Abstract
Intracellular persistence of Chlamydia trachomatis has been implicated in the development of chronic infection that can result in pelvic inflammatory disease and tubal sterility. By inhibition of host cell apoptosis, chlamydiae have evolved a strategy to maintain the intracellular environment for replication and persistence. Both antiapoptotic host cell-derived factors and the chlamydial protease-like activity factor (CPAF) are involved in Chlamydia-mediated apoptosis resistance. Here, we show that in HeLa cells infected with gamma interferon (IFN-γ)-induced persistent C. trachomatis serovar D, the expression of CPAF is downregulated, and proapoptotic protease substrates are not cleaved. Persistent infection protected HeLa cells from apoptosis when they were exposed to staurosporine. Small-interfering RNA-mediated inhibition of myeloid cell leukemia 1 (Mcl-1) protein upregulation sensitized persistently infected cells for apoptosis. The inhibitor of apoptosis protein 2 (IAP-2) seems not to be relevant in this context because IAP-2 protein was not induced in response to IFN-γ treatment. Although apoptosis was inhibited, persistent infection caused cell membrane disintegration, as measured by the increased release of cytokeratin 18 from HeLa cells. Moreover, persistently infected cells released significantly increased amounts of high mobility group box 1 (HMGB1) protein which represents a proinflammatory damage-associated pattern molecule. The data of this study suggest that cells infected with persistent C. trachomatis are protected from apoptosis independently of CPAF but may promote chronic inflammation through HMGB1 release.
INTRODUCTION
As an intracellular bacterial pathogen, Chlamydia trachomatis has evolved the ability to block apoptosis of its host cells even in the presence of strong cell death-inducing agents (13, 14). Inhibition of apoptosis may contribute to the maintenance of the cellular environment for bacterial replication and protect infected cells from elimination by immune or tissue repair mechanisms. However, intracellular Chlamydia replication within an expanding endosome causes massive stress to the host cell and disturbs cell membrane integrity (37, 38). Such cytopathic effects may finally result in necrosis, which would promote inflammation and activation of innate immune mechanisms. In a previous study it has been shown that C. trachomatis prevents an increased release of high mobility group box 1 (HMGB1), a damage-associated molecular pattern (DAMP) molecule that acts as a costimulatory cytokine (28, 38). It can be suggested that Chlamydia is not only able to interfere with apoptosis but also modifies pathways involved in necrotic cell death to limit inflammation and defense responses.
A pathogenicity factor that plays a role in the interference with cell death-associated mechanisms is the chlamydial protease-like activity factor (CPAF), which was first described by Zhong et al. (40). Functional CPAF is organized as an intramolecular dimer of subunits of 35 and 29 kDa generated from a 70-kDa full-length protein (9, 11). The protease is translocated from the chlamydial inclusion into the host cell cytosol and degrades BH3-only proteins such as Bad, Bim, and Puma, which are proapoptotic members of the Bcl-2 protein family (14, 25, 36). Because BH3-only proteins activate Bax and Bak as effectors of cytochrome c release, CPAF-mediated effects could explain the inhibition of the mitochondrial apoptotic pathway (30, 36). On the other hand, it has become clear that additional pathways via induction of antiapoptotic host proteins are involved (30). The upregulation of inhibitor of apoptosis protein 2 (IAP-2) and myeloid cell leukemia 1 (Mcl-1) protein, an antiapoptotic member of the Bcl-2 family, in Chlamydia-infected cells may be sufficient to achieve resistance of host cells against cell death stimuli (26, 27). Because the ability of C. trachomatis to protect IAP-2−/− or Mcl-1−/− cells against apoptosis has also been shown, mechanisms by which Chlamydia interferes with programmed cell death are controversially discussed (35).
CPAF degrades and inactivates poly(ADP-ribose) polymerase 1 (PARP-1), which acts as a regulatory factor in DNA repair, apoptosis, and necrosis (6, 24, 38). Because PARP-1 is involved in the nucleus-to-cytosol translocation of HMGB1, its degradation prevents an increased HMGB1 release from infected cells, indicating that CPAF interferes with cell death regulatory pathways at different target sites (38).
Chlamydiae are able to establish long-lasting infection. During the acute phase, C. trachomatis can cause mucopurulent cervicitis with discharge or symptomatic urethritis, but many infections remain asymptomatic, suggesting that limited inflammatory responses at the primary epithelial target site occur. However, the pathogen frequently ascends into the upper genital tract, especially in women. These infections result in chronic inflammation and are a major risk factor for the development of tubal infertility (21). Intracellular persistence is discussed as the central mechanism underlying chronic disease (29). Chlamydia replicates within an expanding vacuole, the so-called inclusion body, in the host cell. The intracellular replication forms, designated reticulate bodies, asynchronously differentiate into elementary bodies that are released from the host cell and then infect new target cells. In this way several hundred new infectious bacteria can be produced by one infected cell. In the case of C. trachomatis the production of elementary bodies reaches its maximum at 2 days after infection. In response to nutrient deprivation and gamma interferon (IFN-γ), chlamydiae can transform into aberrant intracellular reticulate bodies that will not further replicate but in general retain their viability and the potential to differentiate into elementary bodies (2, 3). Although it has been reported that persistently infected cells are also resistant to apoptosis, underlying mechanisms are less well understood, and the relationship between cell death protection and induction remains unclear (10). Because many chlamydial genes are differentially regulated in replicative and persistent infection, it can be supposed that antiapoptotic and cytopathic effects that may have impact on the maintenance of chronic inflammation are altered during the stage of persistence (5). Therefore, this study was designed to investigate effects of persistent C. trachomatis infection of epithelial cells on apoptosis resistance and HMGB1 release in relation to the activity of CPAF. As a model of persistent infection, IFN-γ-restricted growth of C. trachomatis in HeLa cells was used.
MATERIALS AND METHODS
Cell culture and C. trachomatis propagation.
HeLa cells (ATCC CCL-2) were grown in minimal essential medium (Opti-MEM; Gibco/Invitrogen, Karlsruhe, Germany) with 10% membrane-filtered and heat-inactivated fetal calf serum (FCS) (PromoCell, Heidelberg, Germany). C. trachomatis serovar D strain IC Cal 8 (obtained from the Institute of Ophthalmology, London, United Kingdom) was propagated in buffalo green monkey (BGM) cells as described previously (38). Infectivity titers of chlamydial stocks were quantified by determining the number of inclusion-forming units (IFU) per milliliter in BGM cells. Mycoplasma contaminations in cell cultures were excluded by PCR targeting the 16S rRNA gene of Mycoplasma, Acholeplasma, and Ureaplasma species (38).
Replicative and persistent C. trachomatis infection.
HeLa cells were grown in 35-mm-diameter culture wells (six-well plates) or shell vials to ca. 80% confluence. The cells were inoculated with C. trachomatis at a multiplicity of infection (MOI) of 1 or 5, as indicated below. For mock-infected cultures diluted harvests of uninfected BGM cells were added. After centrifugation at 4,000 × g at 37°C for 45 min, the inoculum was decanted, and the cells were further incubated with Opti-MEM containing 10% FCS.
To establish persistent infections, HeLa cells were treated with 5 ng of IFN-γ per ml for 48 h prior to infection. Cells were infected at an MOI of 1 or 5 as described above and further incubated with Opti-MEM containing 10% FCS and 5 ng of IFN-γ per ml. For determination of the number of chlamydial inclusions, coverslips contained in shell vials were fixed with methanol at 48 h after infection and stained with fluorescein isothiocyanate (FITC)-conjugated antibody to C. trachomatis major outer membrane protein (MOMP; Oxoid). The number of inclusions per coverslip was calculated from determination of inclusions in 10 randomly selected ×200 microscopic fields. To assess the inhibitory effect of different IFN-γ concentrations on the production of infectious chlamydiae, infectivity assays were performed. At 48 h after infection cells were scraped into Opti-MEM with 0.2 M sucrose and 2% FCS and lysed by sonication. The suspensions were centrifuged at 10,000 × g, and the supernatants were frozen at 70°C. The number of IFU was determined by inoculating BGM cells with dilutions of the samples and counting inclusions by immunofluorescence staining.
To induce apoptosis, cells with or without chlamydial infection were treated with 1 μM staurosporine (STS; Roche Applied Science) for 3 h before sampling.
Electron microscopy.
At 48 h after infection, cells were fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer for 1 h at room temperature. The cells were collected from the culture wells using a cell scraper, pelleted by centrifugation at 800 × g for 5 min, and stored at 4°C. Cell pellets were embedded in 2% agarose and sectioned to 1-mm3 cubes. Cubes were postfixed in 2% osmium tetroxide and embedded in araldite Cy212. Semithin sections were prepared to select areas of high cell density for ultramicrotomy. Ultrathin sections of 85 nm were stained with uranyl acetate and lead citrate and examined in a transmission electron microscope (Tecnai 12; FEI, Eindhoven, The Netherlands).
Real-time reverse transcription-PCR (RT-PCR).
Total RNA was isolated using a peqGOLD Total RNA Kit (Peqlab, Erlangen, Germany). A DNase digestion step was included. For each sample, cDNA was reverse transcribed from 1 μg of RNA in a reaction volume of 20 μl, using the Promega reverse transcription system and random hexamer primers (Promega, Mannheim, Germany). Each PCR consisted of 10 μl of 2× Kapa SYBR Fast QPCR Master Mix (Peqlab), 0.4 μl of each primer (10 μM stock solution) (Table 1), 4.2 μl of H2O, and 5 μl of cDNA (diluted 1:10 in H2O prior to use). Standard SYBR green-based real-time PCR was carried out with a Smart Cycler II (Cepheid; BD, Heidelberg, Germany), and cycle threshold (CT) values were determined using Dx software (Cepheid; BD).
Table 1.
Sequences of primers used for real-time RT-PCR
| Primer specificity (gene) | Sequence of primer paira | Amplicon size (bp) | Primer efficiency |
|---|---|---|---|
| cpaf (ct858) | 5′-CGGAGGGTCTTTTCCGCGCT-3′ | 139 | 1.99 |
| 5′-TCCCAAGGAGGCGGTCCTGA-3′ | |||
| ompA | 5′-AATTTCAGATGGGTGCCAAG-3′ | 190 | 1.98 |
| 5′-CCACTGGTGGCTCCTAATGT-3′ | |||
| groEL (ct110) | 5′-CCAGCAAAACTGCTGACAAA-3′ | 101 | 1.82 |
| 5′-TGCTCCAGCTGTTACATTGC-3′ | |||
| trpB | 5′-TGAGTCAGGACGAGCCTTTT-3′ | 121 | 1.89 |
| 5′-ATGTGCGAGAGCATGTGAAG-3′ | |||
| 16S rRNA | 5′-CGGTAATACGGAGGGTGCTA-3′ | 176 | 2.00 |
| 5′-CTACGCATTTCACCGCTACA-3′ | |||
| mcl-1 | 5′-GTACCTTCGGGAGCAGGCCACC-3′ | 157 | 1.99 |
| 5′-GTCCAGTTTCCGAAGCATGCCTTGG-3′ | |||
| gapdh | 5′-TCAAGTGGGGCGATGCTGGC-3′ | 135 | 2.00 |
| 5′-TGGGGGCATCAGCAGAGGGG-3′ |
Sense and antisense, respectively.
Basal gene expression was calculated as ΔCT that represents the difference between the CT value of the test gene and the CT value of the internal standard gene from the same sample. As internal standards, 16S rRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes were used for the expression analysis of chlamydial and host cell genes, respectively. In accordance with Kokab et al. (17), the following formula was applied to calculate the relative expression of the test genes compared to the internal standard: (primer efficiency of test gene)−ΔCT × 100%. Primer efficiencies were determined by analyzing serial dilutions of cDNA. The CT values were plotted against log cDNA concentrations, and the slope was calculated by linear regression. Primer efficiency was expressed as 10(−1/slope) (Table 1).
Immunoblotting.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blot assays were performed according to the protocol of a previous study (38). Blots were incubated with polyclonal rabbit antibodies to indoleamine 2,3-dioxygenase ([IDO] sc-25808; 1:200 dilution; Santa Cruz Biotechnology, Heidelberg, Germany), Bad (9292; 1:1,000 dilution; Cell Signaling, Frankfurt am Main, Germany), Puma (sc-28226; 1:1,000 dilution; Santa Cruz Biotechnology), PARP-1 (VIC5; 1:1,000 dilution; Roche Applied Science, Mannheim, Germany), caspase-3 (9665; 1:1,000 dilution; Cell Signaling), cleaved caspase-3 (Asp175, 9661; 1:1,000 dilution; Cell Signaling), Mcl-1 (sc-20679; 1:200 dilution; Santa Cruz Biotechnology), IAP-2 (sc-7944; 1:200 dilution; Santa Cruz Biotechnology), or GAPDH (sc-25778; 1:1,000 dilution; Santa Cruz Biotechnology). Cytokeratin 18 (CK18) was analyzed using a mouse monoclonal antibody (DC10, 4548; 1:1,000 dilution; Cell Signaling). For the detection of CPAF a 1:750 dilution of a polyclonal mouse anti-CPAF serum was used (38). Chlamydial HSP60 was stained with a mouse monoclonal antibody (SM5080; 1:1,000 dilution; Acris Antibodies, Herford, Germany). Blots were incubated with primary antibodies at 4°C overnight. Alkaline phosphatase-conjugated goat anti-rabbit or rabbit anti-mouse IgG (Dianova, Hamburg, Germany) was used as a secondary antibody at a dilution of 1:1,000. Blots were incubated with the secondary antibody for 2 h at room temperature. Washes between antibody additions were performed with Tris-buffered saline (TBS)-Tween three times for 5 min. The bands were visualized with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium salts (BCIP/NBT; Sigma Fast, Sigma-Aldrich). Molecular weights of proteins and cleaved fragments were calculated from a protein standard curve of log molecular weight versus distance migrated (SDS-PAGE Standards Broad Range; Bio-Rad, Munich, Germany). All results shown are representative of at least two independent experiments.
Laser scanning microscopy.
Mock-infected and infected cells cultured on glass coverslips were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. After cells were washed with PBS, fixed cells were permeabilized with 0.1% saponin in PBS for 10 min and then saturated in blocking buffer (3% bovine serum albumin [BSA] in saponin-PBS) for 1 h at room temperature. For immunostaining of active caspase-3, cells were sequentially incubated with a 1:500 dilution of rabbit polyclonal antibody (pAb) directed against the p19 fragment of caspase-3 (anti-active caspase-3 pAb; G7481; Promega) at 4°C overnight and a 1:500 dilution of Cy3-conjugated donkey anti-rabbit IgG (711-165-152; Dianova) for 2 h at room temperature. Chlamydia inclusions were labeled with an FITC-conjugated MOMP antibody (Trinity Biotech, Lemgo, Germany). Washes between antibody additions were performed with saponin-PBS four times. Coverslips were embedded in ProLong Gold antifade reagent (Invitrogen) containing 4′,6′-diamidino-2-phenylindole (DAPI) for staining of cell nuclei. Images were taken with a confocal laser scanning microscope (Exciter 5; Zeiss, Germany) using Zen 2009 software.
Enzyme immunoassays.
For quantitative determination of apoptosis, cytokeratin 18 fragments containing a neo-epitope (formed by caspase cleavage at Asp396 [CK18Asp396-NE]) (M30) were measured using an M30-Apoptosense enzyme-linked immunosorbent assay (ELISA) (Peviva; Enzo Life Sciences, Lörrach, Germany). To assay total M30 levels in cell culture supernatants and cell extracts, 10% Igepal CA-630 (Sigma-Aldrich) was directly added into the culture wells (50 μl per 1 ml of medium). As a marker of cell membrane disintegration, total soluble full-length CK18 and C-terminal fragments of CK18 were quantified in cell culture supernatants using an M65 ELISA (Peviva; Enzo Life Sciences). HMGB1 levels in culture supernatants were measured with an ELISA from Shino-test (Kanagawa, Japan; purchased from IBL International, Hamburg, Germany). All assays were performed according to the manufacturer's protocols.
RNA interference.
A total of 2 × 105 or 5 × 104 HeLa cells were seeded into 35-mm-diameter wells (six-well plates) or shell vials. HP Validated small interfering RNA (siRNA) targeting Mcl-1 (SI02781205; gene accession number NM_021960) and nonsilencing All Stars Negative Control siRNA were purchased from Qiagen (Hilden, Germany). HeLa cells were transfected with 10 nM siRNA using HiPerfect transfection reagent according to the manufacturer's instructions. Briefly, the transfection complex consisted of 1.2 μl of a 20 μM siRNA stock solution, 100 μl of medium without serum, and 24 μl of HiPerfect reagent. Transfection complex (125 μl) was added to 35-mm-diameter wells (2.3-ml culture medium volume), and 32 μl was added to shell vials (0.5-ml culture medium volume). Transfected cells were incubated in medium containing 5 ng of IFN-γ per ml for 48 h before infection with C. trachomatis. Downregulation of Mcl-1 by specific siRNA was confirmed by RT-PCR and Western Blotting.
Statistical analysis.
Statistical comparisons were made using a Student's t test. P values of <0.05 were considered to be statistically significant.
RESULTS
Establishment of nonproductive infection as a model of persistent C. trachomatis.
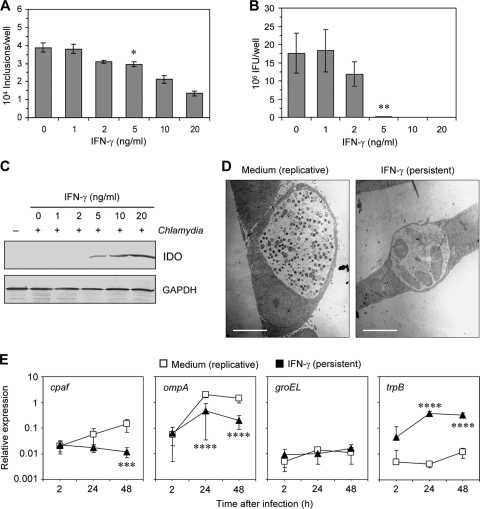
First experiments were performed to evaluate conditions of IFN-γ treatment that induce an infection resembling the main characteristics of persistent-like forms of chlamydial organisms. These criteria included an aberrant morphology of reticulate bodies and significant downregulation of the production of new elementary bodies (2, 3). Moreover, it was important to choose an IFN-γ concentration at which the infectious progeny, but not the number of inclusion-positive cells, was highly reduced to ensure comparability of the experiments with untreated cultures. Figure 1A and B show that the production of elementary bodies of C. trachomatis serovar D was decreased by 99.3% in the presence of 5 ng of IFN-γ per ml although the number of inclusions was reduced by only 20% compared to cells incubated in medium alone. Dose-dependent inhibitory effects of IFN-γ on chlamydial replication corresponded to the amounts of IDO protein detected by Western blotting (Fig. 1C). IDO was induced when the cells were stimulated with 5 ng of IFN-γ per ml. As shown by electron microscopy, inclusions in IFN-γ-treated cells contained few reticulate bodies, which were enlarged, at 48 h after infection, whereas inclusions in untreated cells were filled with elementary bodies (Fig. 1D). Pretreatment of HeLa cells for 48 h prior to infection was necessary to inhibit the recovery of infectious elementary bodies of C. trachomatis serovar D, corresponding to previous findings by Morrison (22). Concentrations of >5 ng of IFN-γ per ml caused a significant decrease in the number of inclusions, and cytopathic effects could be observed. Therefore, in all further experiments a concentration of 5 ng of IFN-γ per ml was used.
Fig 1.
Restriction of C. trachomatis D growth in HeLa cells by IFN-γ and characterization of persistent chlamydiae. (A) Effect of IFN-γ on the number of inclusion-positive cells in infected cultures. *, P < 0.01 (compared to values for untreated cells; n = 4). (B) Recovery of infectious chlamydiae from HeLa cells treated with increasing IFN-γ concentrations. **, P < 0.001 (compared to values for untreated cells; n = 4). (C) IDO induction by IFN-γ. Cell extracts were prepared at 48 h after infection and analyzed by immunoblotting. (D) Inclusion morphologies in untreated and IFN-γ-treated cells. Inclusions were analyzed by transmission electron microscopy at 48 h after infection. Note the numerous small elementary bodies in inclusions of untreated cells and the occurrence of atypical enlarged reticulated bodies in IFN-γ-treated cells. Scale bar, 5 μm. (E) Relative expression of chlamydial genes in IFN-γ-treated and untreated cells determined by real time RT-PCR. Data were normalized to 16S rRNA levels as described in Materials and methods. ***, P < 0.04; ****, P < 0.02 (compared to values for replicative chlamydiae; n = 4).
Real-time RT-PCR was performed to assess altered gene expression in IFN-γ-treated chlamydiae. mRNA signals were normalized against the level of chlamydial 16S rRNA. As shown in Fig. 1E, cpaf and ompA genes were significantly downregulated compared to levels in the replicative infection control (11.9-fold and 7.2-fold, respectively). In contrast, IFN-γ had no significant effect on the amounts of groEL mRNA, which was generally expressed at lower levels than the cpaf and ompA genes. According to Belland et al. the tryptophan synthase B (trpB) gene was selected as a marker gene upregulated in persistence (5). The expression of trpB was 95.2-fold and 26.5-fold increased in the presence of IFN-γ at 24 h and 48 h after infection, respectively (Fig. 1E). The transcriptional response of C. trachomatis to IFN-γ treatment corresponded to previous findings on altered gene and antigen expression of Chlamydia in persistent infection and confirmed the suitability of our cell culture model for further experiments (2, 5).
CPAF substrates are not cleaved upon persistent C. trachomatis infection.
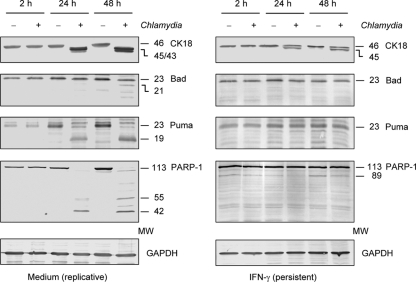
Downregulation of cpaf mRNA by C. trachomatis in response to IFN-γ indicated that in persistently infected cells CPAF activity was diminished. Therefore, CPAF substrates CK18, Bad, Puma, and PARP-1 were examined for degradation (14, 20, 24). The results are summarized in Fig. 2. In infected cells cultured without IFN-γ, the processing of CK18 resulted in the generation of a double band of about 45 and 43 kDa, whereas full-length CK18 disappeared, corresponding to findings by Kumar and Valdivia (20). In persistently infected cells only a partial CK18 processing could be observed. Full-length CK18 and an additional 45-kDa band were detected, indicating the downregulation of CPAF activity. The proapoptotic BH3-only proteins Bad and Puma were not degraded in IFN-γ-treated cells. Upon replicative infection, Bad cleavage bands were detected at 48 h after infection, and Puma was degraded at 24 and 48 h after infection; however, no cleavage bands of either protein was observed upon persistent chlamydial infection of HeLa cells. In a similar manner PARP-1 fragmentation was observed only in cells infected with replicative chlamydiae, whereas in persistently infected cells only the full-length PARP-1 band was present. IFN-γ treatment had a slight effect on the activation of apoptotic pathways because in mock-infected cells a faint 89-kDa PARP-1 cleavage band, which is characteristically generated by caspase-3, was induced (6). However, in infected cells this cleavage band disappeared, indicating that caspase-3 activation was inhibited during the state of persistent C. trachomatis infection.
Fig 2.
Immunoblot analysis of CK18, BH3-only proteins Bad and Puma, and PARP-1 in HeLa cells infected with replicative and persistent Chlamydia bacteria. Cells were incubated in medium alone or with 5 ng of IFN-γ per ml and infected with C. trachomatis at an MOI of 5. GAPDH was stained as a reference band. MW, molecular weights in thousands.
Apoptosis inhibition in persistently infected cells despite CPAF downregulation.
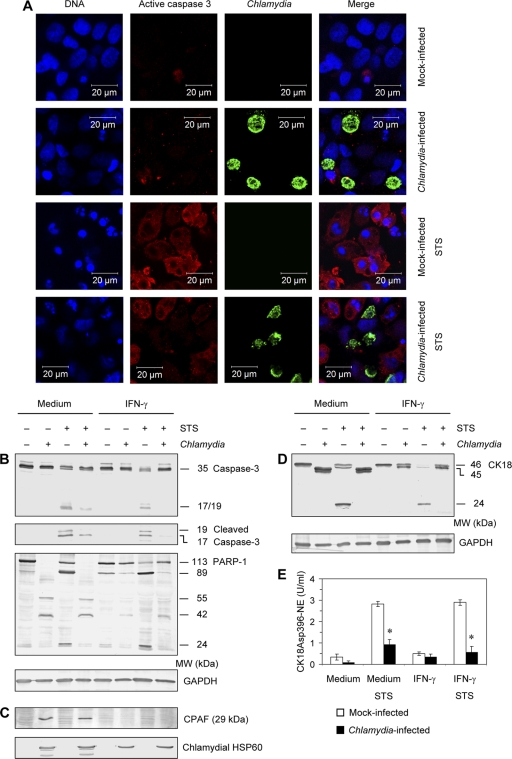
Fan et al. previously showed that Chlamydia blocks STS-induced apoptosis (13). Because proapoptotic BH3-only proteins were not found to be degraded in IFN-γ-treated cells, the question was raised whether HeLa cells were resistant against apoptosis when they were infected by persistent chlamydiae. Cells were exposed to STS and analyzed by staining with DAPI and antibodies against active caspase-3. The results are shown in Fig. 3A. In mock-infected cells incubated with IFN-γ, only a small portion of cells was positively stained by caspase-3 antibody. STS treatment induced active caspase-3 and nuclear fragmentation. However, in infected cultures only cells without chlamydial inclusions were stained positive for caspase-3, whereas inclusion-containing cells were exclusively negative for active caspase-3 and showed no nuclear fragmentation, indicating that Chlamydia-positive cells did not undergo apoptosis. For immunofluorescence assays HeLa cells were infected with C. trachomatis at a lower MOI of 1 instead of an MOI 5 for immunoblotting to demonstrate inclusion-positive as well as -negative cells in the same sample.
Fig 3.
Inhibition of apoptosis by persistent Chlamydia infection. HeLa cells were incubated with 5 ng of IFN-γ per ml and infected with C. trachomatis at an MOI of 1 (A) or 5 (B to E). Samples were collected at 48 h after infection. To induce apoptosis, 1 μM STS was added 3 h before sampling. (A) Cell monolayers were costained with DAPI for DNA (blue), active caspase-3 antibody (probed with a Cy3-conjugated secondary antibody; red), and FITC-conjugated C. trachomatis MOMP antibody (green). (B) Immunoblot analysis of caspase-3 and PARP-1 cleavage patterns in IFN-γ-treated and untreated cells. To examine caspase-3 activation cells were stained with a total-caspase-3 antibody and an antibody that specifically recognizes the large fragments of activated caspase-3. GAPDH was stained as a reference band. (C) Downregulation of CPAF protein production in persistently infected cells. Chlamydial HSP60 was stained as a control. (D) CK18 cleavage patterns in IFN-γ-treated and untreated cells. (E) Apoptotic CK18Asp396-NE was quantified in cell lysates by ELISA. *, P < 0.001 (compared to values for mock-infected cells following STS stimulation; n = 5). MW, molecular weights in thousands.
The inhibition of caspase-3 activation in persistently infected cells was confirmed by immunoblot assays. The addition of STS to mock-infected cells with or without IFN-γ treatment induced the cleavage of full-length caspase-3 (35 kDa) and the formation of 19/17-kDa fragments (Fig. 3B). Similar to cells infected with replicative chlamydiae, the cleavage of full-length caspase-3 and the appearance of 19- and 17 kDa-fragments were inhibited in persistently infected cells in the presence of STS (Fig. 3B). PARP-1 cleavage into two fragments of 89 and 29 kDa represents a sensitive marker to evaluate caspase-3 activation (6, 19). During replicative chlamydial infection PARP-1 was cleaved into multiple fragments independent of the presence or absence of STS (Fig. 3B). This cleavage pattern correlated with the detection of CPAF in Western blotting using an antibody that identified the 29-kDa fragment (Fig. 3C). In persistently infected cells no PARP-1 degradation occurred, corresponding to the downregulation of CPAF protein (Fig. 3C). Chlamydial HSP60 was stained as a control. Apoptotic PARP-1 fragments of 89 and 24 kDa were observed only in mock-infected cells following stimulation with STS or IFN-γ (Fig. 3B). In IFN-γ-treated cells infected with persistent chlamydiae, the cleavage of PARP-1 was inhibited following exposure to STS, confirming the strong antiapoptotic activity of C. trachomatis despite highly reduced CPAF activity (Fig. 3B).
As indicated in Fig. 2, CK18 may be a sensitive target to identify residual CPAF activity. Moreover, CK18 belongs to intermediate filament proteins that are substrates of effector caspases during apoptosis (7). The cleavage sites for caspases in CK18 are located in the central rod domain, whereas the recognition sequence for CPAF has been mapped to the N-terminal head domain. Therefore, CK18 fragmentation patterns could also be used to analyze apoptosis resistance of persistently infected cells. In STS-treated mock-infected cells, CK18 cleavage resulted in the appearance of a characteristic 24-kDa apoptotic fragment (Fig. 3D). Apoptotic CK18 cleavage was clearly inhibited by both replicative and persistent C. trachomatis infection (Fig. 3D). Reduced Chlamydia-mediated CK18 fragmentation in persistently infected cells confirmed the downregulation of CPAF activity in IFN-γ-treated cultures compared to levels in infected cells incubated in medium alone (Fig. 3D). To verify the immunoblot results, levels of apoptotic CK18 fragments (CK18Asp396-NE) were quantified (18). In mock-infected HeLa cells incubated with or without IFN-γ, STS caused a significant increase in CK18Asp396-NE amounts (Fig. 3E). Persistent infection prevented the induction of CK18Asp396-NE just as well as replicative infection (Fig. 3E).
Role of Mcl-1 but not IAP-2 in apoptosis resistance of HeLa cells infected by persistent Chlamydia.
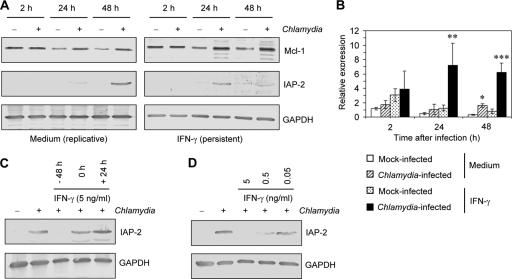
Both Mcl-1 and IAP-2 have been identified as host cell antiapoptotic factors that play a role in inhibition of apoptosis by Chlamydia infection (26, 27, 31). At 2 h after infection no difference in the levels of Mcl-1 protein could be found between cells without or with IFN-γ treatment. However, after 24 and 48 h, the amounts of Mcl-1 were higher in infected cells than in mock-infected controls (Fig. 4A). Immunoblots showed a band of 40 kDa representing Mcl-1 full-length protein. In IFN-γ-exposed cells additional faint bands of 37 and 35 kDa appeared, but a splicing variant of 32 kDa, which has been described as a rather proapoptotic variant, was not identified (Fig. 4A) (1). Analysis of mRNA expression confirmed Mcl-1 upregulation in persistently infected cells. At 2 h after infection no significant differences in Mcl-1 mRNA levels between mock-infected and infected cells could be found. HeLa cells infected with replicative chlamydiae expressed significantly higher levels of Mcl-1 mRNA than mock-infected cells at 48 h after infection (Fig. 4B). In IFN-γ-treated cells the Mcl-1 gene was significantly upregulated at 24 and 48 h after infection compared to levels in the mock-infected controls. Moreover, mRNA expression was significantly increased in persistently infected cells compared to levels in cells infected with replicative chlamydiae (Fig. 4B).
Fig 4.
Effect of persistent Chlamydia infection on Mcl-1 and IAP-2 levels in HeLa cells. (A) Immunoblot analysis of Mcl-1 and IAP-2 protein in IFN-γ-treated and untreated cells. GAPDH was stained as a reference band. (B) Time course of relative mcl-1 mRNA expression in IFN-γ-treated and untreated cells determined by real-time RT-PCR. Data were normalized to gapdh mRNA levels as described in Material and Methods. *, P < 0.02; **, P < 0.05 (both, compared to values for mock-infected cells); ***, P < 0.001 (compared to values for mock-infected cells following IFN-γ treatment and cells infected with replicative chlamydiae). (C) Influence of the time point of IFN-γ addition on IAP-2 protein levels. IFN-γ was added at 48 h before infection, immediately after infection (time point 0), or 24 h after infection. (D) IAP-2 protein levels in response to increasing IFN-γ concentrations. Treatment was started at 48 h before infection.
In contrast to Mcl-1, a differential pattern was observed for IAP-2 protein. Whereas IAP-2 was induced in response to replicative infection at 48 h, in persistently infected cells only a faint band could be detected at 24 h, and IAP-2 was not induced at 48 h after infection (Fig. 4A). To clarify whether this effect was mediated by IFN-γ, we examined IAP-2 levels in infected HeLa cells in response to increasing concentrations of IFN-γ and different incubation periods. As shown in Fig. 4C and D, downregulation of Chlamydia-induced IAP-2 was dependent on prolonged IFN-γ treatment of the cells starting at 48 h before infection and on the concentration that was added. These observations indicate that IFN-γ impairs the production or stabilization of IAP-2 protein. Because HeLa cells were found to be resistant against apoptosis at 48 h after infection with persistent chlamydiae and because IAP-2 protein was not upregulated at the same time point, we could exclude the possibility that IAP-2 is responsible for antiapoptotic mechanisms of persistent C. trachomatis infection.
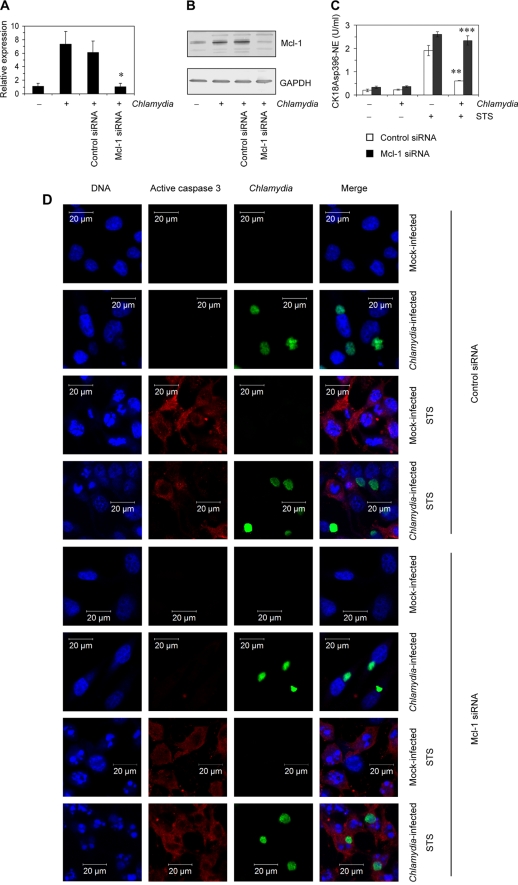
Next, experiments were performed to evaluate whether the increase in Mcl-1 production is responsible for apoptosis inhibition mediated by Chlamydia during the state of persistence. HeLa cells were transfected with specific siRNA to silence Mcl-1 expression. Transfection with 10 nM of the specific siRNA was sufficient to downregulate mRNA levels in infected cells and inhibit Mcl-1 protein production (Fig. 5A and B). Nonsilencing siRNA served as a negative control. Induction of apoptosis following exposure to STS was measured by quantification of CK18Asp396-NE. As shown in Fig. 5C, after STS stimulation HeLa cells transfected with Mcl-1 siRNA and infected with persistent Chlamydia contained high levels of CK18Asp396-NE that did not differ from those measured in mock-infected cells, indicating that Mcl-1 silencing sensitizes persistently infected cells to apoptosis. In contrast, apoptosis inhibition by persistent chlamydial infection was not affected following transfection with control siRNA. Sensitivity of persistently infected cells to apoptosis was further characterized by confocal laser scanning microscopy. Cells were stained with antibodies against active caspase-3 (p18) and C. trachomatis and with DAPI to assess nuclear fragmentation. The results are summarized in Fig. 5D. When infected cell cultures were transfected with control siRNA, STS treatment induced active caspase-3 and nuclear fragmentation in Chlamydia-negative but not in inclusion-positive cells. Mock-infected cells transfected with control siRNA served as controls. In contrast, transfection with Mcl-1-specific siRNA clearly induced apoptosis in HeLa cells containing persistent Chlamydia bacteria. After exposure to STS the inclusion-containing cells were stained positive for active caspase-3 and showed nuclear fragmentation in the same manner as cells in mock-infected cultures.
Fig 5.
Silencing of Mcl-1 in IFN-γ-treated HeLa cells. Cells were transfected with specific or control siRNA for 48 h and then infected with C. trachomatis. IFN-γ stimulation was started at 48 h before infection. Cells were analyzed at 24 h after infection. (A) Downregulation of Mcl-1 mRNA levels was assessed by real-time RT-PCR. Data were normalized to GAPDH mRNA levels as described in Materials and Methods. *, P < 0.02 (compared to values for infected cells with or without control siRNA transfection). (B) Inhibition of Mcl-1 protein production following siRNA transfection. (C) Apoptotic cells in culture with and without STS treatment were determined by quantification of CK18Asp396-NE in cell lysates. **, P < 0.01 (compared to values for mock-infected cells transfected with control siRNA and exposed to STS); ***, P < 0.001 (compared to values for infected cells transfected with control siRNA and exposed to STS; n = 4). (D) Cell monolayers were costained with DAPI for DNA (blue), active caspase-3 antibody (probed with a Cy3-conjugated secondary antibody; red), and FITC-conjugated C. trachomatis MOMP antibody (green). For the experiments shown in panels C and D, 1 μM STS was added 3 h before sampling to induce apoptosis.
Release of HMGB1 from HeLa cells infected with persistent C. trachomatis.
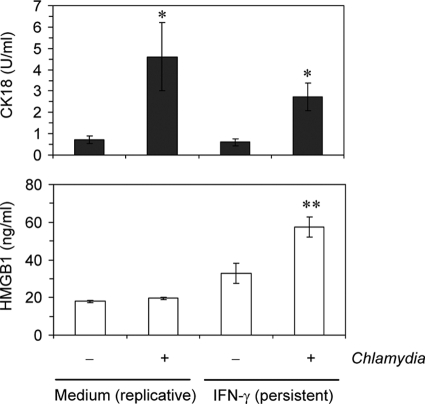
A previous study has shown that CPAF-mediated PARP-1 cleavage interferes with release of the DAMP molecule HMGB1 from infected cells (38). Downregulation of CPAF during persistence may have an impact on HMGB1 release when cell membrane damage occurs. In culture supernatants of infected HeLa cells without or with IFN-γ treatment, increased amounts of total soluble CK18 could be measured, indicating that cell membrane integrity is disturbed in response to chlamydial infection (Fig. 6). Persistently infected HeLa cells also released HMGB1 in significantly increased amounts in contrast to cells infected with replicative Chlamydia bacteria (Fig. 6).
Fig 6.
Release of total CK18 and HMGB1 from C. trachomatis-infected HeLa cells without or with IFN-γ-restricted chlamydial growth. Culture supernatants were collected at 24 h after infection and analyzed by ELISA. *, P < 0.001 (compared to mock-infected cells with or without IFN-γ treatment); **, P < 0.001 (compared to mock-infected cells with IFN-γ treatment and infected cells cultured in medium alone; n = 6).
DISCUSSION
Persistent C. trachomatis infection characterized by aberrant intracellular bacteria and a low replication rate has been reported to play a role in the development of chronic inflammatory disease (2, 3, 29). Here, we show that persistent Chlamydia infection effectively blocks apoptosis but induces the release of HMGB1 from the host cell in contrast to replicative infection.
For the establishment of a persistent state of C. trachomatis infection, IFN-γ treatment of HeLa cells was used. IFN-γ is an essential factor of the cellular immune response against chlamydiae, but it can also contribute to the development of persistent infection (2, 4). The efficacy of IFN-γ-mediated inhibition of C. trachomatis growth differs markedly among serovars (22). For the serovar D strain IC Cal 8, an IFN-γ concentration of 5 ng per ml and treatment of HeLa cells prior to infection were necessary to induce the formation of aberrant intracellular reticulate bodies and block the differentiation into infectious elementary bodies. IDO is known as the main factor that mediates restriction of chlamydial replication in human cells (4). Induction of IDO protein in HeLa cells could be observed in the presence of 5 ng of IFN-γ per ml but not at lower concentrations. This finding corresponds to results of a previous study in which IDO activity and tryptophan catabolism in HeLa cells were induced at IFN-γ concentrations of more than 3 ng per ml (32).
During replicative infection Chlamydia secretes CPAF into the cytosol of the host cell (40). By degradation of BH3-only proteins, CPAF has been implicated in the maintenance of apoptosis resistance of host cells throughout the chlamydial replication cycle (25). In a previous study we have shown that CPAF cleaves and inactivates PARP-1, thereby mediating the suppression of an increase in the release of HMGB1 from infected cells despite an ongoing disturbance of cell membrane integrity (38). By this mechanism CPAF also interferes with pathways involved in necrotic cell death. It has been reported that during persistent infection CPAF is not translocated into the cytoplasm of the host cell but is still present in the chlamydial inclusion (16, 34). This study demonstrates a downregulation of CPAF expression by C. trachomatis during IFN-γ-induced persistence, corresponding to findings by Belland et al. (5). As a consequence, several CPAF substrates are not cleaved in infected cells, with the exception of a partial processing of CK18 that may be caused by small amounts of CPAF at the inclusion. The processing of intermediate filaments has been proposed as a strategy of the pathogen to stabilize the inclusion and increase its flexibility (20). The proapoptotic BH3-only proteins Bad and Puma are not degraded during IFN-γ-induced persistence. Nevertheless, persistently infected cells were found to be fully protected from apoptosis when exposed to STS. This finding supports the hypothesis that the apoptosis resistance of Chlamydia-infected cells does not necessarily depend on CPAF and can be achieved by the upregulation of host cell antiapoptotic factors (30).
Two host cell-derived factors have been implicated in apoptosis protection by Chlamydia: IAP-2 and Mcl-1 (26, 27). IAP-2 belongs to a group of proteins that are known as IAPs or baculovirus IAP repeat (BIR)-containing proteins (BIRCs). IAPs play a role in cell survival in response to stress conditions and in the regulation of cytokine receptor signaling (12). Although IAP-2 has been described to ubiquitinylate caspases 3 and 7 other functions such as modulation of TNFR2 signaling pathways may be more important (12). Following IFN-γ treatment, IAP-2 protein was not induced in infected cells, and therefore the idea that IAP-2 represents an essential factor mediating antiapoptotic effects of persistent chlamydial infection could be excluded. IFN-γ is known to make target cells more susceptible to apoptotic stimuli, and our findings correspond to studies in which an inhibition of TRAIL-induced IAP-2 upregulation in HeLa cells following IFN-γ stimulation could be observed (8, 23).
Mcl-1 is an antiapoptotic member of the Bcl-2 family of proteins. It contains three BH domains, whereas proapoptotic Bcl-2 members such as Bad, Bim, and Puma contain only the BH3 domain (33). A splicing variant of Mcl-1 that encodes a BH3-only protein has been described previously (1). Full-length Mcl-1 blocks apoptosis by binding and sequestering Bax and Bak, factors that are activated by BH3-only proteins to form large oligomeric pores in the mitochondrial membrane (33). Mcl-1 can also bind the BH3-only protein members Bim and Puma (33). In a previous study it has been shown that Mcl-1 is upregulated in Chlamydia-infected cells and plays an important role in apoptosis resistance of host cells during the early stage of the development of a chlamydial inclusion (26). The induction of Mcl-1 expression is consistent with the limitation of apoptosis blockade in Chlamydia-infected cells to a premitochondrial step (30). By degrading BH3-only proteins, CPAF obviously contributes to the apoptosis resistance of cells infected with replicative chlamydia because Chlamydia has been reported to protect Mcl-1−/− cells against apoptosis (35). However, as shown in this study Mcl-1 silencing sensitizes HeLa cells to STS-induced apoptosis when they are infected with persistent Chlamydia bacteria that express only small amounts of CPAF. These findings suggest that the upregulation of Mcl-1 is sufficient to mediate apoptosis inhibition by persistent C. trachomatis infection.
It should be considered that the transcriptional response of Chlamydia varies in different persistence models using iron depletion, beta-lactam, or IFN-γ treatment (15). Such variations may also have impact on the mechanism of apoptosis inhibition. Despite this limitation, the IFN-γ-induced persistence provides a model of the downregulation of CPAF activity.
Apoptosis is designed to eliminate injured cells without eliciting an inflammatory response. Resistance to apoptosis will not necessarily protect host cells from cytopathic effects of chlamydial infection. PARP-1 represents a marker that allows the differentiation of apoptosis and necrosis (6). During apoptosis PARP-1 is cleaved by caspase-3 into 89- and 23-kDa fragments and inactivated. In contrast, PARP-1 overactivation promotes necrosis and mediates the translocation of HMGB1 from the nucleus into the cytosol and subsequent release of HMGB1 from membrane-damaged cells (6). HMGB1 is considered a prototypic DAMP molecule that activates the inflammatory cytokine response via TLR2/TLR4 or RAGE (receptor for advanced glycation end products) signaling (28, 39). Because PARP-1 is multifragmented by CPAF in cells with replicative chlamydial infection, an increased HMGB1 release from the cells is prevented (38). By this mechanism C. trachomatis may have evolved a strategy to limit the epithelial inflammatory response. In persistently infected cells, PARP-1 remains intact, and the cells release increased amounts of HMGB1. It can be suggested that epithelial cells infected with persistent Chlamydia bacteria represent an apoptosis-resistant but inflammatory cell type that may contribute to chronic inflammation and disease.
ACKNOWLEDGMENTS
This work was supported by a grant from the Jena School for Microbial Communication to C.G. and by grant 01KI0726 from the Bundesministerium für Bildung und Forschung, Germany, to E.S. and J.R.
Footnotes
Published ahead of print 24 October 2011
REFERENCES
- 1. Bae J, Leo CP, Hsu SY, Hsueh AJW. 2000. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 275: 25255–25261 [DOI] [PubMed] [Google Scholar]
- 2. Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 90: 3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beatty WL, Morrison RP, Byrne GI. 1995. Reactivation of persistent Chlamydia trachomatis infection in cell culture. Infect. Immun. 63: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. 1994. Trytophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62: 3705–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belland RJ, et al. 2003. Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U. S. A. 100: 15971–15976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouchard VJ, Rouleau M, Poirier GG. 2003. PARP-1, a determinant of cell survival in response to DNA damage. Exp. Haematol. 31: 446–454 [DOI] [PubMed] [Google Scholar]
- 7. Caulín C, Salvesen GS, Oshima RG. 1997. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J. Cell Biol. 138: 1379–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang Y-H, Chao Y, Hsieh S-L, Lin W-W. 2004. Mechanism of LIGHT/interferon-γ-induced cell death in HT-29 cells. J. Cell. Biochem. 93: 1188–1202 [DOI] [PubMed] [Google Scholar]
- 9. Chen D, et al. 2010. Autoprocessing and self-activation of the secreted protease CPAF in Chlamydia-infected cells. Microb. Pathog. 49: 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dean D, Powers VC. 2001. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 69: 2442–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong F, Sharma J, Xiao Y, Zhong Y, Zhong G. 2004. Intramolecular dimerization is required for the Chlamydia-secreted protease CPAF to degrade host transcriptional factors. Infect. Immun. 72: 3869–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubrez-Daloz L, Dupoux A, Cartier J. 2008. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle 7: 1036–1046 [DOI] [PubMed] [Google Scholar]
- 13. Fan T, et al. 1998. Inhibition of apoptosis in Chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer SF, et al. 2004. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 200: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goellner S, et al. 2006. Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect. Immun. 74: 4801–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heuer D, Brinkmann V, Meyer TF, Szczepek AJ. 2003. Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell. Microbiol. 5: 315–322 [DOI] [PubMed] [Google Scholar]
- 17. Kokab A, Jennings R, Eley A, Pacey AA, Cross NA. 2010. Analysis of modulated gene expression in a model of interferon-γ-induced persistence of Chlamydia trachomatis in HEp-2 cells. Microb. Pathog. 49: 217–225 [DOI] [PubMed] [Google Scholar]
- 18. Kramer G, et al. 2004. Differentiation between cell death modes using measurements of different soluble forms of extracellular cytokeratin 18. Cancer Res. 64: 1751–1756 [DOI] [PubMed] [Google Scholar]
- 19. Kroemer G, et al. 2009. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar Y, Valdivia RH. 2008. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4: 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mårdh PA. 2004. Tubal factor infertility, with special regard to chlamydial salpingitis. Curr. Opin. Infect. Dis. 17: 49–52 [DOI] [PubMed] [Google Scholar]
- 22. Morrison RP. 2000. Differential sensitivities of Chlamydia trachomatis strains to inhibitory effects of gamma interferon. Infect. Immun. 68: 6038–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park S-Y, Billiar TR, Seol D-W. 2002. IFN-γ inhibition of TRAIL-induced IAP-2 upregulation, a possible mechanism of IFN-γ-enhanced TRAIL-induced apoptosis. Biochem. Biophys. Res. Commun. 291: 233–236 [DOI] [PubMed] [Google Scholar]
- 24. Paschen SA, et al. 2008. Cytopathicity of Chlamydia is largely reproduced by expression of a single chlamydial protease. J. Cell Biol. 182: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. 2006. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J. Biol. Chem. 281: 31495–31501 [DOI] [PubMed] [Google Scholar]
- 26. Rajalingam K, et al. 2008. Mcl-1 is a key regulator of apoptosis resistance in Chlamydia trachomatis-infected cells. PLoS One 3: e3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajalingam K, et al. 2006. IAP-IAP complexes required for apoptosis resistance of C. trachomatis-infected cells. PLoS Pathog. 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scaffidi P, Mistel T, Bianchi ME. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418: 191–195 [DOI] [PubMed] [Google Scholar]
- 29. Schoborg RV. 2011. Chlamydia persistence—a tool to dissect chlamydia-host interactions. Microbes Infect. 13: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma M, Rudel T. 2009. Apoptosis resistance in Chlamydia-infected cells: a fate worse than death? FEMS Immunol. Med. Microbiol. 55: 154–161 [DOI] [PubMed] [Google Scholar]
- 31. Sharma M, et al. 2011. HIF-1α is involved in mediating apoptosis resistance to Chlamydia trachomatis-infected cells. Cell Microbiol. 13: 1573–1585 [DOI] [PubMed] [Google Scholar]
- 32. Shirey KA, Jung J-Y, Carlin JM. 2006. Up-regulation of gamma interferon receptor expression due to Chlamydia-Toll-like receptor interaction does not enhance signal transducer and activator of transcription 1 signaling. Infect. Immun. 74: 6877–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas LW, Lamm C, Edwards SW. 2010. Mcl-1: the molecular regulation of protein function. FEBS Lett. 584: 2981–2989 [DOI] [PubMed] [Google Scholar]
- 34. Wang J, et al. 2011. Altered protein secretion of Chlamydia trachomatis in persistently infected human endocervical epithelial cells. Microbiology 157: 2759–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ying S, Christian JG, Paschen SA, Häcker G. 2008. Chlamydia trachomatis can protect host cells against apoptosis in the absence of cellular inhibitor of apoptosis proteins and Mcl-1. Microbes Infect. 10: 97–101 [DOI] [PubMed] [Google Scholar]
- 36. Ying S, et al. 2008. Premature apoptosis of Chlamydia-infected cells disrupts chlamydial development. J. Infect. Dis. 198: 1536–1544 [DOI] [PubMed] [Google Scholar]
- 37. Ying S, et al. 2006. Characterization of host cell death induced by Chlamydia trachomatis. Infect. Immun. 74: 6057–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu H, et al. 2010. Role of high-mobility group box 1 protein and poly(ADP-ribose) polymerase 1 degradation in Chlamydia trachomatis-induced cytopathicity. Infect. Immun. 78: 3288–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu M, et al. 2006. HMGB1 signals through Toll-like receptor (TLR)4 and TLR2. Shock 26: 174–179 [DOI] [PubMed] [Google Scholar]
- 40. Zhong G, Fan P, Ji H, Dong F, Huang Y. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193: 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]