Abstract
Enterohemorrhagic Escherichia coli O157:H7 (EHEC O157) is an important cause of food and waterborne illness in the developed countries. Cattle are a reservoir host of EHEC O157 and a major source of human exposure through contaminated meat products. Shiga toxins (Stxs) are an important pathogenicity trait of EHEC O157. The insertion sites of the Stx-encoding bacteriophages differentiate EHEC O157 isolates into genogroups commonly isolated from cattle but rarely from sick humans (bovine-biased genotypes [BBG]) and those commonly isolated from both cattle and human patients (clinical genotypes [CG]). Since BBG and CG share the cardinal virulence factors of EHEC O157 and are carried by cattle at similar prevalences, the infrequent occurrence of BBG among human disease isolates suggests that they may be less virulent than CG. We compared the virulence potentials of human and bovine isolates of CG and BBG in newborn conventional pig and weaned Dutch Belted rabbit models. CG-challenged piglets experienced severe disease accompanied by early and high mortality compared to BBG-challenged piglets. Similarly, CG-challenged rabbits were likely to develop lesions in kidney and intestine compared with the BBG-challenged rabbits. The CG strains used in this study carried stx2 and produced significantly higher amounts of Stx, whereas the BBG strains carried the stx2c gene variant only. These results suggest that BBG are less virulent than CG and that this difference in virulence potential is associated with the Stx2 subtype(s) carried and/or the amount of Stx produced.
INTRODUCTION
Enterohemorrhagic Escherichia coli O157:H7 (EHEC O157) is a major cause of food- and waterborne illnesses characterized by bloody diarrhea, hemorrhagic colitis (HC), and life-threatening hemolytic uremic syndrome (HUS) in developed nations across the globe (31, 42, 49). The lack of specific treatment leads to 2 to 5% mortality among HUS patients. The annual incidence of domestically acquired food-borne EHEC O157 infection is estimated to be 63,153 cases in the United States (59). The CDC surveillance data show that the number of EHEC O157 outbreaks increased recently from 27 in 2006 to 39 in 2007 (9, 10). In 2008, the CDC reported 1.12 cases of food-borne EHEC O157 infections per 100,000 population in the United States (8). The infectious dose of EHEC O157 is estimated to be very low (10 to 100 bacteria) (62), and transmission occurs primarily via contaminated food or water or direct contact with infected animals (21, 23, 29, 53). Cattle are considered a major reservoir of EHEC O157 due to the frequent association of EHEC O157 outbreaks with the consumption of contaminated beef and dairy products, as supported by molecular epidemiological evidence of identical strain types (4, 6). Cattle colonized with EHEC O157 remain asymptomatic while shedding up to 105 CFU/g of feces typically for periods of days to weeks (2). EHEC O157 is defined as an adulterant when identified in foods, and therefore, in addition to its public health significance, this organism is responsible for significant economic losses to the food industry due to mandatory recalls of contaminated meat and meat products.
Nearly all EHEC O157 isolates harbor two cardinal virulence factors: the production of one or more antigenically distinct Shiga toxins (Stxs) and the presence of a chromosomal pathogenicity island referred to as the locus of enterocyte effacement (LEE) (38, 49). Stxs are encoded by late genes of one or more lambdoid bacteriophages that are integrated into the EHEC O157 bacterial chromosome at specific sites (12, 25, 35, 36, 47, 51, 54). Shiga toxins are important virulence factors associated with the systemic effects of EHEC O157 infection, including HC and HUS (32, 64). The LEE encodes a type III secretion system (T3SS), including an effector molecule Tir (translocated intimin receptor), and intimin, an EHEC O157 surface-expressed adhesin that interacts with Tir (50). The LEE is required for EHEC O157 colonization and formation of attaching and effacing (A/E) lesions on intestinal epithelium (16, 45).
Diverse genotyping systems for EHEC O157 strains (5, 33, 34, 41, 47, 60, 72, 74, 76) emphasize the role of bacteriophage, including Stx-encoding bacteriophages, in generating genetic diversity in EHEC O157. Several of these methods have identified genotypes that occur at different frequencies in cattle and human hosts (5, 33, 41, 74). Recent studies comparing some of these genotyping methods suggest that they result in generally concordant classifications (37, 70). Using Stx-encoding bacteriophage insertion (SBI) sites as a marker for strain differentiation, EHEC O157 isolates can be classified into two genogroups: bovine-biased genotypes (BBG, predominantly SBI genotypes 5 and 6, found primarily in cattle and rarely isolated from sick humans) and clinical genotypes (CG, predominantly SBI genotypes 1 and 3, found in both cattle and humans) (5, 60, 71). Both BBG and CG carry and express the cardinal virulence factors of EHEC O157 (5). Despite the similar prevalences of both genogroups (CG and BBG) in cattle, indicating approximately equal levels of human exposure, the relatively rare occurrence of BBG as a cause of human disease suggests that BBG may be less virulent than CG. Therefore, we hypothesized that BBG strains are less pathogenic than CG strains. To test this hypothesis, we evaluated the virulence potentials of human and bovine strains belonging to BBG and CG, using the newborn conventional pig and Dutch Belted rabbit models. We show that strains of EHEC O157 belonging to BBG are less virulent and cause less severe disease in both animal models. This difference in virulence potential is associated with the presence of different Stx2 subtype(s) and/or the quantity of Stx produced by BBG strains.
MATERIALS AND METHODS
Bacterial strains for piglet challenge.
Twenty EHEC O157 strains (Table 1) were randomly selected from the bank of SBI genotypes of EHEC O157 at Washington State University. EHEC O157 strain Sakai (CG-3, kindly provided by Thomas S. Whittam, NFSTC-MSU) isolated from an outbreak in Japan served as the positive control. To generate spontaneous nalidixic acid-resistant variants (Nalr), each EHEC O157 strain was inoculated into brain heart infusion (BHI) broth and incubated at 37°C overnight (∼16 h), and 1 ml was plated on sorbitol MacConkey agar plates (Hardy Diagnostics, Santa Maria, CA) supplemented with nalidixic acid (30 μg/ml) (SMAC-N). For animal challenge studies, Nalr strains were inoculated in 200 ml of Trypticase soy broth (TSB) and incubated at 37°C overnight with shaking at 200 rpm. Aliquots of these cultures were then stored in glycerol (10% [vol/vol]) stocks at −80°C until use. Before challenge, bacteria in the aliquots were pelleted by centrifugation, the supernatant was discarded, and the bacterial pellet was resuspended in a sterile 10-ml portion of TSB.
Table 1.
EHEC O157 strains used in this study and their origin
| Strain | SBI genotypea | Yearb | Source of strainc | Cohort(s)d |
|---|---|---|---|---|
| CDC EDL 933 | CG–3 | 1982 | Raw hamburger meat implicated in hemorrhagic colitis outbreak | R1, R2, and R3 |
| E2325 | CG–1 | 1995 | Bovine fecal, WA, USA | R1 |
| E3046 | CG-3 | 1995 | Bovine fecal, WA, USA | R3 |
| E5252 | CG-3 | 1998 | Bovine fecal, OR, USA | R3 |
| E5880 | BBG-5 | 1999 | Water, WA, USA | R2 |
| E6996 | BBG-6 | 2000 | Bovine fecal, WA, USA | R2 |
| E12000 (Sakai) | CG-3 | 1996 | Sakai City, Osaka Pref. Japan | NA |
| E12053 | BBG-5 | 1993 | Bovine fecal, WA, USA | P3 |
| E12056 | BBG-5 | 1999 | Water, WA, USA | P1 |
| E12057 | BBG-6 | 1999 | Bovine fecal, WA, USA | P2 |
| E12058 | BBG-6 | 2000 | Bovine fecal, WA, USA | P1 |
| E12059 | BBG-5 | 2001 | Bovine fecal, WA,USA | P2 |
| E12061 | CG-1 | 2004 | Human clinical, WADOH, WA, USA | P2 |
| E12062 | CG-1 | 2004 | Human clinical, WADOH, WA, USA | P5 |
| E12063 | CG-3 | 2004 | Human clinical, WADOH, WA, USA | P2 |
| E12064 | CG-1 | 2005 | Human clinical, WADOH, WA, USA | P1 |
| E12065 | CG-3 | 2005 | Human clinical, WADOH, WA, USA | P3 |
| E12066 | CG-1 | 2006 | Human clinical, WADOH, WA, USA | P3 |
| E12067 | CG-3 | 2006 | Human clinical, WADOH, WA, USA | P1 |
| E12068 | BBG-5 | 1996 | Bovine fecal, TX, USA | P5 |
| E12149 | BBG-6 | 1995 | Bovine fecal, WA, USA | P4 |
| E12153 | BBG-6 | 2001 | Bovine fecal, WA, USA | P4 |
| E12154 | CG-1 | 2004 | Human clinical, WADOH, WA, USA | P5 |
| E12155 | CG-3 | 2006 | Human clinical, WADOH, WA, USA | P5 |
| E12156 | BBG-5 | 1994 | Bovine fecal, WA, USA | P5 |
| E12157 | BBG-6 | 1995 | Bovine fecal, OR, USA | P3 |
| E12377 | CG-3 | 2004 | Human clinical, WADOH, WA, USA | P5 |
CG, clinical genotypes; BBG, bovine-biased genotypes.
Year of the strain isolation or time deposited in WSU EHEC bank.
WADOH, Washington State Department of Health; Pref., Prefecture.
P, litter cohorts of piglets; R, experimental cohorts of rabbits; NA, not assigned.
Piglet husbandry and experimental challenges.
All piglet challenge experiments were conducted according to the protocols approved by the WSU Institutional Animal Care and Use Committee (IACUC). Piglet challenges were conducted as described previously (14) with some modifications. Piglets were obtained from the WSU swine center at 4 h after birth, housed in separate cages to avoid direct contact, and confirmed as EHEC O157 free by testing fecal swabs obtained prior to challenge by using immunomagnetic separation (Dynabeads anti-E. coli O157; Invitrogen Dynal AS, Oslo, Norway) and plated on SMAC-CT (SMAC agar supplemented with cefixime at 50 ng/ml and potassium tellurite at 2.5 μg/ml). Piglets were orally treated with nalidixic acid (25 mg) followed by sodium bicarbonate (10 ml, 10% [wt/vol]) 1 h later. Immediately after sodium bicarbonate treatment, piglets were orally challenged with nalidixic acid-resistant EHEC O157 (∼1010 CFU, Nalr, resuspended in sterile TSB) (Table 1). Piglets were fed Enfamil infant milk formula (Mead Johnson & Company, Evansville, IN) three times daily beginning at 210 ml/day and gradually increasing to 450 ml/day on days 6 and 7 postinfection (p.i.). Nalidixic acid (25 mg every 8 h) was orally administered to each piglet throughout the experiment. To avoid confounding by potential genetic factors, approximately equal numbers for CG and BBG strain challenges were included in each litter of 4 to 6 piglets (piglet cohort) with a random assignment of piglets to the challenge strain. Piglets were observed daily for EHEC O157-associated clinical signs (Table 2) for up to 7 days p.i. Piglets were treated with oral fluids and electrolytes (Enterolyte H.E., oral powder; Pfizer) and flunixin meglumine (Banamine, 1 mg/lb of body weight [BW] intramuscularly [i.m.] every 24 h [q24 h]; Schering-Plough) when they showed signs of diarrhea and moderate central nervous system (CNS) disease (shivering, tremors, or mild seizures). Piglets exhibiting severe clinical signs (severe seizures, lateral recumbency, or paresis) were humanely euthanized. At 7 days p.i., all surviving piglets were humanely euthanized, complete necropsies were performed, and internal organs were collected and processed for bacteriological and histopathological examination. All clinical and pathological assessments were conducted by veterinarians blind to the challenge genotype.
Table 2.
Scoring system and categories for clinical signs applied to neonatal piglets
| Score | Clinical sign for indicated categorya |
||||
|---|---|---|---|---|---|
| Diarrhea (consistency, frequency, straining) (0–3) | CNS signs (0–5) | Vomiting (0–1) | Loss of appetite (0–2) | Vocalization (0–1) | |
| 0 | Normally formed feces | None | None | None | Normal |
| 1 | Loose (unformed), but no straining or blood | Lethargy | Vomiting | Partial | Abnormal |
| 2 | Thick liquid, mild straining and staining of hind limbs with feces | Splayleg, hind-limb weakness, thinner appearance | Complete | ||
| 3 | Watery and/or bloody, severe straining and staining of hind limbs with feces | Shivering, tremors | |||
| 4 | Seizures, convulsions | ||||
| 5 | Grand mal seizures or lateral recumbency or paralysis of legs | ||||
Numbers in parentheses are ranges of scores used for scoring criteria.
Bacteriological culture for piglet challenge.
Intestinal tissue samples (2 to 4 cm long) consisting of ileum, cecum, spiral colon, distal colon, and rectoanal junction (RAJ) were collected separately in sterile tubes. The segments of intestine were cut open and washed gently in phosphate-buffered saline (PBS) to remove grossly visible intestinal contents. The size of each segment was measured in cm2. The samples were then homogenized in 10 ml of PBS by using a stomacher (Stomacher 80 laboratory blender; Seward Laboratory Systems Inc., Bohemia, NY) at a normal setting for 5 min. The homogenized contents were filtered through a sterile gauze and used to quantify the bacterial load. EHEC O157 in fecal samples and intestinal tissues of each piglet was detected and enumerated by decimal dilution plating on SMAC-N agar plates by using a spiral plater (Whitley automated spiral plater; Don Whitley Scientific Ltd., Shipley, United Kingdom). Ten non-sorbitol-fermenting colonies from each specimen were tested for lactose fermentation (MacConkey agar; Becton, Dickinson and Company, Sparks, MD) and for beta-glucuronidase activity (EC-MUG agar; Hardy Diagnostics, Santa Maria, CA). Presumptive EHEC O157 colonies (sorbitol negative, lactose positive, and beta-glucuronidase negative) were confirmed by an O157 latex agglutination test (E. coli PRO O157; Hardy Diagnostics). One confirmed, the EHEC O157 colonies recovered from each sample were stored (18% buffered glycerol in BHI, −80°C) for subsequent comparisons with their respective challenge strains by SBI genotyping (71) and by pulsed-field gel electrophoresis (PFGE) following XbaI restriction digestion (7, 13). A method described by Tenover et al. (66) was used to identify the PFGE profiles of reisolated strains that were different from the challenge strain.
Histopathological examination of piglet tissues.
Duodenum, jejunum, ileum, cecum, spiral colon, distal colon, RAJ, kidney, and brain collected at necropsy were immediately fixed in 10% neutral buffered formalin and subsequently paraffin embedded, sectioned, and stained with hematoxylin and eosin for histopathological examination. Brain tissue sections were also stained with Luxol-fast blue stain (69). Histopathological scoring was based on observed bacterial attachment (score range, 0 to 3), necrosis (score range, 0 to 3), inflammation (score range, 0 to 3), vasculitis (score range, 0 or 1), and the presence or absence of fibrin (score, 0 or 1). Scoring was performed independently by two ACVP board-certified pathologists (K.A.P. and K.K.L., intestine, kidney, and brain) and by S.S. (intestine), all blinded to the identity of the challenge strain.
Rabbit husbandry and experimental challenges.
All rabbit challenge experiments were approved by the IACUC at the Massachusetts Institute of Technology. Three challenge experiments were performed using weaned 7- to 8-week-old Dutch Belted rabbits. Fecal samples collected from rabbits before inoculation were cultured in SMAC, and bacterial DNA extracted from cultures was subsequently tested by PCR for eae, stx1, and stx2 (18, 73). Each challenge experiment included five rabbits for positive controls inoculated with EHEC O157 strain EDL933 (CG-3) and four or five rabbits for negative controls inoculated with PBS, along with six rabbits each inoculated with the CG or BBG challenge strains of EHEC O157. Five challenge strains were selected for the rabbit infection studies (Table 1). In each experiment, rabbits were fasted overnight and sedated prior to blood collection and orogastric intubation. Experimental and control rabbits were inoculated with 10 ml of 10% sterile sodium bicarbonate to neutralize gastric acidity. Experimental rabbits received 1 × 109 to 2 × 109 CFU of EHEC O157 resuspended in sterile PBS, and control rabbits received sterile PBS (vehicle only). Food and water were provided ad libitum following inoculation. Rabbits were monitored for clinical signs including lethargy, abnormal stools, decreased appetite, and dehydration. The body weight (BW) of most experimentally infected rabbits in all three experiments was recorded daily from the day of inoculation (day 0) to the day of euthanasia (day 6 p.i. for experiment 1 and day 7 p.i. for experiments 2 and 3). BW was not recorded for one of the groups (E2325) on day 2 p.i. BW was also recorded daily for sham-inoculated rabbits in all three experiments, but data were not available for 3 days (days 1 to 3 p.i.) in experiment 1 and for 1 day (day 4 p.i.) in experiment 3. Fecal samples from inoculated rabbits were collected on days 1 and 3 p.i., 10-fold serial dilutions of each fecal sample were cultured overnight on SMAC agar, and bacterial colonies on culture-positive plates were counted to estimate the number of CFU per gram of feces. Bacterial colonies recorded as sorbitol positive were not included in the count. Rabbits exhibiting severe clinical signs were euthanized before the end of the study (day 4 p.i.). Prior to euthanasia, blood samples were collected for clinical pathological evaluation. Complete necropsies were performed on all the rabbits, and tissue samples from various organs including cecum and kidneys were collected for histopathological evaluation.
Histopathological examination of rabbit tissues.
Tissues were fixed in formalin, embedded in paraffin, sectioned at 4 μm, and stained with hematoxylin and eosin. Kidney sections were also stained with Carstairs' to detect fibrin deposition. Kidney and intestinal (cecal) sections were evaluated by a board-certified veterinary pathologist (S.M.) blinded to the identity of the challenge strain. Scoring criteria for histopathological evaluation included renal glomerular (capillary thickening, luminal constriction, fibrin thrombi, white blood cell (WBC) infiltration, mesangial deposits, changes in Bowman's capsule and space) and vascular (intimal swelling, mural degeneration, perivascular edema, and red blood cell fragmentation) criteria as well as assessment of intestinal vasculopathy (20). One rabbit exhibited a lesion unrelated to EHEC O157 infection (multifocal chronic-active tubulo-interstitial nephritis) and was excluded from the study.
CRP analysis.
The concentration of C-reactive protein (CRP) was measured in serum samples from rabbits in experiment 3 by using a commercially available rabbit CRP enzyme-linked immunosorbent assay (ELISA) (Immunology Consultants Laboratory, Inc., Newberg, OR) following the manufacturer's directions. Rabbit serum samples were diluted 1:400 in assay diluent buffer for testing. Samples were adjusted to a 1:10,000 dilution and retested if they did not fall within the working range of the assay's standard curve. The normal range of rabbit CRP is 0 to 31 μg/ml (22).
stx2 gene typing.
The stx2 gene variants present in the challenge strains were determined using PCR-restriction fragment length polymorphism (RFLP) as described previously (15). In this method, restriction patterns generated by HaeIII differentiate stx2 from stx2c, while restriction patterns generated by PvuII differentiate stx2 and stx2c from other stx2 variant types.
Shiga toxin quantitation.
Shiga toxin production by 20 Nalr EHEC O157 strains used for piglet challenge study and their nalidixic acid-susceptible wild-type (WT-Nals) parents was assessed using ELISA (Premier EHEC test kit; Meridian Bioscience, Cincinnati, OH). Briefly, for each strain, a single colony from blood agar plates was inoculated into 10 ml of TSB incubated at 37°C overnight with shaking at 200 rpm. Approximately 500 μl of overnight culture was transferred into 10 ml of fresh TSB. This subculture was processed for bacterial counts, and three aliquots (1 ml each) were transferred into three wells of 96-well culture plates followed by the addition of (i) carbadox (0.5 μg/ml; Sigma-Aldrich, St. Louis, MO), (ii) nalidixic acid (30 μg/ml), or (iii) no inducing agent. The plates were incubated overnight at 37°C with shaking at 250 rpm. Overnight cultures were diluted 1:100 in sterile TSB immediately followed by a 1:2 dilution in sample diluent provided in the ELISA kit. Aliquots (100 μl) of diluted samples were tested by ELISA according to the manufacturer's protocol. The number of bacteria in the initial subculture used for induction by nalidixic acid or carbadox or left uninduced was ∼107 CFU/ml (range, 1.6 × 107 CFU/ml to 7.8 × 107 CFU/ml). There was no significant difference in the bacterial counts (mean ± standard error of the mean [SEM] log10 CFU/ml) of BBG (7.54 ± 0.06) compared with CG (7.65 ± 0.03) at the time of induction.
Statistical analysis.
Statistical analyses for the piglet challenge study were performed using NCSS 2007 version 07.1.19 (26), and the outcomes of challenges with different genogroups were compared using the following statistical tests. The clinical scores for each of the five clinical scoring categories (diarrhea, CNS disease, appetite, vomiting, and abnormal vocalization) and scores for histopathological lesions in the intestinal tract (a total score of ileum, duodenum, jejunum, cecum, spiral colon, distal colon, and RAJ) were analyzed using Kruskal-Wallis one-way analysis of variation (ANOVA) followed by Dunn's multiple comparison test. Survival data were compared using Kaplan-Meier survival analysis and the log rank test. Bacterial loads recovered from five segments of intestine (ileum, cecum, spiral colon, distal colon, and RAJ) of piglets were compared using a two-way generalized linear model (GLM). The association of the development of brain lesions in the piglets with the development of CNS disease or with the genogroup of the EHEC O157 strain used to challenge the piglets was assessed using Fisher's exact test. In the rabbit challenge study, statistical analyses were performed using GraphPad Prism version 5.03 for Windows (GraphPad Software, San Diego, CA). One-way ANOVA with Tukey's multiple comparison test was used to analyze the percentage change in BW data (expressed as mean ± standard deviation [SD]) and clinical pathological parameters. Two-tailed t tests were also used to analyze clinical pathological data. The Mann-Whitney two-tailed test was used to compare the lesion scores of rabbits infected with specific CG or BBG strains against EDL933- or sham-inoculated rabbits. The Kruskal-Wallis test with Dunn's multiple comparison test was also used to analyze the lesion scores of rabbits infected with specific CG and BBG strains against sham-inoculated and EDL933-infected rabbits. The combined lesion scores of all the rabbits infected with CG strains (E2325, E3046, and E5252), of all the rabbits infected with BBG strains (E5880 and E6996), and of EDL933- and sham-inoculated rabbits were also compared by Mann-Whitney two-tailed test and Kruskal-Wallis test with Dunn's multiple comparison test. Statistical analyses were performed by using the combined scores of all EDL933-infected rabbits (positive controls) and the combined scores of all sham-inoculated rabbits (negative controls). The association of the development of serosal hemorrhages near the cecocolic junction with the challenge group of the rabbit was assessed using Fisher's exact test. The association between stx gene typing and genogroup of the challenge strain was assessed using chi-square test. Stx ELISA optical density at 450 nm (OD450) scores were analyzed using a three-way (growth condition, nalidixic acid susceptibility, and genogroup) GLM with a Tukey-Kramer multiple comparison test. Multiple comparisons were done only when the overall statistical model was significant. Each statistical test was conducted using a significance level of P ≤ 0.05.
RESULTS
Piglet challenge.
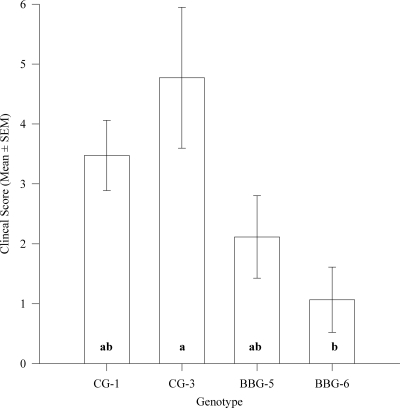
The clinical signs of all the piglets were scored for disease severity by using a standard scoring system (Table 2). Piglets challenged with the clinical genotype strains (CG-1 and CG-3) exhibited significantly high clinical scores compared with the piglets challenged with bovine-biased genotype strains (BBG-5 and BBG-6) (P = 0.006) (Table 3). Piglets challenged with CG strains and BBG strains had clinical scores that did not differ significantly from the positive-control Sakai strain- and sham-inoculated piglets, respectively. In individual genotype comparisons, CG-3-challenged animals had significantly higher clinical scores than BBG-6 inoculated piglets (P = 0.033) (Fig. 1), whereas CG-1- and BBG-5-challenged piglets showed intermediate clinical scores that were not significantly different from those seen with CG-3 or BBG-6. The clinical scores of the CG-3-challenged piglets did not differ significantly from piglets challenged with the positive-control Sakai strain. Diarrhea, CNS symptoms, appetite, vomiting, and abnormal vocalizations were scored separately. The results revealed that CNS disease scores were significantly high for piglets challenged with CG strains compared with the piglets challenged with BBG strains (P = 0.005) (Table 3). However, no other scoring category differed significantly between CG- and BBG-challenged piglets. The clinical scores of cohorts of piglets (litters) did not differ significantly, suggesting that the cohorts of the piglet did not affect the outcome of the challenge experiments.
Table 3.
Disease outcomes in negative (sham-inoculated)- and positive (Sakai-inoculated)-control group and experimental (CG or BBG)-group piglets
| Disease outcome | Value (mean ± SEM) for piglet groupa |
|||
|---|---|---|---|---|
| Negative control (n = 4) | Positive control (n = 4) | CG (n = 10) | BBG (n = 10) | |
| Clinical scorec | 0.98 ± 0.34A | 3.96 ± 0.26AB | 4.12 ± 0.66B | 1.5 ± 0.45A |
| CNS disease scorec | 0.07 ± 0.07A | 2.35 ± 0.28B | 2.39 ± 0.46B | 0.69 ± 0.25A |
| Survival time (days) | 7.00 ± 0.00A | 4.25 ± 0.63AB | 3.70 ± 0.06B | 6.20 ± 0.55A |
| Intestinal bacterial load (log10 CFU)b | 1.13 ± 0.47C | 5.85 ± 0.17B | 5.09 ± 0.19B | 4.30 ± 0.18A |
| Histopathological score for intestine | 1.00 ± 0.39 | 2.88 ± 0.79 | 2.01 ± 0.49 | 1.08 ± 0.37 |
Groups not sharing specific letters differed significantly (P ≤ 0.05).
Average intestinal bacterial load (log10 CFU) from five segments of the intestine (ileum, cecum, spiral colon, distal colon, and RAJ).
Clinical score is the daily mean of the total clinical score based on all five categories in Table 2, whereas CNS disease score is the daily mean score of the CNS category only.
Fig 1.
Clinical scores of piglets challenged with EHEC strains belonging to different genotypes. Five animals were challenged with each genotype (CG-1, CG-3, BBG-5, and BBG-6). Groups not sharing the specific letters shown in bars differed significantly (P = 0.023, Kruskal-Wallis one-way ANOVA with Dunn's multiple comparison).
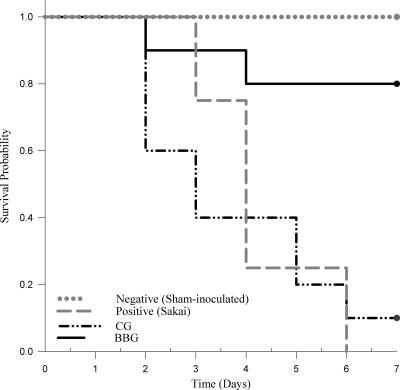
Challenge with CG strains resulted in earlier and higher piglet mortality than challenge with BBG strains (P = 0.002) (Fig. 2 and Table 3). All the sham-inoculated piglets survived until the end of the experiment. CG-challenged piglets showed mortality similar to that of the piglets challenged with positive-control strain Sakai. Specifically, mortality shown by the CG-3-challenged piglets did not differ significantly from that of piglets challenged with positive-control strain Sakai. Out of 10 piglets challenged with CG strains, only 1 piglet (challenged with strains E12064) survived until the end of the experiment. In contrast, out of 10 piglets challenged with BBG strains, only 2 piglets (challenged with strains E12053 and E12156) died, on days 2 and 4 p.i., respectively.
Fig 2.
Kaplan-Meier survival curves for piglet groups challenged with different EHEC O157 genogroups. CG strains differ significantly from BBG strains according to the log rank test (P = 0.002).
The mean ± SEM log10 CFU/cm2 of EHEC O157 recovered from intestine (ileum, cecum, spiral colon, distal colon, and RAJ) were significantly high for piglets challenged with CG strains compared with the piglets challenged with BBG strains (P = 0.003) (Table 3). The mean ± SEM log10 CFU/cm2 of EHEC O157 recovered from intestine of piglets challenged with positive-control strain Sakai did not differ significantly from that of CG-challenged piglets in general but was significantly higher than that of the CG-3-challenged group. However, it should be noted that these CFU were determined at different days p.i., since most of the CG-challenged piglets died prior to day 7 p.i., while most of the BBG-challenged piglets were euthanized on day 7 p.i. The mean ± SEM log10 CFU/cm2 of EHEC O157 recovered were 3.86 ± 0.40 (ileum), 4.92 ± 0.32 (cecum), 4.56 ± 0.22 (spiral colon), 5.00 ± 0.26 (distal colon), and 5.14 ± 0.27 (RAJ). Although the number of bacteria recovered from large intestine was high compared with that recovered from small intestine, statistically significant differences were observed only between RAJ and ileum.
Negative-control piglets were housed in separate cages within the same room as EHEC-challenged piglets. EHEC O157 was recovered from 2 out of 4 negative-control piglets, albeit in small numbers, indicating the likely cross-transmission of EHEC O157 between challenged and sham-inoculated animals. Therefore, SBI genotypes and PFGE profiles of challenge strains were compared with the strains recovered from each piglet to determine the level of cross-transmission. Five to 9 isolated colonies per animal were recovered from 20 piglets and tested for cross-transmission. The genotypes of all the isolated colonies (n = 70) recovered from the 10 CG-challenged piglets and 57 out of 65 isolated colonies recovered from the 10 BBG-challenged piglets were identical to the genotype of the original challenge strain used in each piglet. Eight out of 65 isolated colonies for which the genotype did not match with the challenge strain were recovered from two BBG-challenged piglets. Therefore, in all but two cases, only the challenge strain was recovered from each experimental piglet, indicating that cross-transmission was uncommon. Both of the cross-contaminated piglets were challenged with a BBG-6 strain. In the first BBG-6-challenged piglet, 1 out of 6 recovered isolates belonged to a clinical genotype (CG-3), whereas in the second BBG-6-challenged piglet, all 7 isolates recovered belonged to non-BBG-6 genotypes (BBG-17 [described below], 1 isolate; BBG-5, 1 isolate; CG-1, 5 isolates). However, it is important to note that despite the low-level cross-contamination with CG strains, the clinical scores of these two BBG-6-challenged piglets were lower than those of the piglets that were challenged with the cross-contaminating CG strains in the same cohort, suggesting that this transmission had no measurable effect on the clinical outcomes. Three out of a total of 135 recovered isolates belonged to either genotype 9 or genotype 17 (5, 71), neither of which was used in the study as challenge strain, and these were isolated from two piglets. Nevertheless, the PFGE profiles of these recovered isolates were similar to the challenge or the contaminating BBG-5 strain (characterized by the presence of the stx2 gene and stx1-encoding bacteriophage inserted in the yehV gene) and exhibited a Nalr phenotype, confirming their origin. These changes in genotypes from BBG-5 to BBG-9 or BBG-17 suggest a loss of the stx2 gene (BBG-17 strain) and a loss of both the stx2 gene and the stx1-encoding bacteriophage inserted in the yehV gene (BBG-9 strain) either in vivo or during the in vitro isolation process.
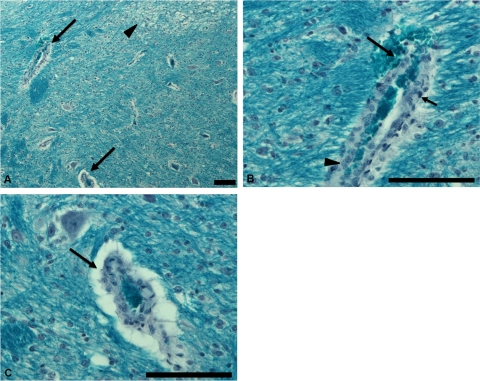
The histopathological lesions in the intestine were characterized by varying degrees of bacterial attachment to the intestinal epithelium, neutrophilic infiltration, and necrosis of intestinal mucosa. In general, CG-challenged piglets showed higher bacterial attachment, increased neutrophilic infiltration, and severe necrosis of intestinal mucosa. As a result, the histopathological scores for intestinal tract in CG- and Sakai-challenged piglets were higher than BBG and sham-inoculated piglets; however, this difference did not reach statistical significance (Table 3). The histopathological scores for intestinal tract in CG-3-challenged piglets did not differ significantly from those for the positive-control Sakai strain-challenged animals. Brain lesions were scored as present or absent (Fig. 3). The number of piglets in which brain lesions were observed by at least one observer were 5/10, 2/10, 2/4, and 0/4 in CG, BBG, and positive- and negative-control groups, respectively. The number of piglets showing brain lesions did not differ significantly between CG- and BBG-challenged piglets. Nevertheless, 7 out of 11 piglets that died or were euthanized following severe CNS signs exhibited brain lesions, while 9 piglets that did not exhibit severe CNS signs or mortality did not develop observable brain lesions (P < 0.003). No kidney lesions were observed for any of the piglets included in this study.
Fig 3.
Tissue from a piglet challenged with the positive-control Sakai strain exhibiting mortality on day 6 p.i. (A) Section of the brain stained with Luxol-fast blue stain showing affected blood vessel (arrows, magnified in panels B and C) with vacuolation of adjacent tissues (arrowhead) caused by focal myelin degeneration. (B) Affected blood vessel in brain showing microhemorrhage (long arrow), pyknosis (short arrow), and hyperplasia (arrowhead). (C) Affected blood vessel in brain showing perivascular edema. Bar = 100 μm.
Rabbit challenge.
Three challenge experiments were performed to compare the virulence potentials of CG and BBG strains. On day 4 p.i., 2 of 17 (∼12%) CG-infected rabbits and 1 of 15 (∼7%) EDL933-infected rabbits were euthanized based on their clinical condition and diarrheal disease; however, none of the sham-inoculated or BBG-infected rabbits developed clinical signs that prompted euthanasia before the end of the experiment. Overall, a significant increase in body weight (BW) was observed in BBG-inoculated rabbits relative to CG-, EDL933-, or sham-inoculated rabbits during days 1 to 6 p.i. (P < 0.05 to P < 0.001). However, on day 3 p.i., there was no significant difference in BW between BBG- and sham-inoculated rabbits. The percentage changes in BW of rabbits on day 4 p.i. were 41.3% ± 18.5% (BBG), 17.7% ± 11.9% (CG), 14.6% ± 9.67% (EDL933), and 20.8% ± 6.21% (sham inoculated). The estimated mean bacterial shedding on days 1 and 3 p.i., respectively, was lower in BBG-infected rabbits (2.05 × 105 and 2.29 × 105 CFU/g of feces) than in CG-infected rabbits (24.7 × 105 and 137 × 105 CFU/g of feces) and EDL933-infected rabbits (12.5 × 105 and 134 × 105 CFU/g of feces); however, there were no significant differences in bacterial shedding on day 1 and day 3 p.i. between these groups.
There were no significant differences observed in preinoculation clinical pathological parameters including hematocrit, white blood cell count, percent heterophils, platelet count, blood urea nitrogen, total protein, and albumin between groups; however, significant differences in postinoculation parameters were observed in white blood cell count, percent heterophils, and platelets (Table 4). Significant differences in hematocrit observed postinoculation were due to an increase observed in sham-inoculated controls relative to CG- and BBG-infected groups (data not shown). The increased numbers of platelets observed in the BBG group were not associated with increased production of new platelets based on our finding of no significant differences in mean platelet volume between the groups in preinoculation and postinoculation samples. Rabbits that were euthanized before the end of the experiment due to severe disease exhibited increased serum concentration of CRP (451.4 μg/ml [EDL933], 162.1 μg/ml [E3046], and 399.5 μg/ml [E5252]) relative to the means for each individual group (108.8 ± 191.6 μg/ml [EDL933], 53.0 ± 56.1 μg/ml [E3046], and 109.7 ± 147.9 μg/ml [E5252], respectively). The mean value for sham-inoculated rabbits was 29.2 ± 44.1 μg/ml (highest value, 95.2 μg/ml), and there were no significant differences between groups. Interestingly, the CRP values of clinically affected animals were higher than those observed in controls and were consistent with published rabbit values (65 to 350 μg/ml), indicative of an acute-phase response (22).
Table 4.
Clinical pathological parameters in negative (sham-inoculated)- and positive (EDL933)-control group and experimental (CG or BBG)-group rabbits
| Clinical pathological parameter | Pre- or postinoculationb | Value (mean ± SEM) for rabbit groupa |
|||
|---|---|---|---|---|---|
| Negative control (n = 14) | Positive control (n = 15) | CG (n = 17) | BBG (n = 12) | ||
| WBC count (×103/μl) | Pre | 3.2 ± 0.2 | 3.0 ± 0.3 | 3.1 ± 0.2 | 3.2 ± 0.2 |
| Post | 4.8 ± 0.6AB | 6.1 ± 0.5A | 6.0 ± 0.5A | 4.4 ± 0.2B | |
| Heterophils (%) | Pre | 53.1 ± 4.1 | 53.9 ± 3.9 | 45.7 ± 2.0 | 51.3 ± 3.2 |
| Post | 50.0 ± 3.4 | 61.6 ± 2.8c | 51.0 ± 1.5 | 45.5 ± 3.6 | |
| Platelets (×103/μl) | Pre | 747.6 ± 54.8 | 673.7 ± 67.6 | 654.2 ± 37.4 | 675.8 ± 52.9 |
| Post | 626.4 ± 70.8 | 728.3 ± 65.2 | 761.0 ± 44.3 | 1,043 ± 81.0c | |
Groups not sharing specific letters differ significantly (P < 0.02, two-tailed t test).
Pre, preinoculation; Post, postinoculation (p.i.). p.i. data represent samples collected on day 6 or 7 p.i. (negative control), day 4, 6, or 7 p.i. (positive control and CG), or day 7 p.i. (BBG).
Value significantly different from others within same row according to Tukey's multiple comparison test.
On gross necropsy, infected rabbits exhibited serosal hemorrhages near the cecocolic junction (Fig. 4). There was a difference in the numbers of rabbits showing serosal hemorrhages among CG-, BBG-, EDL933-, and sham-inoculated rabbits. Specifically, hemorrhages were observed in 35% (6/17) of CG-infected rabbits and in none of the BBG-infected rabbits (P = 0.028). Of the six CG-infected rabbits with intestinal serosal hemorrhages, five were infected with E5252 and one with E3046. Approximately 33% (5/15) of EDL933-infected rabbits and none of the sham-inoculated rabbits exhibited serosal hemorrhages (P = 0.042). Some EHEC-infected and control rabbits exhibited focal pale areas on the liver on gross examination.
Fig 4.
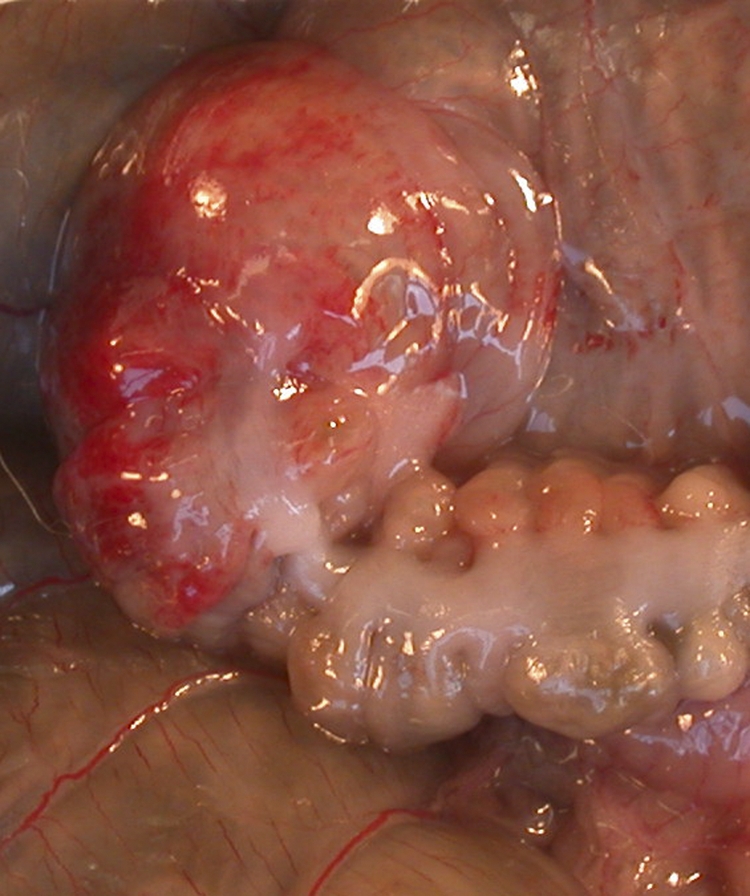
Serosal hyperemia and hemorrhage in the area of the distal cecum adjacent to the junction with the proximal colon in a Dutch Belted rabbit infected with EHEC O157 strain EDL933 on day 7 p.i.
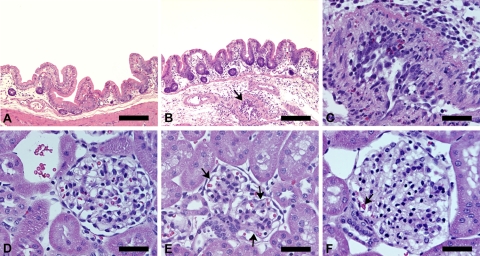
Significant differences in intestinal vasculopathy (acute type) (Fig. 5B and C) were found in all rabbits infected with specific CG strains (E5252, E3046, and E2325) and in one group infected with a specific BBG strain (E6996) relative to sham-inoculated controls but not relative to EDL933-infected rabbits (Table 5). EDL933-infected rabbits also developed significant intestinal vasculopathy relative to sham-inoculated controls. Significant differences in renal lesions were found in rabbits infected with specific CG strains (E5252 and E3046) relative to sham-inoculated and/or EDL933-infected rabbits (Table 5). Specifically, rabbits infected with CG strain E5252 developed significant renal glomerular lesions, including capillary thickening (Fig. 5E), luminal constriction (Fig. 5F), and red blood cell fragmentation (Fig. 5F), as well as renal vascular lesions, including intimal swelling and perivascular edema, relative to EDL933-infected and sham-inoculated rabbits or relative to sham-inoculated rabbits only. In addition, rabbits infected with CG strain E3046 developed significant renal vascular lesions similarly to EDL933-infected and sham-inoculated rabbits. One group infected with a specific BBG strain (E5880) developed significant red blood cell fragmentation within the glomeruli relative to sham-inoculated controls. White blood cell infiltration within the glomeruli was significantly increased in EDL933-infected rabbits relative to BBG strain (E5880)-infected rabbits. Overall, comparing the combined scores of all the rabbits infected with CG strains (E5252, E3046, and E2325) with the combined scores of all the rabbits infected with BBG strains (E6996 and E5880) revealed that CG-infected rabbits and BBG-infected rabbits developed significant intestinal vasculopathy relative to sham-inoculated controls (Table 5). In addition, analyses of the combined scores revealed significant differences in perivascular edema between CG-, BBG-, EDL933-, and sham-inoculated rabbits (P < 0.05). Specifically, perivascular edema was significantly different between CG- and EDL933-infected rabbits (P < 0.01). CG-infected rabbits developed significant luminal constriction and intimal swelling relative to BBG-, EDL933-, and sham-inoculated rabbits. BBG-infected rabbits exhibited significant red blood cell fragmentation relative to sham-inoculated rabbits (Table 5).
Fig 5.
(A) Sham-inoculated normal rabbit cecum. (B) EHEC O157 strain E5252 (CG-3)-infected rabbit cecum with submucosal edema and acute necrotizing heterophilic vasculitis (arrow). (C) Higher magnification of submucosal vascular lesion in B showing heterophilic vasculitis and perivasculitis with fibrinoid vascular degeneration/necrosis and intimal proliferation. (D) Normal rabbit glomerulus. (E) Glomerulus of a rabbit infected with EHEC O157 strain E5252 demonstrating mild capillary thickening (arrows) and few heterophils in distended capillaries. (F) Glomerulus of a rabbit infected with EHEC O157 strain E5252 showing global edematous swelling, luminal constriction, decreased numbers of erythrocytes (“bloodless glomerulus”), and fragmentation of erythrocytes (arrow). All tissue sections from rabbits on day 7 p.i. Bars = ∼150 μm (A and B) and ∼50 μm (C, D, E, and F).
Table 5.
Renal and intestinal lesion scores in Dutch Belted rabbits infected with CG or BBG EHEC O157 strainse
| Group | Lesion score (mean ± SEM)d |
||||||
|---|---|---|---|---|---|---|---|
| Renal glomerular |
Renal vascular |
Cecal | |||||
| Capillary thickening | Luminal constriction | WBC infiltration | RBC fragmentation | Intimal swelling | Perivascular edema | Vasculopathy | |
| Negative control (sham) | 0.32 ± 0.12 | 0.68 ± 0.17 | 1.11 ± 0.20 | 0.46 ± 0.12 | 0.29 ± 0.10 | 0.43 ± 0.15 | 0.11 ± 0.06 |
| Positive control (EDL933) | 0.73 ± 0.17 | 0.73 ± 0.15 | 1.33 ± 0.14 | 0.77 ± 0.21 | 0.13 ± 0.06 | 0.30 ± 0.10 | 0.80 ± 0.19a*† |
| E5252 (CG-3) | 1.0 ± 0.26a* | 1.58 ± 0.15a**,b** | 1.25 ± 0.11 | 1.42 ± 0.30a** | 1.17 ± 0.21a**†,b***† | 1.17 ± 0.21a*,b**† | 1.17 ± 0.42a**† |
| E3046 (CG-3) | 0.67 ± 0.36 | 1.0 ± 0.18 | 1.08 ± 0.24 | 0.92 ± 0.37 | 0.83 ± 0.21a*,b** | 0.92 ± 0.15a*,b** | 0.75 ± 0.25a** |
| E2325 (CG-1) | 0.20 ± 0.12 | 0.80 ± 0.34 | 1.30 ± 0.37 | 0.20 ± 0.12 | 0.10 ± 0.10 | 0.20 ± 0.12 | 0.50 ± 0.16a* |
| E6996 (BBG-6) | 0.42 ± 0.20 | 0.58 ± 0.15 | 1.58 ± 0.42 | 0.67 ± 0.21 | 0.08 ± 0.08 | 0.33 ± 0.11 | 0.67 ± 0.31a* |
| E5880 (BBG-5) | 0.33 ± 0.17 | 0.83 ± 0.28 | 0.67 ± 0.21b* | 1.17 ± 0.21a* | 0.25 ± 0.11 | 0.58 ± 0.24 | 0.33 ± 0.17 |
| All CG | 0.65 ± 0.17 | 1.15 ± 0.15a,b,c | 1.21 ± 0.14 | 0.88 ± 0.20 | 0.74 ± 0.15a*,b**†,c**† | 0.79 ± 0.14b** | 0.82 ± 0.18a***† |
| All BBG | 0.38 ± 0.13 | 0.71 ± 0.16 | 1.13 ± 0.26 | 0.92 ± 0.16a* | 0.17 ± 0.07 | 0.46 ± 0.13 | 0.50 ± 0.17a* |
Sham-inoculated rabbits.
EDL933-inoculated rabbits.
CG relative to BBG.
Superscript letters indicate comparisons within each CG or BBG column relative to sham- and EDL933-inoculated rabbits and for CG relative to BBG. Asterisks indicate significance levels as follows: *, P ≤ 0.05; **, P < 0.01; ***, P < 0.001. Unless otherwise indicated, group differences were identified using the Mann-Whitney two-tailed test; “†” indicates use of the Kruskal-Wallis test with Dunn's multiple comparison test. Unless otherwise indicated, the statistical difference was identified by the Mann-Whitney two-tailed test.
Rabbits were euthanized on day 6 or 7 p.i. (sham), day 4, 6, or 7 p.i. (EDL933), day 4 or 7 p.i. (E5252 and E3046), day 6 p.i. (E2325), and day 7 p.i. (E6996 and E5880). Abbreviations: WBC, white blood cells; RBC, red blood cells.
stx2 gene typing.
All the CG strains tested in this study carried stx2, and two of the six CG-1 strains also carried stx2c. In contrast, all BBG strains carried stx2c alone (P < 0.001).
Shiga toxin quantitation.
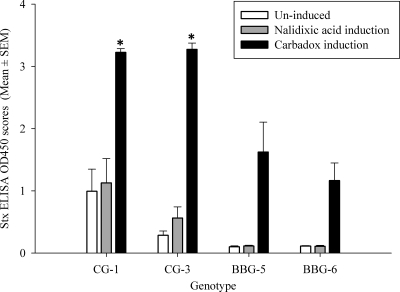
CG strains produced significantly high Stx ELISA OD450 scores compared to BBG strains under noninducing and carbadox-inducing conditions (P ≤ 0.05). In individual genotype comparisons, challenge strains belonging to CG-1 and CG-3 produced significantly high Stx ELISA OD450 scores compared to BBG-5 and BBG-6 under carbadox-inducing conditions (P ≤ 0.05) (Fig. 6). However, under noninducing conditions, challenge strains belonging to CG-1 produced Stx ELISA OD450 scores significantly higher than those of BBG-5 and BBG-6. Nalidixic acid induction resulted in no significant difference between genotypes. Induction with carbadox resulted in a significant increase in the Stx ELISA OD450 scores irrespective of the genogroups, i.e., CG or BBG (P ≤ 0.05), while induction with nalidixic acid did not result in a significant increase in Stx ELISA OD450 scores in any genogroups, probably due to the Nalr phenotype of all challenge strains. There was no difference in production of Stx between Nals and Nalr strains within the genotypes under uninduced and carbadox-inducing conditions, suggesting that the use of nalidixic acid to suppress the normal flora of the experimental piglets did not affect in vivo Stx production during these challenge experiments.
Fig 6.
Stx ELISA OD450 scores by EHEC O157 genotypes following uninduced, nalidixic acid-induced, or carbadox-induced growth in broth enrichment culture. Five EHEC O157 strains were tested in each genotype (CG-1, CG-3, BBG-5, and BBG-6). All strains are nalidixic acid resistant. *, significantly different from all the other values (P ≤ 0.05), three-way (growth conditions, nalidixic susceptibility, and genogroup) GLM with a Tukey-Kramer multiple comparison test.
DISCUSSION
The differences in the clinical signs, symptoms, and lesions induced by BBG versus CG strains in both animal models support the hypothesis that different EHEC O157 SBI genotypes are not equally virulent. Specifically, CG-challenged piglets experienced higher clinical disease severity, earlier and higher mortality, and more-severe histopathological lesions, and CG-challenged rabbits exhibited significantly lower BW gain, higher white blood cell (WBC) counts, and more-severe gross and microscopic histopathological lesions. In contrast, the BBG strains generally showed little virulence in either animal model. Our observations are consistent with the previously published study where EHEC strains from healthy cattle were, on average, less virulent in gnotobiotic piglets than EHEC strains isolated from human disease outbreaks (3), although the stx genotypes of the isolates were not reported. However, our piglet model differed in several ways from that report (3). In contrast to the gnotobiotic piglet model, in which 24-h-old piglets were inoculated with 109 CFU of EHEC O157, we used 4-h-old, nalidixic acid-treated, conventional piglets that were pretreated with 10% sodium bicarbonate before challenge with 1010 CFU of EHEC O157. In the present study, we can refine the previous observations to show that virulence in animal models is associated with specific EHEC O157 genotypes and that these differences in virulence correlate with the frequency of occurrence of these genotypes among clinical isolates. Further, we show that the CG and BBG genotypes tested in these animal models are characterized by consistent differences in their Stx2 variant gene content, with stx2 consistently present in the CG strains (with or without stx2c), while stx2c is consistently present and stx2 is consistently absent in the BBG strains.
The piglet model closely reproduces some aspects of EHEC O157-induced systemic disease of human infection. One of the important clinical manifestations of EHEC O157 infection in piglets is CNS disease, which is often associated with brain lesions (3). In this case, the animal model demonstrates lesions in a tissue (brain) which is also affected in infected humans suffering from severe HUS (44, 46, 58, 67). Nevertheless, the brain lesions and CNS disease are arguably analogous to Stx-mediated renal disease in humans, as both are exhibited when Stx is absorbed in circulation, resulting in damage to the Stx receptor-expressing endothelial cells characterized by microangiopathy and target tissue damage (14, 64). CG strains induced severe CNS symptoms (seizures, convulsions, paresis, and lateral recumbency) compared with BBG strains. In addition, there was a significant association between CNS disease and the presence of histopathological lesions in the brain.
Several recent studies have shown that EHEC O157-infected piglets also develop kidney lesions that appear to be associated with Stx production (3, 24, 52). Kidney lesions were not observed in the current study for piglets infected with either BBG or CG strains. Perhaps due to the rapid onset of severe CNS disease in piglets, renal disease was only minimally expressed before death (3). Genetic differences (the breed of pigs), the age at the time of infection, and the time of euthanasia after the appearance of first CNS signs may have contributed to the observed differences in the severity of brain lesions and lack of kidney lesions in our study compared with the previous studies using the piglet model.
We also observed some degree of cross-transmission by CG strains in our piglet challenge studies. This may suggest that CG strains may be readily transmissible or may have a low infectious dose compared with BBG strains. Further, the severe clinical signs exhibited by the CG-challenged piglets may have facilitated increased aerosol formation and thus the cross-transmission to other piglets. However, the direction and degree of cross-transmission were not sufficient to confound the strong bacterial genotype effects on virulence observed here. Cross-transmission of EHEC O157 from infected to uninfected piglets was also described earlier by Cornick and Vukhac (11), who suggested that EHEC O157 was readily transmitted among swine via contaminated aerosols with a low infectious dose. Similarly, the differences in bacteriophage content seen in a very small proportion of isolates recovered following the animal passage observed in the current study have also been reported previously, for example in experimental cattle infections (75). The infrequency of these observations in our study, however, demonstrates the overall stability of genotypes during piglet challenges.
The rabbit challenge model of EHEC O157 infection exhibits renal lesions that mimic HUS in humans (20). Therefore, we used this model to evaluate the relative virulences of BBG and CG strains and their capacity to induce intestinal and renal lesions. Rabbits experimentally infected with different EHEC O157 genotypes developed a wide range of intestinal and renal histopathological changes, suggesting that the virulence of EHEC O157 varies depending on the strain and/or genotype. More specifically, CG-3 strains E5252 and E3046 appeared to be the most pathogenic based on the extent and severity of the renal and intestinal lesions. Furthermore, the increased serum CRP concentration in the most clinically affected rabbits challenged with CG-3 strains suggested that this acute-phase protein may represent a potential biomarker of severe EHEC infection and disease. CRP has been considered a predictive factor for HUS development in humans with EHEC O157 infection (27). Overall, our findings in the rabbit challenge study suggest that CG strains are more pathogenic than BBG strains based on their capacity to induce renal disease with concurrent intestinal vascular lesions that are relatively more severe. These findings are consistent with Stx-mediated systemic disease after initial vascular damage to the intestine.
Several studies have found an association between the amount of Stx produced and/or the type of stx gene present and the virulence of EHEC O157 strains (3, 17, 39, 43, 48). The results presented here suggest that the presence of the stx2 gene and/or production of higher amounts of Stx may contribute to the increased virulence of CG strains. These results are consistent with a previous study documenting increased basal and inducible production of Stx in HUS-associated Shiga-toxin producing E. coli (STEC) relative to bovine-associated STEC (56). Although a similar relationship between the amount of Stx produced and virulence has been reported earlier (3, 17, 43, 48), the quantification of Stx depends on the specificity of antibody used in the kit, which detects Stx1 or Stx2 and Stx2c with different sensitivities (30, 48). Therefore, a careful interpretation is needed while using ELISA to compare the amounts of Stx produced among EHEC O157 strains. In our study, higher Stx ELISA scores for the CG strains could explain the severe CNS signs that were observed in CG-challenged piglets compared with the BBG-challenged piglets. However, it was not possible for us to attribute this difference solely to the amounts of toxin produced by these strains, as they also differ in the stx2 gene subtype.
HUS and severe disease in humans have been linked to strains that carry stx2 and stx2c alone or in combination (1, 19, 48). Nine out of 10 piglets died after challenge with strains carrying stx2, whether alone or in combination with stx1 or stx2c. However, only 2 out of 10 piglets challenged with the strains carrying stx2c alone died. These results suggest that the EHEC O157 strains that carry stx2c alone are likely to be less pathogenic than the strains that carry stx2 alone or in combination with stx2c or stx1. These results are supported by a recent study (19) showing that purified Stx2 is more potent than Stx2c against primary human kidney cell lines and in mouse models. Other researchers have observed histopathological lesions in the brain and kidney and have attributed these to the systemic effect of Stx (3, 24, 52). The effects of the relative in vivo toxicities of stx2 variants need further investigation (19, 48).
Recent studies using in vitro cell culture and mouse models suggest that besides inducing systemic disease, Stx2 production can also promote the colonization of epithelial cells by enhancing the expression of host cell intimin receptors (40, 57). In contrast to this, similar studies conducted in cattle and rabbits suggested no effect of Stx on colonization on intestine (55, 61). Thus, the effect of the stx2 gene and more specifically its subtype on intestinal colonization needs more investigation.
CG strains induced severe intestinal histopathological lesions and had high bacterial load in the intestine compared with the BBG strains or sham-inoculated controls in the piglet and rabbit models, suggesting differences in the strains' abilities to colonize the intestinal mucosa. Recently, a whole-genome expression microarray was used to compare the differential gene expression between BBG and CG strains (68). The important virulence factors, including the genes contained on chromosomal LEE and several genes contained on the pO157 plasmid (the enterohemolysin ehxA, toxB, and etp genes necessary for T2SS) showed increased expression in the CG compared with BBG strains. In contrast, the genes essential for survival in the environment, such as acid resistance and stress response, were upregulated in the BBG compared with the CG strains (28, 63, 65, 68). These observations suggest that differential expression of LEE-contained virulence genes in vivo may have led to the differences in the magnitudes of intestinal bacterial load and histopathological lesions in the intestines between the piglets and rabbits challenged with CG and BBG strains. This initial damage to the intestinal epithelium may facilitate the absorption of Stx into the bloodstream, where it may cause more-severe systemic damage.
In conclusion, we have demonstrated that BBG and CG EHEC O157 differ in virulence potential by using two animal models, consistent with the hypothesis that not all EHEC O157 strains are equally pathogenic. The amount of Stx and/or the subtype of stx2 gene present is correlated to the virulence differences of EHEC O157 genotypes, and more studies are needed in order to clarify the role of these bacterial factors in the pathogenesis and virulence of specific EHEC O157 genotypes.
ACKNOWLEDGMENTS
This work was funded in part by NIAID NIH contract N01-AI-30055, R21AI073803, T32 RR007036-24, and a Poncin fellowship (S.S.) and by the Agricultural Animal Health Program, Washington State University College of Veterinary Medicine, Pullman, WA.
We thank Rebecca A. King, Melissa W. Mobley, and Amanda Potter for their technical assistance with studies involving the rabbit model.
Footnotes
Published ahead of print 24 October 2011
REFERENCES
- 1. Abu-Ali GS, et al. 2010. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One 5: e10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bach SJ, McAllister TA, Veira DM, Gannon VPJ, Holley RA. 2002. Transmission and control of Escherichia coli O157:H7—a review. Can. J. Anim. Sci. 82: 475–490 [Google Scholar]
- 3. Baker DR, et al. 2007. Differences in virulence among Escherichia coli O157:H7 strains isolated from humans during disease outbreaks and from healthy cattle. Appl. Environ. Microbiol. 73: 7338–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Besser RE, Griffin PM, Slutsker L. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50: 355–367 [DOI] [PubMed] [Google Scholar]
- 5. Besser TE, et al. 2007. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 73: 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borczyk AA, Karmali MA, Lior H, Duncan LM. 1987. Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet i: 98. [DOI] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention 1999. One-day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli serotype O157 by pulsed field gel electrophoresis (PFGE). PulseNet, the National Molecular Subtyping Network for Foodborne Disease Surveillance, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 8. Centers for Disease Control and Prevention 2009. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food–10 states, 2008. MMWR Morb. Mortal. Wkly. Rep. 58: 333–337 [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention 2010. Surveillance for foodborne disease outbreaks—United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 59: 973–979 [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention 2009. Surveillance for foodborne disease outbreaks—United States, 2006. MMWR Morb. Mortal. Wkly. Rep. 58: 609–615 [PubMed] [Google Scholar]
- 11. Cornick NA, Vukhac H. 2008. Indirect transmission of Escherichia coli O157:H7 occurs readily among swine but not among sheep. Appl. Environ. Microbiol. 74: 2488–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Creuzburg K, et al. 2005. Genetic structure and chromosomal integration site of the cryptic prophage CP-1639 encoding Shiga toxin 1. Microbiology 151: 941–950 [DOI] [PubMed] [Google Scholar]
- 13. Davis MA, Hancock DD, Besser TE, Call DR. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41: 1843–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dean-Nystrom EA, Pohlenz JF, Moon HW, O'Brien AD. 2000. Escherichia coli O157:H7 causes more-severe systemic disease in suckling piglets than in colostrum-deprived neonatal piglets. Infect. Immun. 68: 2356–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Baets L, et al. 2004. Genetic typing of Shiga toxin 2 variants of Escherichia coli by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70: 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donnenberg MS, et al. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Invest. 92: 1418–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eklund M, Leino K, Siitonen A. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40: 4585–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedrich AW, et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185: 74–84 [DOI] [PubMed] [Google Scholar]
- 19. Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. 2011. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79: 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. García A, et al. 2006. Renal injury is a consistent finding in Dutch Belted rabbits experimentally infected with enterohemorrhagic Escherichia coli. J. Infect. Dis. 193: 1125–1134 [DOI] [PubMed] [Google Scholar]
- 21. García A, Fox JG, Besser TE. 2010. Zoonotic enterohemorrhagic Escherichia coli: a one health perspective. ILAR J. 51: 221–232 [DOI] [PubMed] [Google Scholar]
- 22. Gentry PA. 1999. Acute phase proteins, p 336–398 In Loeb WF, Quimby FW. (ed), The clinical chemistry of laboratory animals, 2nd ed. Taylor and Francis, Philadelphia, PA [Google Scholar]
- 23. Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13: 60–98 [DOI] [PubMed] [Google Scholar]
- 24. Gunzer F, et al. 2002. Gnotobiotic piglets develop thrombotic microangiopathy after oral infection with enterohemorrhagic Escherichia coli. Am. J. Clin. Pathol. 118: 364–375 [DOI] [PubMed] [Google Scholar]
- 25. Hayashi T, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8: 11–22 [DOI] [PubMed] [Google Scholar]
- 26. Hintzee J. 2007. NCSS 2007. NCSS, LLC; Kaysville, UT. [Google Scholar]
- 27. Ikeda K, et al. 2000. Predictors for the development of haemolytic uraemic syndrome with Escherichia coli O157:H7 infections: with focus on the day of illness. Epidemiol. Infect. 124: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kailasan Vanaja S, Bergholz TM, Whittam TS. 2009. Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J. Bacteriol. 191: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karmali MA. 2004. Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26: 117–122 [DOI] [PubMed] [Google Scholar]
- 30. Karmali MA, Petric M, Bielaszewska M. 1999. Evaluation of a microplate latex agglutination method (Verotox-F assay) for detecting and characterizing verotoxins (Shiga toxins) in Escherichia coli. J. Clin. Microbiol. 37: 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karmali MA, et al. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151: 775–782 [DOI] [PubMed] [Google Scholar]
- 32. Karmali MA, Steele BT, Petric M, Lim C. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i: 619–620 [DOI] [PubMed] [Google Scholar]
- 33. Kim J, Nietfeldt J, Benson AK. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. U. S. A. 96: 13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim J, et al. 2001. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, beta-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 183: 6885–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koch C, Hertwig S, Appel B. 2003. Nucleotide sequence of the integration site of the temperate bacteriophage 6220, which carries the Shiga toxin gene stx(1ox3). J. Bacteriol. 185: 6463–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kotewicz ML, Mammel MK, LeClerc JE, Cebula TA. 2008. Optical mapping and 454 sequencing of Escherichia coli O157:H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology 154: 3518–3528 [DOI] [PubMed] [Google Scholar]
- 37. Laing CR, et al. 2009. In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Law D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88: 729–745 [DOI] [PubMed] [Google Scholar]
- 39. Lefebvre B, Diarra MS, Vincent C, Moisan H, Malouin F. 2009. Relative cytotoxicity of Escherichia coli O157:H7 isolates from beef cattle and humans. Foodborne Pathog. Dis. 6: 357–364 [DOI] [PubMed] [Google Scholar]
- 40. Liu B, et al. 2010. Verotoxin 2 enhances adherence of enterohemorrhagic Escherichia coli O157:H7 to intestinal epithelial cells and expression of {beta}1-integrin by IPEC-J2 cells. Appl. Environ. Microbiol. 76: 4461–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Manning SD, et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105: 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mead PS, Griffin PM. 1998. Escherichia coli O157:H7. Lancet 352: 1207–1212 [DOI] [PubMed] [Google Scholar]
- 43. Muniesa M, et al. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71: 4554–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nathanson S, et al. 2010. Acute neurological involvement in diarrhea-associated hemolytic uremic syndrome. Clin. J. Am. Soc. Nephrol. 5: 1218–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Naylor SW, et al. 2005. Escherichia coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151: 2773–2781 [DOI] [PubMed] [Google Scholar]
- 46. Oakes RS, Siegler RL, McReynolds MA, Pysher T, Pavia AT. 2006. Predictors of fatality in postdiarrheal hemolytic uremic syndrome. Pediatrics 117: 1656–1662 [DOI] [PubMed] [Google Scholar]
- 47. Ohnishi M, et al. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. U. S. A. 99: 17043–17048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Orth D, et al. 2007. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn. Microbiol. Infect. Dis. 59: 235–242 [DOI] [PubMed] [Google Scholar]
- 49. Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11: 450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perna NT, et al. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66: 3810–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perna NT, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529–533 [DOI] [PubMed] [Google Scholar]
- 52. Pohlenz JF, Winter KR, Dean-Nystrom EA. 2005. Shiga-toxigenic Escherichia coli-inoculated neonatal piglets develop kidney lesions that are comparable to those in humans with hemolytic-uremic syndrome. Infect. Immun. 73: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 11: 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Recktenwald J, Schmidt H. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage phiP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. 2003. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect. Immun. 71: 7129–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ritchie JM, Wagner PL, Acheson DW, Waldor MK. 2003. Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 69: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robinson CM, Sinclair JF, Smith MJ, O'Brien AD. 2006. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. U. S. A. 103: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robson WL, Leung AK, Montgomery MD. 1991. Causes of death in hemolytic uremic syndrome. Child Nephrol. Urol. 11: 228–233 [PubMed] [Google Scholar]
- 59. Scallan E, et al. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shaikh N, Tarr PI. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185: 3596–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74: 4685–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shimizu K, et al. 2003. Development of a lethal Shiga toxin-producing Escherichia coli-infection mouse model using multiple mitomycin C treatment. Microb. Pathog. 35: 1–9 [DOI] [PubMed] [Google Scholar]
- 63. Shin S, et al. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41: 1133–1150 [DOI] [PubMed] [Google Scholar]
- 64. Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365: 1073–1086 [DOI] [PubMed] [Google Scholar]
- 65. Tatsuno I, et al. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71: 2598–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33: 2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tzipori S, Chow CW, Powell HR. 1988. Cerebral infection with Escherichia coli O157:H7 in humans and gnotobiotic piglets. J. Clin. Pathol. 41: 1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vanaja SK, Springman AC, Besser TE, Whittam TS, Manning SD. 2010. Differential expression of virulence and stress fitness genes between Escherichia coli O157:H7 strains with clinical or bovine-biased genotypes. Appl. Environ. Microbiol. 76: 60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Watanabe M, et al. 2004. Oral therapeutic agents with highly clustered globotriose for treatment of Shiga toxigenic Escherichia coli infections. J. Infect. Dis. 189: 360–368 [DOI] [PubMed] [Google Scholar]
- 70. Whitworth J, et al. 2010. Diverse genetic markers concordantly identify bovine origin Escherichia coli O157 genotypes underrepresented in human disease. Appl. Environ. Microbiol. 76: 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Whitworth JH, et al. 2008. International comparison of clinical, bovine, and environmental Escherichia coli O157 isolates on the basis of Shiga toxin-encoding bacteriophage insertion site genotypes. Appl. Environ. Microbiol. 74: 7447–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187: 1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wieler LH, et al. 1996. Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J. Clin. Microbiol. 34: 2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang Z, et al. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70: 6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yoshii N, et al. 2009. Pulsed-field gel electrophoresis profile changes resulting from spontaneous chromosomal deletions in enterohemorrhagic Escherichia coli O157:H7 during passage in cattle. Appl. Environ. Microbiol. 75: 5719–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang Y, et al. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]