Abstract
Streptococcus pneumoniae pilus 1 is present in 30 to 50% of invasive disease-causing strains and is composed of three subunits: the adhesin RrgA, the major backbone subunit RrgB, and the minor ancillary protein RrgC. RrgB exists in three distinct genetic variants and, when used to immunize mice, induces an immune response specific for each variant. To generate an antigen able to protect against the infection caused by all pilus-positive S. pneumoniae strains, we engineered a fusion protein containing the three RrgB variants (RrgB321). RrgB321 elicited antibodies against proteins from organisms in the three clades and protected mice against challenge with piliated pneumococcal strains. RrgB321 antisera mediated complement-dependent opsonophagocytosis of piliated strains at levels comparable to those achieved with the PCV7 glycoconjugate vaccine. These results suggest that a vaccine composed of RrgB321 has the potential to cover 30% or more of all pneumococcal strains and support the inclusion of this fusion protein in a multicomponent vaccine against S. pneumoniae.
INTRODUCTION
The human pathogen Streptococcus pneumoniae is responsible for upper respiratory tract infections, such as pneumonia and acute otitis media, and invasive diseases, such as meningitis, bacteremia, and endocarditis. Pathogenic strains are generally encapsulated and can be classified into >90 different capsular types (serotypes) (4a, 32). The capsule represents one of the major S. pneumoniae virulence factors; it is crucial for bacterial survival in the bloodstream, and it protects the bacteria from serum-dependent opsonophagocytic killing (5).
Vaccines based on the use of a limited number of conjugated capsular polysaccharides have been developed and licensed. The first licensed pneumococcal glycoconjugate vaccine was PCV7 (Prevnar; Wyeth), which includes the seven serotypes most prevalent in the United States (4, 6B, 9V, 14, 18C, 19F, and 23F), each conjugated to CRM197 (a nontoxic derivative of diphtheria toxin) with alum phosphate used as an adjuvant (16). Given the well-established correlation between antibody titers raised by glycoconjugate vaccines and protection (20, 31), according to the recommendations of the World Health Organization (WHO), the vaccine efficacy can be estimated by measuring the capsular serotype-specific IgG concentration (by enzyme-linked immunosorbent assay) in vaccinees; in addition, the opsonophagocytosis assay (OPKA) is used to evaluate the functionality of the antibodies (10).
Despite a significant effect of PCV7 on the burden of disease (4, 22, 39) in the 10 years since its introduction, the emergence of nonvaccine serotypes due to serotype replacement and capsular switching, two phenomena important in remodeling the epidemiology of pneumococcal disease over time, has partially reduced the benefits of vaccination (8, 11, 38).
Second-generation glycoconjugate vaccines that include 3 to 6 additional serotypes (1, 5, 7F, 3, 6A, and 19A) have been developed to attempt to address this issue (29, 34). However, the efficacy of these vaccines against mucosal disease remains unclear, and further serotype replacement will very likely occur over the next few years. Given these issues, the development of a serotype-independent vaccine composed of conserved protein antigens would be desirable.
To date, a number of candidate protein antigens, including PspA, PspC, and pneumolysin, have been explored, but their variability or toxicity has limited their effective use in the development of a vaccine (23, 30, 33, 36). Among the others, antigens proposed as potential vaccine candidates include proteins identified by reverse vaccinology (41), the StkP and PcsB proteins, which were identified through the antigenome approach (14), the histidine triad Pht proteins (15), and the pilus 1 subunits (13). A major issue associated with the development of a protein-based vaccine is the lack of an accepted correlate of protection for protein antigens (10).
S. pneumoniae pilus 1 is encoded by a genetic islet (PI-1) present in 30 to 50% of the pneumococcal strains and is implicated in adhesion to epithelial cells, lung infection, and virulence (1, 3, 26, 27). Pilus 1 is composed of the backbone subunit RrgB, the adhesin RrgA, and the minor component RrgC (17, 18). Molecular analysis has revealed the existence of three different variants of the pilus backbone RrgB (RrgB clade I, RrgB clade II, and RrgB clade III) which have a degree of protein homology of 48 to 60%. On the other hand, RrgB proteins expressed by strains belonging to the same clade share >99% sequence identity and are therefore virtually identical (27). In line with this, antisera generated against each of three RrgB variant prototypes were able to recognize the pili of different strains belonging to the homologous clade both by Western blotting (WB) and by fluorescence-activated cell sorting (FACS) (28). Furthermore, in animal models, each of the RrgB variants was protective against challenge with strains of the same clade, but no cross-protection was observed between clades (reference 13 and our unpublished data). On this basis, in this study we constructed a fusion protein comprising all three RrgB variants, called RrgB321. Here we report on the immunogenicity and protective efficacy of this fusion protein and the ability of antibodies against RrgB321 to induce opsonophagocytic killing activity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The sources of the bacterial strains used in this study were as follows: TIGR4 was from Gianni Pozzi (University of Siena, Italy), 6B Finland12 was from Lesley McGee (Centers for Disease Control and Prevention, Georgia) and Stanley Falkow (Stanford University, California), SPEC 6B was from Moon Nahm (University of Birmingham, Alabama), and 35B SME15 was from Birgitta Henriques-Normark (Karolinska Institutet, Sweden). The strains were routinely grown at 37°C in 5% CO2 on tryptic soy agar plates (TSA; Becton Dickinson) supplemented with 10 mg/liter colistin, 5 mg/liter oxolinic acid, and 5% defibrinated sheep blood (vol/vol) or in Todd-Hewitt broth supplemented with 0.5% (wt/wt) yeast extract (THYE) (Becton Dickinson). For animal experiments, bacteria grown overnight (ON) on the plates were inoculated in tryptic soy broth (TSB; Difco) and grown to an A600 of 0.25. Bacteria were then harvested by centrifugation, resuspended in TSB–20% glycerol (vol/vol)–10% fetal bovine serum (vol/vol), and frozen in aliquots at −80°C. The frozen stock was titrated by plating culture aliquots at serial dilutions and counting CFU. Immediately prior to intraperitoneal challenge, frozen aliquots were thawed and diluted in saline to reach the working concentration. For intravenous challenge, bacteria were freshly harvested from THYE liquid cultures at an A600 of 0.5 and brought to the working concentration before administration. The challenge input was titrated by plating bacterial suspensions as described above immediately after challenge.
Cloning and protein expression and purification.
Standard recombinant DNA techniques were used to construct plasmids expressing the three RrgB variants and the RrgB321 chimera, consisting of the three variants in a head-to-tail organization and separated by a six-amino-acid linker (Gly-Ser-Gly-Gly-Gly-Gly). Briefly, rrgB open reading frames (nucleotides corresponding to the N-terminal signal sequence and C-terminal cell wall sorting signal motif were excluded from the cloning) were amplified by PCR from chromosomal DNAs of S. pneumoniae TIGR4 (rrgB clade I), 6B SPEC (rrgB clade II), and 35B SME 15 (rrgB clade III) by using specific primers listed in Table S1 in the supplemental material. The PCR fragments obtained were digested with the appropriate restriction enzymes and ligated into the C-terminal 6×His-tag expression vector pET21b+ (Novagen). The resulting plasmids were confirmed by DNA sequencing and then transformed into competent Escherichia coli BL21(DE3) Star (Invitrogen). Protein expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma) at a 1 mM final concentration to a bacterial culture at an A600 of 0.4 to 0.5 (LB medium supplemented with 100 μg/ml ampicillin), and the proteins were purified by metal chelate affinity chromatography on His-Trap HP columns (GE Healthcare). Pooled fractions containing the purified protein were dialyzed ON against phosphate-buffered saline (PBS) and stored at −80°C until further use. Purified recombinant proteins were subsequently used to immunize either CD1 or BALB/c mice or rabbits for antibody generation (Charles River Laboratory) and CD1 or BALB/c mice to evaluate their protective efficacy.
SDS-PAGE and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was performed using Nu-PAGE 3 to 8% Tris-acetate gradient gels (Invitrogen) according to the manufacturer's instructions. A Hi-Mark prestained high-molecular-weight protein standard (Invitrogen) served as the protein standard. Gels were processed for Western blot analysis by using standard protocols. Rabbit antisera raised against recombinant RrgB321 chimera were used at a 1:10,000 dilution. Secondary goat anti-rabbit IgG alkaline phosphatase-conjugated antibodies (Promega) were used at 1:5,000, and the signal was developed by using Western Blue stabilized substrate for alkaline phosphatase (Promega).
Flow cytometry on entire bacteria.
Bacteria were grown in THYE to exponential phase (A600 = 0.25) and stained with rabbit primary antibodies (final dilution, 1:300) and then with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (final dilution, 1:100) (Jackson Laboratories). Finally, bacteria were fixed with 2% paraformaldehyde, and bacterial staining was analyzed by using a FACSCalibur cytometer (Becton Dickinson). Sera from mice immunized with PBS plus adjuvant and with RrgA served as negative and positive controls, respectively.
Generation of a TIGR4 PI-1 deletion mutant.
TIGR4ΔPI-1, an isogenic mutant, was generated by allelic exchange, as briefly described below. Fragments of approximately 500 bp upstream and downstream the target gene were amplified by PCR (oligonucleotides are listed in Table S2 in the supplemental material) and spliced to a kanamycin resistance cassette by using overlap extension PCR; the PCR fragment obtained was cloned into pGEMt (Promega), and the resulting plasmid was transformed in TIGR4 with conventional methods (2). Bacteria were plated on selective blood agar plates (kanamycin, 500 μg/ml). The presence of the isogenic mutation was confirmed by PCR and the absence of pilus 1 expression as confirmed by Western blot analysis (data not shown).
Specific IgG measurement using protein-coupled microspheres.
A Luminex assay was developed to determine antibody titers in mouse and rabbit antisera. Specifically, 20 μg of recombinant protein was coupled to the carboxyl groups of 2.5 million MicroPlex microspheres (Luminex Corp), according to the manufacturer's instructions. The coupling reaction was confirmed by incubating 5,000 antigen-coupled microspheres with eight serial twofold dilutions of a hyperimmune antiserum used as a reference. To determine serum titers, the beads were incubated with mouse- or rabbit-specific sera (dilution, 1:10,000 or 1:20,000, respectively, in PBS), washed twice in 200 μl of PBS, and then incubated with phycoerythrin-conjugated secondary antibodies (1:200; Jackson ImmunoResearch) for 15 min in a dark room onto an orbital shaker.
Since sera raised against the three RrgB variants were not cross-reactive, the IgG content specific for recombinant RrgB clade I, II, and III in sera raised against RrgB321 was measured using a triplex assay in which the different antigen-coupled microspheres were mixed to a working concentration of 3,500 beads per color per well.
IgG measurements were determined on the Luminex 200 analyzer using Bio-Plex Manager 5.0 software (Bio-Rad, Hercules, CA). Tests were performed in duplicate, and the mean fluorescence intensity (MFI) was determined. The limit of quantification (LOQ) of the assay was determined at an MFI of 100 and was considered the threshold for positive results.
Formulation of the His-tagged RrgB321 chimera.
The RrgB321 chimera was formulated under sterile conditions under a flow hood; recombinant protein was adsorbed onto aluminum hydroxide (alum) at 0.1 mg/ml protein, 2 mg/ml aluminum hydroxide, and 9 mg/ml NaCl in 10 mM histidine, pH 6.5. Water for injection and histidine buffer were premixed. Sodium chloride was added to give a final formulation osmolality of 308 mosmol/kg. Alum addition was calculated based on the concentration of the alum stock to obtain a final concentration of 3 mg/ml. The antigen was added to the mix and left for 15 min with stirring at room temperature and then stored ON at 4°C before the immunization. Final formulations were isotonic and at physiological pH.
All formulations were characterized soon after immunization; antigen adsorption was ≥95%, and the adsorption profile was similar for all antigens tested.
Antisera.
To generate sera against the specific proteins, purified recombinant proteins were used to immunize either BALB/c or CD1 mice (as detailed in “Animal Experiments” below) or New Zealand White rabbits with a body weight of 2.5 kg (100 μg with Freund's adjuvant (FA), three doses administered subcutaneously on day 0, 21 and 35) (Charles River Laboratory). To generate antipolysaccharide antibodies to be used as positive controls in an in vitro OPKA (see below), BALB/c mice were immunized either with PCV7 (Prevnar [Wyeth]; single administration of 1/5 human dose) or with heat-inactivated 35B SME15 (serotype 35B; three administrations of 2 × 108 CFU); New Zealand White rabbits were immunized with a single administration of a PCV7 human dose. A rabbit polysaccharide multivalent antiserum (Omniserum) was purchased from Statens Serum Institut (Copenhagen, Denmark).
Animal experiments.
Animal studies were done in compliance with the current law, approved by the local Animal Ethics Committee, and authorized by the Italian Ministry of Health.
Female, 6-week-old, specific-pathogen-free BALB/c or CD1 mice (Charles River) received three intraperitoneal (i.p.) immunizations, 2 weeks apart. Each dose was composed of 20 μg of recombinant protein, along with 400 μg of aluminum hydroxide as an adjuvant, in a final volume of 200 μl of saline. Negative controls received the same course of saline plus adjuvant. Ten days after the third immunization, serum samples were obtained for serological and functional studies. Two weeks after the third immunization, BALB/c mice were challenged i.p., while CD1 mice were challenged intravenously (i.v.) via the tail vein. The pneumococcal strains and the challenge doses are reported in Table S3 in the supplemental material. Challenge doses were previously set in naïve mice in order to achieve a mortality of ≥80%. The three strains were selected for challenge experiments in a large screening in which >20 different piliated pneumococcal strains were tested in vivo for their ability to infect mice and cause >80% mortality. In particular, for each tested strain, naïve mice were infected either i.p. or i.v. with increasing doses of bacteria, and mortality was monitored for 10 (i.p.) or 15 (i.v.) days. Only strains for which >80% mortality could be reproducibly achieved were selected for further in vivo experiments. Bacteremia was evaluated in blood samples taken 24 h (i.p. challenge) or 48 h (i.v. challenge) postchallenge and plated on blood agar plates at serial dilutions. After 24 h of culture, CFU were counted and the CFU/ml of blood calculated. Bacteremia was expressed as log10 (log) of the CFU/ml value. After challenge, the animals were monitored for 10 (i.p. challenge) or 15 (i.v. challenge) days. Mice were euthanized when they exhibited defined endpoints for humane killing that had been preestablished for the study in agreement with Novartis Animal Welfare Policies, and the day was recorded. Survival rates were calculated according to the following formula: % survival = [1 − (% dead vaccinated animals/% dead controls)] × 100.
For the passive protection experiments, 15 min before TIGR4 i.p. challenge, each mouse received either 100 μl of rabbit antiserum or 50 μl of pooled mouse antisera to RrgB321 i.p. Controls received either 100 μl of normal rabbit serum or 50 μl of pooled mouse sera obtained from the negative controls immunized with saline plus adjuvant. The challenge dose was 140 CFU/mouse.
Statistical analysis.
GraphPad Prism software (version 5.0) was used for statistical analyses. For immunogenicity, the two-tailed Mann-Whitney U test was used, and MFI values were analyzed. For the in vivo experiments, the following one-tailed tests were applied: Mann-Whitney U test to analyze data for bacteremia, log rank (Mantel-Cox) test for survival curves, and chi square test for survival rates. P values of ≤0.05 were considered and referred to as significant.
Opsonophagocytosis killing assay (OPKA) using HL60 cells.
Human HL-60 promyelocytes (ATCC CCL240) were maintained in enriched medium (RPMI 1640 plus Glutamax [Invitrogen], 10% fetal calf serum [HyClone], 1% penicillin/streptomycin [Gibco]) and differentiated into phagocytes using 0.8% N,N-dimethylformamide (DMF; Sigma). After 5 days of treatment with DMF, cells were ready to be used in the OPKA (15). Following heat inactivation (30 min at 56°C), rabbit and mouse antisera were prediluted 1:3 in opsonization buffer (OPB; Hanks' balanced salt solution [HBSS] with Ca2+ and Mg2+ [Invitrogen], supplemented with 0.1% gelatin and 10% inactivated FC1 [HyClone]) and subsequently threefold serially diluted in a 96-well plate (BD Corning). Frozen bacteria were thawed, washed once in OPB (3,000 rpm for 5 min), and then incubated with sera (1,200 CFU/well) at room temperature for 30 min. Baby rabbit complement (BRC) was added at 12%, and differentiated HL-60 cells were distributed at 4 × 105 per well (HL-60–bacterium ratio, 400:1). Plates with reaction mixtures (final volume, 80 μl) were incubated at 37°C and 5% CO2 for 1 h on a shaking platform (400 rpm, Orbit 300 shaker; LabNet). Phagocytosis was stopped by resting plates on ice for 15 min. Five microliters of reaction mixture was tilt plated onto TH agar-yeast extract. Seeded bacteria were allowed to dry, included in TH agar-yeast extract supplemented with triphenyl tetrazolium chloride dye (100 mg/liter, TTC; Sigma), and incubated overnight at 37°C and 5% CO2 (21). Bacterial survival was detected with a ProtoCOL colony counter (Synbiosis). Results were expressed as percent killing, calculated as the percentage of bacteria that were killed in samples with bacteria, phagocytes, and active complement plus sera (BPC′+S), compared to those in samples containing bacteria, phagocytes, and active complement only (BPC′): [(BPC′+S)/BPC′] × 100.
Bacteria to be used in the OPKA were selected from a panel of 30 strains representative of the three clades based on pilus type (I, II, and III), previously screened for complement activity and toxicity. The strains were considered suitable for the in vitro assay if they showed nonspecific killing (NSK) lower than 20%, where NSK defines the effect of active complement (BPC′) with respect to heat-inactivated complement (BPCh.i.) on bacterial viability [1 − (BPC′/BPCh.i.)]. Working stocks were prepared by growing pneumococci in THYE to a final A600 of 0.25, and bacterial aliquots were stored at −80°C until the experiment was performed.
Competitive OPKA.
Rabbit antiserum raised against RrgB321 (1:3 in OPB) was incubated with 0.55 mg/ml of either RrgB321 recombinant protein or an unrelated protein (UCP, a Streptococcus agalactiae pilus protein) in an 80-μl final volume. After ON incubation at 4°C with horizontal shaking, 30 μl of serum was tested in duplicate in the OPKA as described above.
RESULTS
RrgB321 is immunogenic and elicits antibodies against the three RrgB variants.
In order to test the specificity of the antibody response to the three RrgB variants, groups of mice were immunized with purified RrgB proteins of organisms belonging to clade I, clade II, and clade III. The antisera raised against each of the three single recombinant RrgB variants were clade specific and not able to recognize the other two variants when tested by Luminex (Fig. 1).
Fig 1.
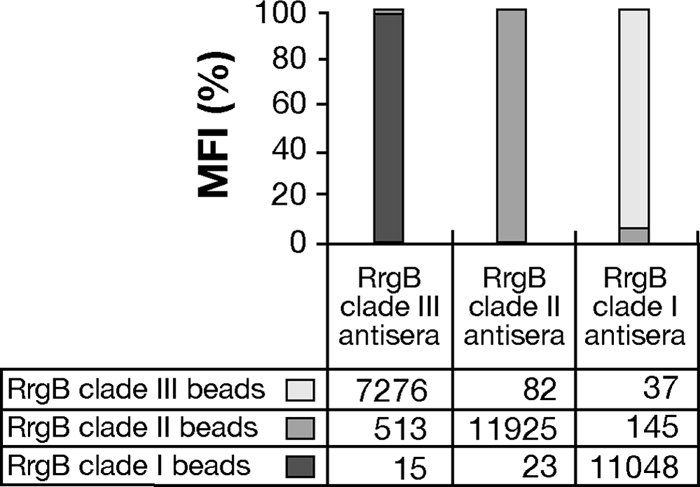
Mouse antisera raised against the three RrgB variants are not cross-reactive. Luminex beads were coated with recombinant RrgB of clade I, II, or III. The cross-reactivity of BALB/c mouse sera raised against the three RrgB variants with aluminum hydroxide as an adjuvant was analyzed by probing the sera on the mix of beads. Mean fluorescence intensity (MFI) values are reported below the bars. The MFI as indicated by the histograms is calculated by considering the contribution of the single RrgB variant to the total across the clades, multiplied by 100.
To generate an antigen able to elicit an immune response directed against RrgB from each of the three clades, five fusion proteins that included the three variants in different reciprocal positions (RrgB123, RrgB231, RrgB321, RrgB312, and RrgB213) were expressed and purified. The fusion proteins displayed comparable immunogenic properties; therefore, the subsequent decision to conduct further studies by using the RrgB321 chimera was made on the basis of expression levels and stability of the final product (data not shown).
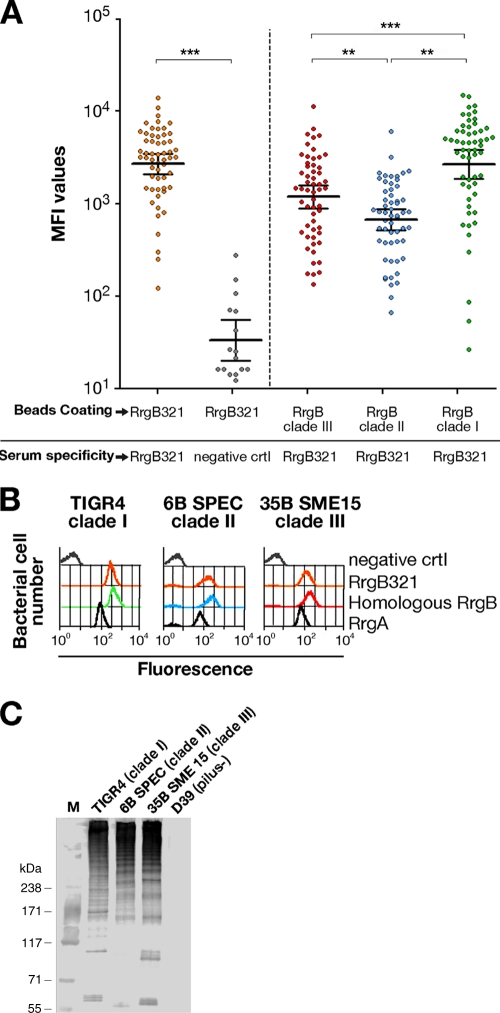
To evaluate the immunogenic activity of the RrgB321 chimera, single mouse RrgB321 antisera were analyzed for specific IgG and IgM content. When RrgB321-coated beads were incubated with RrgB321 antisera (serum dilution, 1:10,000), high MFI (mean fluorescence intensity) signals were observed, significantly different from those detected for the negative control (PBS plus alum) (P < 0.0001) (Fig. 2A), indicating a good antibody response in immunized animals. RrgB321 antisera were also tested against the single recombinant RrgB proteins (clade I, II, and III). High levels of specific IgG against each of the three RrgB variants were detected; RrgB clade I protein in particular was the most recognized by specific Abs in RrgB321 antisera, followed by clade III and clade II. IgM levels were not significant (data not shown).
Fig 2.
RrgB321 antisera recognize the three native and recombinant RrgB variants. (A) Antisera derived from a group of 56 BALB/c mice immunized with RrgB321 chimera with alum as the adjuvant were analyzed by Luminex using beads coated with RrgB321 or single RrgB variants; 16 mice immunized with PBS plus alum served as negative controls. Recognition of RrgB321 by specific IgG, expressed as mean fluorescence intensity (MFI), was highly significant compared to negative-control values (P < 0.0001). The response to single RrgB proteins was highest for clade I, followed by clade III and clade II. Samples were run in duplicate. Each circle represents the MFI value for a single animal, and horizontal bars represent the geometric mean with 95% confidence interval. Statistically significant differences based on a Mann-Whitney two-sample rank test are indicated. *, **, and ***, P ≤ 0.05, 0.01, and 0.0001, respectively. (B) Rabbit polyclonal antisera raised against RrgB321 were tested by FACS analysis on strains expressing the pili of clade I (TIGR4), clade II (6B SPEC), and clade III (35B SME15), along with sera generated against the homologous RrgB. RrgA and PBS+alum antisera were used as positive and negative controls, respectively. (C) WB analysis of TIGR4, 6B SPEC, and 35B SME15 whole lysates performed with rabbit RrgB321 antisera (1:10,000 dilution) (D39, which is not piliated, was used as negative control).
Furthermore, the RrgB321 antisera were able to recognize the native pilus as efficiently as sera generated against the three homologous recombinant proteins, as demonstrated by FACS analysis on whole bacteria and by Western blotting of bacterial lysates of the S. pneumoniae strains TIGR4 (clade I), 6B SPEC (clade II), and 35B SME15 (clade III) (Fig. 2B and C). Strains that did not express the pilus were consistently negative by FACS analysis when probed with the same panel of sera (data not shown).
RrgB321 protects against challenge with pneumococcal strains expressing each of the three RrgB variants.
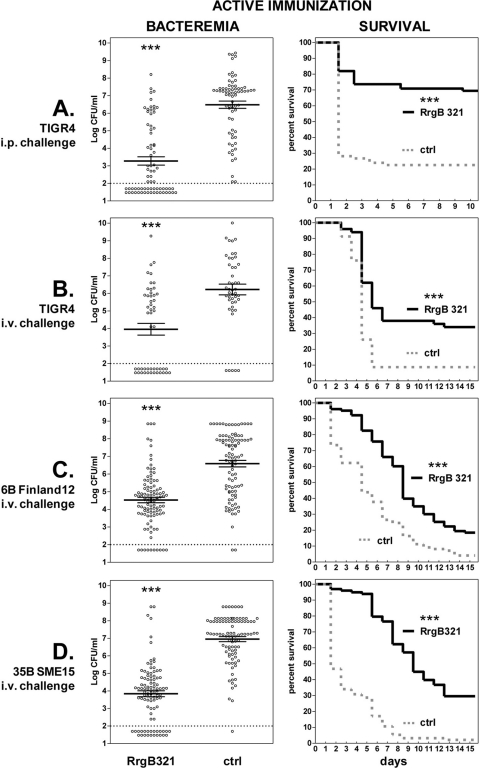
To assess the protective efficacy of RrgB321, the protein was tested in mouse models of sepsis in which mice were immunized intraperitoneally (i.p.) with the recombinant protein and then challenged either i.p. or intravenously (i.v.) with strains representative of the three pilus clades (Fig. 3; also, see Table S3 in the supplemental material). As shown in Fig. 3A, RrgB321 immunization induced protection against intraperitoneal challenge with TIGR4; the bacteremia in the immunized group was >3 log lower than that of the control group (3.28 versus 6.48; P < 0.0001), the mean survival time was 8.1 versus 3.6 days (P < 0.0001), and a highly significant survival rate was observed at 10 days (P < 0.0001).
Fig 3.
Protective efficacy afforded by active RrgB321 immunization against intraperitoneal (i.p,) or intravenous (i.v.) challenge. The challenge strain and route are indicated on the left. Mice were either immunized with RrgB321 or given alum plus saline (ctrl). (Left) Bacteremia. Circles represent values for the blood of single animals, horizontal bars show the mean ± standard error of the mean for each group, and the dashed line indicates the detection limit (i.e., no CFU were detected in samples below the dashed line). (Right) Survival. The solid line and the dashed line represent the survival course for RrgB321 and the control group, respectively. *, **, and ***, P ≤ 0.05, 0.01, and 0.0001, respectively.
The protective efficacy of RrgB321 was also evaluated in the intravenous challenge model (Fig. 3B to D; also, see Table S3 in the supplemental material). Highly significant reduction of bacteremia (P < 0.0001) was observed for each of the three strains tested. The mean log CFU values for the immunized group versus for the control group were 3.96 versus 6.22 with TIGR4 challenge, 4.53 versus 6.59 with 6B Finland12, and 3.87 versus 6.96 with 35B SME15. Consistent with the reduction of bacteremia, the mean survival was significantly increased (P < 0.0001) in immunized animals versus the corresponding control groups: 8.8 versus 5.3 days with TIGR4 challenge, 9.1 versus 5.4 for 6B Finland12, and 10.2 versus 3.4 for 35B SME15. Immunization with RrgB321 also resulted in significant (P < 0.05) survival rates at the end of the observation.
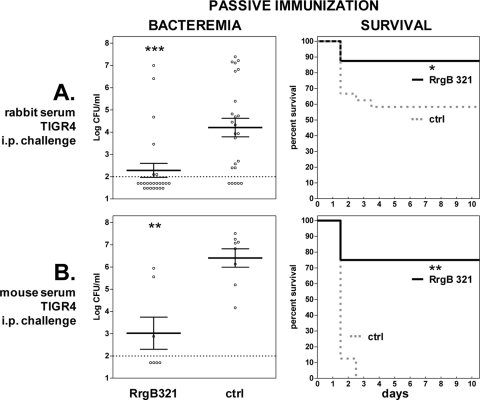
Consequently, passive immunization of naïve mice was performed to test the functionality of RrgB321 antibodies. Mice were immunized with a rabbit RrgB321 antiserum prior to TIGR4 challenge, resulting in a significant decrease of bacteremia by 1.86 log (mean log CFU, 2.35 for immunized versus 4.21 for control group; P = 0.0001), an increase of mean survival time by 2.5 days (9.4 versus 6.9 days; P < 0.05), and a significant survival rate (P < 0.05) (Fig. 4A; also, see Table S3 in the supplemental material). Similar protection was observed with the passive transfer of mouse serum against RrgB321, with a reduction of bacteremia by >3 log (mean log CFU, 3.20 versus 6.41; P < 0.01), an increase of mean survival time by 6.3 days (7.9 versus 1.6 days; P < 0.01), and a significant survival rate (P < 0.01) (Fig. 4B; also, see Table S3). These results indicate that specific antibodies are sufficient to confer protection under the experimental conditions tested.
Fig 4.
Protective efficacy afforded by passive RrgB321 immunization against intraperitoneal (i.p.) challenge with TIGR4. Mice received rabbit (A) or mouse (B) anti-RrgB321 serum before challenge; the corresponding control groups (ctrl) received normal rabbit serum (A) or serum from mice immunized with alum plus saline (B). Symbols are as described in the legend to Fig. 3.
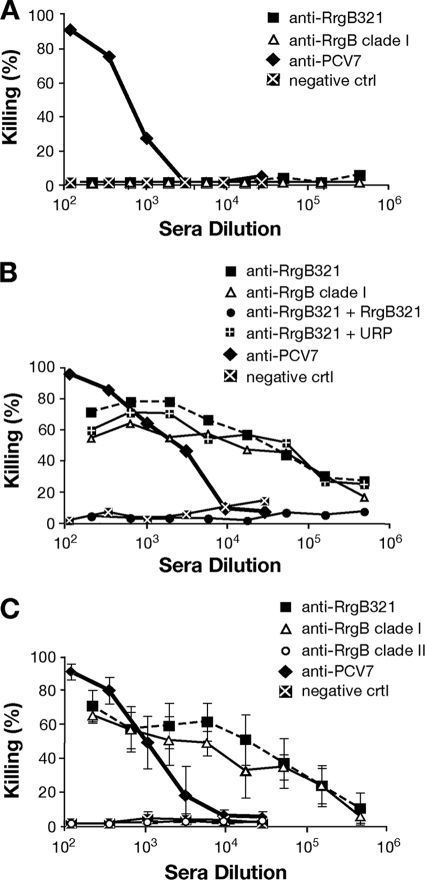
Polyclonal RrgB321 antisera elicit killing of pneumococci in vitro in the OPKA.
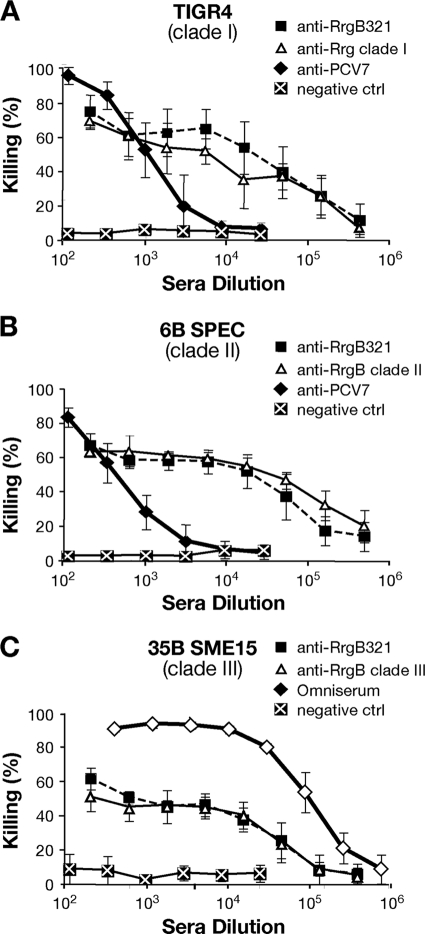
To assess whether antibodies against RrgB321 were functional in vitro, polyclonal antisera obtained from immunized rabbits were tested in the OPKA against strains representative of the three RrgB variants, according to the protocol developed by Burton and Nahm for polysaccharide antigens (6). Following a large screening, one strain from each of the three pilus variants (TIGR4, clade I; 6B SPEC, clade II; 35B SME15, clade III) was finally selected on the basis of the low nonspecific killing (<20%) exerted by complement. When the experimental assay was performed by incubating TIGR4 with rabbit RrgB321 antisera, the antibodies killed up to 80% of the bacteria in a concentration-dependent manner (sera were serially diluted from 1:200 to 1:437,400), with a trend similar to that obtained with RrgB clade I-specific antibodies (Fig. 5A). Antisera derived from PCV7 immunization, used as positive controls, showed an efficacy close to 100% at the lowest dilution (sera were serially diluted from 1:12 to 1:26,244), but the killing effect decreased faster at higher dilutions. No relevant bacterial killing was detected with sera from animals immunized with adjuvant only. Notably, when the RrgB321 antisera were used at the same dilution as the positive-control sera (from 1:12 to 1:26,244), the percentage of bacterial killing remained almost unchanged (around 80%) until the highest dilution used (1:26,244) (data not shown). Only by increasing the antiserum dilution (from 1:200 to 1:437,400) did we observe a decrease in percent killing to the background levels, similar to what was observed with the PCV7 antisera.
Fig 5.
RrgB321 chimera elicits opsonic antibodies able to kill S. pneumoniae strains TIGR4 (clade I) (A), 6B SPEC (clade II) (B), and 35B SME15 (clade III) (C). Serial dilutions of specific antisera generated in rabbits were incubated with the bacteria in the presence of BRC and HL-60 phagocytes. All values are the averages from three experiments; error bars represent standard deviations. Negative-control sera were obtained by immunizing rabbits with PBS plus FA. Positive controls were PCV7 antiserum (A and B) and Omniserum (C).
The in vitro killing elicited by RrgB321 antisera with strains of clade II and clade III was comparable to that obtained with TIGR4, although the highest killing percentages varied among the strains (about 70% for 6B SPEC and about 60% for 35B SME15, at the lowest dilution). In both cases, the curves obtained with RrgB clade II- and clade III-specific antisera overlapped with the curve obtained with RrgB321 antiserum. PCV7 antiserum and Omniserum (a positive-control rabbit polyclonal antiserum) both reached 90% bacterial killing at the lowest dilution used against 6B SPEC and 35B SME15, respectively (Fig. 5B and C).
Data obtained with rabbit sera were also confirmed using pooled and single mouse anti-RrgB321 sera (from BALB/c and CD1 mice) against TIGR4 (see Fig. S1 and S2 in the supplemental material), 6B SPEC, and 35B SME15 (data not shown). Notably, the OPKA values obtained with single BALB/c mouse antisera showed a low variability, overlapping the trend obtained with pooled sera (see Fig. S2).
In vitro killing is specifically due to antibodies against RrgB321.
The specificity of RrgB antibodies was confirmed by performing the OPKA with an isogenic knockout mutant of TIGR4 deprived of the pilus islet (TIGR4ΔPI-1). As shown in Fig. 6A, the in vitro killing activity was abrogated with both RrgB clade I and RrgB321 antisera, in contrast to what was previously observed with the wild-type (wt) strain (Fig. 5A). On the other hand, PCV7 and PBS+FA antisera (positive- and negative-control sera) showed results similar to those obtained with the TIGR4ΔPI-1 and TIGR4 wt strains.
Fig 6.
The functional antibodies induced by RrgB321 chimera are specific, and pilus clades are non-cross-reactive in OPKA. (A) RrgB321 antisera tested in OPKA do not kill the isogenic knockout mutant TIGR4ΔPI-1, which does not contain the pilus islet 1. (B) Preincubation of RrgB321 antiserum with the purified RrgB321 protein abolishes the killing activity against wt TIGR4 (clade I) in competitive OPKA. (C) Serum obtained from animals immunized with RrgB of clade II did not induce any OPKA killing when tested against TIGR4 strain (pilus of clade I). Negative-control sera were obtained by immunizing rabbits with PBS plus FA. The positive control was PCV7 antiserum. Values are from single experiments (A and B) or averages from three experiments (C); error bars represent standard deviations.
The RrgB321 antiserum was then preincubated with the purified recombinant RrgB321 protein. As expected, no opsonic killing was observed. In contrast, the preincubation of the RrgB321 serum with an unrelated protein (URP, a Streptococcus agalactiae pilus protein) did not affect the killing activity, as the percent killing measured was comparable to that obtained with RrgB clade I and RrgB321 antisera that were not preabsorbed (Fig. 6B). Moreover, when sera obtained from immunization with a heterologous RrgB variant (clade II) were tested against TIGR4 (clade I), no killing was observed, i.e., it was at the same level as the negative control (Fig. 6C), supporting our previous results showing no cross-reaction among RrgB variants (Fig. 1).
DISCUSSION
The three allelic variants of the RrgB backbone pilus subunit (RrgB from clades I, II, and III) are protective in vivo against homologous challenge with Streptococcus pneumoniae strains but not cross-protective; thus, their combination could broaden the efficacy of a pilus-based vaccine. Recently, Spraggon et al. (37) and Gentile et al. (12) demonstrated that the RrgB protein is organized into four independently folded domains (D1 to D4). Experiments performed in vivo with the four single recombinant domains showed that multiple protective epitopes located along the entire molecule contribute to its immunogenicity and that the overall protective effect of RrgB is not due solely to a single antigenic domain.
On this basis, in the present study the RrgB321 fusion protein, containing the three full-length RrgB variants in a head-to-tail organization, was constructed. We demonstrated that RrgB321 is immunogenic and protective in both active and passive immunization studies. In addition, it elicits antibodies that recognize the native pili of clade I, II, and III organisms with the same efficacy as sera raised against the homologous recombinant RrgB proteins. However, the immune response elicited by RrgB321 to the single recombinant variants was different, with RrgB of clade II always being the least well recognized. To investigate whether this lower immunogenicity could be due to the central position of RrgB clade II within the RrgB321 protein construct, sera obtained from the immunization with different fusion proteins where RrgB clade II was at either the N or C terminus (RrgB213 and RrgB312) were analyzed. The clade II RrgB was the least well recognized by sera against RrgB213 and by sera against RrgB312 (data not shown), indicating that variant-specific antibody content in the antisera was independent of the order of the three RrgB variants in the fusion proteins.
It is noteworthy that in spite of the lower IgG response to the RrgB clade II protein, the opsonophagocytic killing efficacy exerted by RrgB321 antisera against the variant II strain (6B SPEC) was comparable to that obtained against the variant I strain (TIGR4). This finding suggests that the quality rather than the quantity of antibodies is important for eliciting functional activity.
Indeed, the three strains considered in this study showed similar OPKA trends with respect to RrgB321 antisera, even though their susceptibility to killing was different, with 35B SME15 being the least susceptible. Published studies report how the characteristic of the capsule of S. pneumoniae strains can affect the site and quantity of complement deposition (19, 40, 42), and this could account for the different responses to in vitro killing obtained here. Furthermore, a variable concentration of capsular polysaccharide-specific antibodies is required to obtain similar OPKA levels, suggesting that these antibodies could also have different functional properties (24, 25). In light of this, we cannot exclude the possibility that the different OPKA results observed for the three pilus-positive strains could be due to different functionalities of the antibodies directed against the three variants constituting the fusion protein.
In addition, the observation that the RrgB321-mediated killing never reached 100%, in contrast to what was observed with antisera against conjugated polysaccharides, is supported by the recent report of pneumococcal pilus biphasic expression (9). In fact, two bacterial phenotypes that express or do not express the pilus are present in PI-1-positive strains. Since the opsonic activity of RrgB321 antiserum is directed only toward the pilus-positive subpopulation, this eventually results in a lower opsonophagocytic efficacy. Animal experiments are ongoing to evaluate the influence of pilus expression on the protective efficacy exerted by RrgB321 immunization.
The OPKA data were also analyzed to define the antibody functionality as an opsonophagocytic titer, meaning the minimum serum dilution needed to elicit 50% bacterial killing. The values obtained with RrgB321 rabbit antisera against pneumococcal strains with variants I and II were 18,528 and 26,316, respectively. This is the first report of pneumococcal protein antigens being able to elicit opsonophagocytic killing comparable to that obtained with conjugated polysaccharides. This result could be due to the pilus protruding beyond the capsule, allowing greater accessibility to the antibodies in comparison to other protein antigens. In addition, the number of ligands for the antibodies directed against the RrgB epitopes is increased by the presence of several RrgB subunits covalently linked to form the pilus shaft, thus enhancing the opsonophagocytic killing.
To confirm that the killing observed was not simply a function of the OPKA established in our laboratory, we sent rabbit sera obtained from immunization with all the RrgB constructs (RrgB321, RrgB213, RrgB312, RrgB231, and RrgB123) to our collaborators at the WHO Pneumococcal Reference Laboratory (Institute of Child Health, London, United Kingdom), who have an established an OPKA used for the analysis of human-derived sera. Their assay has recently been shown to be comparable to other OPKAs in an interlaboratory study (6, 35).
Sera from rabbits immunized with all constructs showed relevant functional activity with a serotype 6B strain and lower activity with a serotype 14 and another serotype 6B strain but no opsonic killing of strains of serotypes 18C, 19A, and 23F, which were not piliated. Unexpectedly, no functional activity was observed against a serotype 4 piliated strain. Although sequence analysis confirmed that the RrgB sequence in this strain is >99% identical to the TIGR4 counterpart, by FACS we found that this strain expresses very small amounts of pilus in vitro (our unpublished data). Further experiments are ongoing to investigate the mechanism of pilus expression in vivo.
In summary, the OPKA has the potential to evaluate the efficacy of pilus protein-based vaccines and in the future could be used as a correlate of protection. Preliminary data generated with sera from healthy adults indicate that all tested subjects displayed a measurable immune response to the pilus RrgB backbone protein (our unpublished data). However, to assess the real potential of this vaccine candidate, it would be crucial to perform a seroepidemiological study on an age-stratified human sera collection to verify if a negative correlation exists between the anti-RrgB pilus backbone antibody content (and subclasses) and the susceptibility to invasive disease. While these studies are ongoing, attempts to evaluate a correlation between OPKA titers and either immune response in the animals (antibody content) or protection in mouse models of infection have been performed. Data collected so far indicate that the measured OPKA titers do not correlate with the total IgG antibody responses or with protection in vivo.
In conclusion, the data reported here highlight the capacity of the RrgB321 chimera to elicit in vivo protection against piliated pneumococcal strains and to induce functional specific antibodies against the pneumococcal pilus, thus supporting the validity of this candidate as a potential antigen for the generation of a multicomponent protein-based vaccine against S. pneumoniae.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gianni Pozzi, Lesley McGee, Stanley Falkow, Moon Nahm, and Birgitta Henriques Normark for providing the pneumococcal strains used in this study (TIGR4, 6B Finland12, 6B SPEC, and 35B SME15, respectively) and Richard Moxon and Steven Black for support and critical revision of this study. We acknowledge Marco Tortoli, Elena Amantini and Luigi Manganelli for the animal treatments.
We declare a potential conflict of financial interest as employees and/or consultants of Novartis Vaccines and Diagnostics. We have no other competing financial interests.
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Aguiar SI, Serrano I, Pinto FR, Melo-Cristino J, Ramirez M. 2008. The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol. 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alloing G, Martin B, Granadel C, Claverys JP. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29: 75–83 [DOI] [PubMed] [Google Scholar]
- 3. Basset A, et al. 2007. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J. Clin. Microbiol. 45: 1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Black S, et al. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr. Infect. Dis. J. 19: 187–195 [DOI] [PubMed] [Google Scholar]
- 4a. Bratcher PE, Kim KH, Kang JH, Hong JY, Nahm MH. 2010. Identification of natural pneumococcal isolates expressing serotype 6D by genetic, biochemical and serological characterization. Microbiology 156: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown EJ, Hosea SW, Frank MM. 1983. The role of antibody and complement in the reticuloendothelial clearance of pneumococci from the bloodstream. Rev. Infect. Dis. 5 Suppl. 4: S797–S805 [DOI] [PubMed] [Google Scholar]
- 6. Burton RL, Nahm MH. 2006. Development and validation of a fourfold multiplexed opsonization assay (MOPA4) for pneumococcal antibodies. Clin. Vaccine Immunol. 13: 1004–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reference deleted.
- 8. Dagan R. 2009. Serotype replacement in perspective. Vaccine 27(Suppl. 3): C22–C24 [DOI] [PubMed] [Google Scholar]
- 9. De Angelis G, et al. 2011. The Streptococcus pneumoniae pilus 1 displays a biphasic expression pattern. PLoS One 6: e21269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feavers I, Knezevic I, Powell M, Griffiths E. 2009. Challenges in the evaluation and licensing of new pneumococcal vaccines, 7–8 July 2008, Ottawa, Canada. Vaccine 27: 3681–3688 [DOI] [PubMed] [Google Scholar]
- 11. Flasche S, et al. 2011. Effect of pneumococcal conjugate vaccination on serotype-specific carriage and invasive disease in England: a cross-sectional study. PLoS. Med. 8: e1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gentile MA, et al. 2011. Structural and functional characterization of the Streptococcus pneumoniae RrgB pilus backbone D1 domain. J. Biol. Chem. 286: 14588–14597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gianfaldoni C, et al. 2007. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75: 1059–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giefing C, et al. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205: 117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Godfroid F, Hermand P, Verlant V, Denoel P, Poolman JT. 2011. Preclinical evaluation of the Pht proteins as potential cross-protective pneumococcal vaccine antigens. Infect. Immun. 79: 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hausdorff WP, Bryant J, Paradiso PR, Siber GR. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30: 100–121 [DOI] [PubMed] [Google Scholar]
- 17. Hilleringmann M, et al. 2008. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog 4: e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hilleringmann M, et al. 2009. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28: 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hostetter MK. 1986. Serotypic variations among virulent pneumococci in deposition and degradation of covalently bound C3b: implications for phagocytosis and antibody production. J. Infect. Dis. 153: 682–693 [DOI] [PubMed] [Google Scholar]
- 20. Jodar L, et al. 2003. Serological criteria for evaluation and licensure of new pneumococcal conjugate vaccine formulations for use in infants. Vaccine 21: 3265–3272 [DOI] [PubMed] [Google Scholar]
- 21. Kim KH, Yu J, Nahm MH. 2003. Efficiency of a pneumococcal opsonophagocytic killing assay improved by multiplexing and by coloring colonies. Clin. Diagn. Lab Immunol. 10: 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lexau CA, et al. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294: 2043–2051 [DOI] [PubMed] [Google Scholar]
- 23. McDaniel LS, Sheffield JS, Delucchi P, Briles DE. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59: 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Melin M, et al. 2009. Streptococcus pneumoniae capsular serotype 19F is more resistant to C3 deposition and less sensitive to opsonophagocytosis than serotype 6B. Infect. Immun. 77: 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melin M, et al. 2010. Serotype-related variation in susceptibility to complement deposition and opsonophagocytosis among clinical isolates of Streptococcus pneumoniae. Infect. Immun. 78: 5252–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moschioni M, et al. 2010. Prevalence of pilus encoding islets among acute otitis media Streptococcus pneumoniae isolates from Israel. Clin. Microbiol. Infect. 16: 1501–1504 [DOI] [PubMed] [Google Scholar]
- 27. Moschioni M, et al. 2008. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197: 888–896 [DOI] [PubMed] [Google Scholar]
- 28. Moschioni M, et al. 2010. The two variants of Streptococcus pneumoniae pilus 1 RrgA adhesin retain the same function and elicit cross-protection in vivo. Infect. Immun. 78: 5033–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nunes MC, Madhi SA. 2011. Review on the immunogenicity and safety of PCV-13 in infants and toddlers. Expert Rev. Vaccines 10: 951–980 [DOI] [PubMed] [Google Scholar]
- 30. Ogunniyi AD, Grabowicz M, Briles DE, Cook J, Paton JC. 2007. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect. Immun. 75: 350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paradiso P. 2009. Essential criteria for evaluation of pneumococcal conjugate vaccine candidates. Vaccine 27(Suppl. 3): C15–C18 [DOI] [PubMed] [Google Scholar]
- 32. Park IH, et al. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45: 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paton JC, Lock RA, Hansman DJ. 1983. Effect of immunization with pneumolysin on survival time of mice challenged with Streptococcus pneumoniae. Infect. Immun. 40: 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prymula R, Schuerman L. 2009. 10-valent pneumococcal nontypeable Haemophilus influenzae PD conjugate vaccine: Synflorix. Expert Rev. Vaccines 8: 1479–1500 [DOI] [PubMed] [Google Scholar]
- 35. Rose CE. 2011. Multilaboratory comparison of Streptococcus pneumoniae opsonophagocytic killing assays and their level of agreement for the determination of functional antibody activity in human reference sera. Clin. Vaccine Immunol. 18: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosenow C, et al. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25: 819–829 [DOI] [PubMed] [Google Scholar]
- 37. Spraggon G, et al. 2010. Supramolecular organization of the repetitive backbone unit of the Streptococcus pneumoniae pilus. PLoS One 5: e10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinberger DM, Malley R, Lipsitch M.Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011. doi: 10.1016/S0140-6736(10)62225–8. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 39. Whitney CG, et al. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348: 1737–1746 [DOI] [PubMed] [Google Scholar]
- 40. Winkelstein JA, Bocchini JA, Jr., Schiffman G. 1976. The role of the capsular polysaccharide in the activation of the alternative pathway by the pneumococcus. J. Immunol. 116: 367–370 [PubMed] [Google Scholar]
- 41. Wizemann TM, et al. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae. Infection. Infect Immun. 69: 1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yuste J, et al. 2008. Impaired opsonization with C3b and phagocytosis of Streptococcus pneumoniae in sera from subjects with defects in the classical complement pathway. Infect. Immun. 76: 3761–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.