam PM. 1998. Platelet microbicidal proteins and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanisms of action. J. Clin. Invest. 101: 178–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yeaman MR, Gank KD, Bayer AS, Brass EP. 2002. Synthetic peptides that exert antimicrobial activities in whole blood and blood-derived matrices. Antimicrob. Agents Chemother. 46: 3883–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yeaman MR, Puentes SM, Norman DC, Bayer AS. 1992. Partial characterization and staphylocidal activity of thrombin-induced platelet microbicidal protein. Infect. Immun. 60: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]