Abstract
Like several other bacterial pathogens, Anaplasma marginale has an outer membrane that induces complete protection from infection and disease. However, the proteins that confer protective immunity and whether protection requires interacting proteins and/or linked T-cell and immunoglobulin G epitopes are not known. Our goal is to target the conserved type IV secretion system (T4SS) to identify conserved, immunogenic membrane proteins that are interacting and linked recognition candidates. Linked recognition is a process by which a B cell is optimally activated by a helper T cell that responds to the same, or physically associated, antigen. A. marginale T4SS proteins VirB2, VirB4-1, VirB4-2, VirB6-1, VirB7, VirB8-2, VirB9-1, VirB9-2, VirB10, VirB11, and VirD4 were screened for their ability to induce IgG and to stimulate CD4+ T cells from outer membrane-vaccinated cattle. VirB9-1, VirB9-2, and VirB10 induced the strongest IgG and T-cell responses in the majority of cattle, although three animals with major histocompatibility complex class II DRB3 restriction fragment length polymorphism types 8/23, 3/16, and 16/27 lacked T-cell responses to VirB9-1, VirB9-1 and VirB9-2, or VirB9-2 and VirB10, respectively. For these animals, VirB9-1-, VirB9-2-, and VirB10-specific IgG production may be associated with T-cell help provided by responses to an interacting protein partner(s). Interacting protein partners indicated by far-Western blotting were confirmed by immunoprecipitation assays and revealed, for the first time, specific interactions of VirB9-1 with VirB9-2 and VirB10. The immunogenicity and interactions of VirB9-1, VirB9-2, and VirB10 justify their testing as a linked protein vaccine against A. marginale.
INTRODUCTION
Anaplasma marginale is an obligate intraerythrocytic Gram-negative bacterium causing bovine anaplasmosis. Immunization of cattle with A. marginale outer membranes (OM) and cross-linked OM proteins can induce complete protection against disease and infection (10, 43, 57). However, the well-characterized immunodominant major surface proteins 1 to 5 (MSP1 to MSP5), which include antigenically variant MSP2 and MSP3, do not induce equivalent protection against A. marginale infection (1, 46, 47, 49, 50). Because immunodominant MSPs are not protective, we have focused on identifying subdominant and conserved surface proteins, including those that associate within the membrane that may be important components of the protective OM vaccine. Among the subdominant antigens identified in the OM are type IV secretion system (T4SS) proteins. Several T4SS proteins induced CD4 T cell responses, including gamma interferon (IFN-γ) production, and were recognized by IgG2 in cattle immunized with the OM vaccine (32–34, 56). These type 1 immune responses are associated with protection against A. marginale infection in cattle immunized with OM (9, 10, 50).
The T4SS is a 1.1-MDa protein complex that spans the outer and inner bacterial membranes and has been most widely studied in Agrobacterium tumefaciens (13, 17). Typically, the T4SS is made up of 12 interacting VirB/D membrane proteins, several of which are likely to be surface exposed (14) and are thus targets for neutralizing and protective immune responses. The T4SS core complex is made up of 14 copies of interacting VirB7, VirB9, and VirB10, which form a complex that was crystallized and studied with Escherichia coli VirB homologues TraN (VirB7), TraO (VirB9), and TraF (VirB10) (13, 17). The Agrobacterium VirB proteins also assemble into a pilus made up of VirB2, VirB5, and VirB7 (28, 29, 53, 54). There are three nucleotide triphosphate (NTP)-utilizing T4SS proteins—VirB4, VirB11, and VirD4—that supply energy for substrate translocation, and VirD4 specifically acts as the substrate-coupling protein (3, 11). VirB6 and VirB8 assemble the T4SS apparatus at the cell pole and also make up the core complex (24, 25, 27). The T4SS proteins harbored by A. marginale and other members of the family Anaplasmataceae are unique because they are encoded by two copies of virB4, virB8, and virB9, four or more copies of virB6, and multiple copies of virB2. There are single copies of virB3, virB7, virB10, virB11, and virD4, whereas homologues of virB1 and virB5 have not been identified (7, 19, 20). The function of the T4SS in A. marginale has not been determined, but in a pathogen that has undergone reductive evolution, retention of these genes indicates their requirement for invasion and survival within erythrocytes and/or tick cells (19, 20). Because Anaplasmataceae lack lipopolysaccharides (7, 20), several T4SS proteins may be surface exposed where they could be targeted by neutralizing antibody. Furthermore, T4SS proteins interact within the bacterial membrane so associated proteins could provide linked recognition for T-cell–B-cell interactions.
Linked recognition was described in the 1970s by Mitchison, who revealed that both T cells and B cells must recognize antigenic determinants on the same molecule for B-cell activation to occur (39–41). T cells help B cells to undergo somatic hypermutation and isotype switching through cognate interaction and cytokine secretion. However, linked recognition can also occur with two associated proteins, where one has B cell epitopes and a second provides T-cell epitopes. For A. marginale, this was suggested in studies with MSP1, a heteromer consisting of covalently associated MSP1a and MSP1b (35, 60). In these studies, MSP1a-specific T cells provided help to B cells specific for MSP1b to promote increased IgG production (8, 36). Immunization with physically associated B- and T-cell antigens achieves long-lasting immunological memory to the B-cell antigen and is a common strategy used for vaccination against bacterial polysaccharides that lack T-cell epitopes (26, 52).
In the present study, we test the hypothesis that subdominant T4SS proteins of A. marginale that induce an IgG response but no CD4+ T-cell response in an OM-vaccinated individual interact with a T4SS protein(s) that does stimulate T-cell proliferation which could provide T-cell help. We present evidence for the linked recognition of VirB9-1, VirB9-2, and VirB10, which are naturally associated in A. marginale, and a basis for testing these linked T4SS proteins as a vaccine for anaplasmosis.
MATERIALS AND METHODS
Immunization of six haplotype-diverse cattle with A. marginale OM.
St. Maries strain A. marginale OM were isolated as previously described from splenectomized bovine C31919 blood with 40.4% bacteremia (34, 43, 48). Five age-matched Holstein steers and one Holstein cow with various major histocompatibility complex (MHC) class II haplotypes were purchased from the Washington State University (WSU) dairy. Bovine lymphocyte antigen (BoLA) MHC class II DRB3 types were determined by restriction fragment length polymorphism (RFLP) analysis of exon 2 of the DRB3 gene (44, 51, 55, 59). Animals (DRB3 RFLP types) used in the present study were cared for according to an approved Institutional Animal Care and Use Center (IUCAC) protocol for WSU and were designated by number (type) as follows: 35113 (11/22), 35141 (22/24), 35160 (3/16), 35280 (16/27), 35287 (16/22), and 583 (8/23). It is important to note that the haplotypes represented by the six OM-immunized cattle are the most abundant haplotypes of Holsteins in Washington and Canada (unpublished data and reference 55). Cattle were immunized four times subcutaneously with 60 μg of OM emulsified in saponin, at 0, 2, 4, and 8 weeks as previously described (34). Four immunizations were used to effectively prime cattle to these subdominant antigens so that T-cell and antibody responses would be detected (33, 34). All six cattle produced specific antibody, determined by immunoblotting, and had statistically significant peripheral blood mononuclear cell (PBMC) proliferative responses to A. marginale OM, compared to preimmunization responses (see Fig. S1 in the supplemental material).
Expression of recombinant proteins.
Full-length recombinant T4SS proteins were expressed from the A. marginale St. Maries strain. Genes encoding VirB9-1 (AM097), VirB9-2 (AM1315), and VirB10 (AM1314) were cloned into pBAD/TOPO ThioFusion vector and expressed as previously described with C-terminal FLAG and 6×His tags (33). Recombinant VirB7 (AM306) with just a C-terminal 6×His tag was also cloned into the pBAD/TOPO ThioFusion vector and expressed and purified as previously described (56). Genes encoding VirB2 (AM030), VirB4-1 (AM814), VirB4-2 (AM1053), the first fragment of VirB6-1 (AM813 F1), VirB8-2 (AM1316), VirB11 (AM1313), and VirD4 (AM1312) were cloned into the pEXP1-DEST vector (Invitrogen), with N-terminal 6×His and C-terminal FLAG tags, and expressed in BL-21 DE3 pLysS One Shot E. coli (Invitrogen). Primers used for all of these proteins were described previously (56). Plasmid DNA was extracted from E. coli and sequencing was performed using the BigDye kit and ABI Prism automated sequence (Applied Biosystems) as described previously (56).
The negative control protein Babesia bovis merozoite surface antigen 1 (MSA1) (31) was expressed using the amplified gene from Mo7 genomic DNA with the forward primer (GCCGATACTTCAATCGTCCTTCC) and reverse primer without a stop codon (TTTGTCGTCGTCGTCTTTATAGTCTGTACCCTGTTGTCCTTGGAGG) encoding a C-terminal FLAG epitope (DYKDDDDK). The thermal cycling parameters consisted of 40 cycles of melting at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 2 min. The plasmid DNA was excised from agarose gels and purified using a Genelute agarose column (Sigma-Aldrich). Amplicons were cloned into the pBAD/TOPO ThioFusion expression vector (Invitrogen) according to the manufacturer's protocol, and plasmid DNA was extracted from E. coli using the Wizard Plus SV Miniprep DNA purification system (Promega) and sequenced.
Colonies with the correct sequence were grown to mid log phase in LB broth containing 50 μg of carbenicillin/ml and induced using 0.2% arabinose for pBAD/TOPO ThioFusion constructs or 1 to 3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for the pEXP1-DEST constructs. LB broth used for the expression of rVirB8-2 contained 50 μg of carbenicillin/ml and 34 μg of chloramphenicol/ml. Induced E. coli was centrifuged at 3,800 × g, and the pellets were resuspended in denaturing lysis buffer (6 M guanidine HCl, 20 mM sodium phosphate, 500 mM NaCl [pH 7.8]), sonicated twice on maximum strength for 3 min, and frozen overnight at −80°C.
Purification of recombinant proteins.
All proteins were purified using the hybrid conditions according to the Probond nickel purification kit instructions (Invitrogen). Protein concentrations were determined with the Quick Start Bradford protein assay (Bio-Rad), and purity was assessed using 1 or 10 μg/well separated by using Coomassie blue-stained precast sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 4 to 20% gradient gels (Bio-Rad).
MS/MS analysis of rVirB9-1, rVirB9-2, and rVirB10.
Bands corresponding to 20 and 40 kDa for rVirB7, 50 and 100 kDa for rVirB9-1 and rVirB9-2, and 65 and 130 kDa for rVirB10 were excised from Coomassie blue-stained gels, destained (50% methanol, 5% acetic acid), dehydrated with 100% acetonitrile, reduced with 10 mM dithiothreitol (DTT), quenched with 50 mM iodoacetamide, and digested with 20 ng of trypsin (Promega)/μl. The peptides were subjected to tandem mass spectrometric (MS/MS) fragmentation on a high-performance liquid chromatography (HPLC)-coupled quadruple-time of flight (Q-TOF) MS instrument located at University of Idaho, Environmental Biotechnology Institute. Fragment ion lists and the identified peptide sequences were searched against the Mascot databases that contain A. marginale, E. coli, or National Center for Biotechnology Information all entries. Identification of the protein was based upon the Mascot score, the probability, and the mass. One missed trypsin cleavage, fixed carbamidomethyl modifications, and variable oxidation were allowed during the search. A probability of ≥95% showed the peptide match was not a random occurrence, and the individual ion score is reported as −10 log10 P, where P is the probability. An ion score greater than 19 has significant identity.
Determination of serological responses to T4SS proteins by immunoblotting.
Purified recombinant T4SS proteins (1 to 2 μg) were loaded onto a 4 to 20% precast gradient SDS-polyacrylamide gel (Bio-Rad) with denaturing sample buffer (8.5 mM Tris, 15% SDS, 50% glycerol, 12% β-mercaptoethanol, 0.1% bromophenol blue), electrophoresed at 100 V for 1.5 h, and transferred to a nitrocellulose membrane (Bio-Rad) at 100 V for 1 h. The membrane was blocked overnight with blocking buffer (10 mM Tris-HCl, 150 mM NaCl, 0.2% Tween 20, 1% PVP40). OM-immune bovine sera that had been extensively adsorbed with protease-inhibited and sonicated BL21 or Top10 E. coli (Invitrogen) cells that contained either no vector or a vector encoding MSA1 were diluted 1:100 and added to the membrane for 1 h. The membrane was washed three times with wash buffer (10 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20, 1% PVP40). A 1:20,000 dilution of horseradish peroxidase (HRP)-labeled recombinant protein G (Zymed) was added for 1 h, and blots were then washed extensively with wash buffer. ECL Western blotting reagent substrate (Thermo Scientific) was applied for 2 min. Preimmune, adsorbed sera from all animals were also used at a 1:100 dilution on the blots containing all of the T4SS proteins to rule out nonspecific IgG binding. After positive IgG responses were identified for a given animal and protein, the same procedure was followed as described above, except sera were diluted 1:300 to 1:10,000.
T-cell proliferation assays.
To establish T-cell lines, PBMC were depleted of CD8+ cells and γδ T cells prior to antigen stimulation by incubation with sodium azide-free monoclonal antibodies (MAbs; 3 μg/ml), followed by complement lysis (33). The antibodies were anti-CD8α IgG2a (7C2B), anti-CD8α IgM (BAQ111A), anti-TCRδ IgG2b (GB21A), and anti-TCRδ IgM (CACT61A) obtained from the WSU Monoclonal Antibody Center. Two-week cell lines were generated from CD8- and γδ-T-cell-depleted PBMC by stimulating for 1 week with 5 μg of OM/ml and rested for 1 week as previously described (33). The efficiency of depletion was confirmed by fluorescence-activated cell sorting analysis, which showed the T cells were composed of 93.4 to 96.7% CD4+ T cells, 1.1 to 2.4% CD8+ T cells, and 2.2 to 2.4% γδ T cells. Proliferation assays were performed in triplicate with 1 and 10 μg of recombinant T4SS proteins/ml, as described previously (33, 56). Positive controls included A. marginale OM and 10% T-cell growth factor and negative controls included uninfected red blood cell membranes (URBC) and B. bovis MSA1 also used at 1 and 10 μg/ml. T-cell proliferation was quantified by incorporation of 0.25 μCi/well [3H]thymidine. The results are presented as stimulation indices (SI), calculated as the mean counts per minute (cpm) of cells cultured with antigen/mean cpm of cells cultured with medium. The SI for the different T4SS antigens were compared to the SI for the matching concentration of MSA1 using Dunnett's test, a one-way multiple-comparison test. Statistically significant T-cell stimulation by an antigen was set at a P value of <0.05.
Preparation of antibodies specific for VirB7, VirB9-1, VirB9-2, and VirB10.
Rabbits were used in the present study in compliance with the WSU IUCAC. Purified recombinant proteins were excised from the SDS-PAGE gels and trypsin-digested for Q-TOF MS analysis to confirm that the purified protein was the protein of interest. Recombinant VirB7, rVirB9-1, rVirB9-2, and rVirB10 were emulsified with 50% phosphate-buffered saline (PBS) and 50% TiterMax Gold adjuvant (Sigma-Aldrich) to make a final concentration of 200 μg of protein/ml, and 100 μg of protein was inoculated subcutaneously into rabbits at 0, 3, 5, 7, 9, and 11 weeks (4 to 6 times) until the immunization was successful, which was determined by Western blotting with pre- and postimmunization sera diluted 1:500. A negative control rabbit antiserum was made by Pacific Immunology, using Freund complete and incomplete adjuvants and a fragment of a B. bovis nuclear-encoded protein, acyl carrier protein (ACP). The ACP fragment from amino acids 68 to 148 was expressed in pTrcHIS TOPO TA expression vector (Invitrogen) and purified using denaturing conditions on a nickel column, followed by electroelution of the gel-embedded protein.
Prior to purification of rabbit IgG, 5 ml of each rabbit serum was incubated with 3 ml of crude Top10 E. coli lysate expressing MSA1 for 1 h in binding buffer (20 mM sodium phosphate [pH 7.0]) at room temperature and centrifuged to remove nonspecific antibodies. The adsorbed antisera were buffer-exchanged twice by diluting with binding buffer and centrifuging using 15-ml Amicon 10k filters (Millipore). Subsequently, a HiTrap protein A column (GE Healthcare Biosciences) was used to purify IgG from the sera according to the manufacturer's protocol. Purified IgG was buffer exchanged with sterile PBS and concentrated with an Amicon 10-kDa cutoff filter, and the final concentration of IgG was determined with a Bio-Rad Bradford assay. IgGs were purified on protein A columns, the purity was assessed by SDS-PAGE, and the specificity was determined on immunoblots with 1 μg of each recombinant T4SS protein, 10 μg of A. marginale OM, and 2 μg of adsorbed and purified IgG/ml. Secondary antibody was alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG(H+L) (Invitrogen) diluted 1:10,000, and blots were developed with the Western Star chemiluminescence kit (Applied Biosystems).
Far-Western blotting.
Far-Western blotting was performed as previously described (4, 12, 62) with the following modifications and under conditions of protein denaturation and renaturation. SDS-PAGE was performed with 1 μg of each recombinant protein loaded per well on 4 to 20% gradient gels and electrophoresis at 100 V for 1.5 h. After transfer to nitrocellulose membranes at 100 V for 1 h, the nitrocellulose membrane with recombinant T4SS protein was first incubated with 6 M guanidine hydrochloride–20 mM HEPES (pH 7.5)–50 mM KCl–10 mM MgCl2–1 mM DTT–0.1% Nonidet P40–2% milk (GuHCl buffer) for 30 min at room temperature, then successively incubated for 30 min each with 3 M and 1.5 M GuHCl buffer at room temperature, then incubated with 0.6 M GuHCl buffer at 4°C for 30 min, and finally incubated with buffer lacking GuHCl overnight at 4°C. The membrane was blocked with 20 mM Tris-HCl (pH 7.5)–150 mM NaCl (TBS) with 5% milk for 3 h. Two far-Western membranes were run in tandem. One membrane was incubated with a 1:2,000 dilution of HRP-conjugated anti-His MAb (Qiagen) to confirm proteins on the membrane after denaturing (see Fig. S2 in the supplemental material). The second membrane was used to identify a target recombinant protein interacting with another recombinant protein. To determine whether the target recombinant protein (VirB9-1, VirB9-2, or VirB10) bound to a prey recombinant T4SS protein(s), the membrane containing 1 μg of each transferred prey protein was incubated with 10 μg of target protein in 5 ml of TBS with 5% milk overnight at 4°C. The membrane was then washed three times with TBS containing 0.05% Tween 20 (TBST). Rabbit antiserum, adsorbed with Top10 E. coli lysate expressing MSA1 and directed against the target protein (rVirB9-1, rVirB9-2, or rVirB10), was added to the membrane at a 1:500 dilution in TBS plus 5% milk, and the excess was washed off. The membrane was incubated with a 1:10,000 dilution of AP-conjugated goat anti-rabbit IgG(H+L) for 1 h and then washed extensively with TBST and developed using the Western-Star chemiluminescent substrate.
Determining the binding coefficients of interacting rVirB9-1, rVirB9-2, and rVirB10.
Serial dilutions (0.5 to 5.0 μg) of rVirB9-2 and rVirB10 were applied to a dot blot apparatus and used to find the binding coefficient to rVirB9-1 (12). Recombinant VirB9-1 was also applied to the dot blot as a positive control, and a higher concentration of (0.5 to 10.0 μg) rMSA1 was used as a negative control for background subtraction. The dot blot was developed using the same procedure as for far-Western blotting described above using 10 μg of rVirB9-1 to probe the blot and a 1:500 dilution of adsorbed polyclonal anti-VirB9-1 rabbit serum to detect rVirB9-1. Densitometry analysis of the spots containing interacting proteins or controls was performed with ImageJ 1.43 software from National Institutes of Health. The data were fit to a standard one-site saturation binding curve Y = Ymax (X)/(KD + X), where Y is the intensity of the spot, X is concentration of the protein on the membrane, and KD is the binding association constant using SigmaPlot 10.0. From this equation of the line the binding association constants were found for rVirB9-1 binding to rVirB9-2 and rVirB10.
Production of MAbs.
Mice were used in the present study in compliance with the WSU IUCAC. Two female BALB/c mice were immunized subcutaneously with 20 μg of rVirB9-1, rVirB9-2, or rVirB10 emulsified in an equal volume of complete Freund adjuvant for a total volume of 150 μl. Mice received three booster immunizations at 2-week intervals with 20 μg of antigen in incomplete Freund adjuvant. Pre- and postimmunization sera were used to probe blots to detect specific protein in native OM. A final immunization with 10 μg of protein in 100 μl of PBS was given intravenously, and spleens were removed 3 days later for hybridoma fusion. Fusion and limiting dilution cloning were performed as described previously (63). Hybridomas were screened for a positive reaction on immunoblots of all three recombinant proteins, as well as the OM. Hybridomas that reacted specifically with the recombinant protein of interest, as well as native protein, were selected for cloning. Supernatants from clones were similarly screened, and three clones were selected for MAb production. These are 133/248.14.1.28 specific for VirB9-1 (IgG1), 137/774.8.7 specific for VirB9-2 (IgG2b), and 138/481.3.9 specific for VirB10 (IgG1).
Immunoprecipitation.
A high concentration of OM was used for immunoprecipitation because the T4SS proteins are not abundant (34). The nonionic detergent n-dodecyl β-d-maltoside (DM) was used for solubilization to help maintain native protein conformation and the natural hydrophobic interaction or hydrogen bonding of the membrane proteins. A. marginale OM were solubilized for at least 4 h in DM (Sigma-Aldrich) at 4°C with rotation to yield a final concentration of 2.5 mg/ml in 2% DM, and cell debris was removed by centrifugation at 10,000 × g for 10 min at 4°C. A 350-μl aliquot of supernatant was used for each immunoprecipitation reaction. Protein A-purified polyclonal rabbit IgG (2 μg) specific for VirB9-1, VirB9-2, VirB10, or B. bovis ACP (negative control) was incubated with the solubilized OM proteins for 2 h at 4°C with rocking. Protein A-agarose (20 μl; Santa Cruz Biotechnology) was added, followed by incubation for 1 h at 4°C, with rocking. The native protein complexes were centrifuged at 660 × g for 5 min, and the pellet was washed four times with 400 μl of 50 mM Tris-HCl (pH 8)–150 mM NaCl–0.1% Nonidet P40 and once with sterile PBS. For analysis, the pellet was immediately resuspended in 0.3 M Tris-HCl–5% SDS–50% glycerol–100 mM dithiothreitol–tracking dye (pH 6.8 [Lane Marker Sample Buffer; Thermo Scientific]) to yield a total volume of 95 μl in 1× sample buffer, and 10 μl was loaded per well on 4 to 20% gradient gels and electrophoresed at 100 V for 1.5 h. After transfer to nitrocellulose membranes at 100 V for 1 h, the interacting proteins were determined on Western blots probed with 1 μg/ml of polyclonal rabbit IgG specific for a candidate interacting protein partner or for the target protein. After extensive washing in I-block containing 0.3% Tween 20, blots were incubated with HRP-conjugated Clean-Blot (Thermo Scientific) that only binds to intact antibodies, according to the manufacturer's protocol, washed with I-block containing 0.3% Tween 20, and developed with the ECL Western blotting reagent (Thermo Scientific). MAbs used at 2 μg/ml specific for MSP5 (AnaF16C1) and MSP2 (AnaR49a) (38) were also used to probe the blots for additional specificity controls. The blots probed with anti-MSP5 were incubated with HRP-Clean-Blot and developed as described above, and the anti-MSP2 probed blots were incubated with AP-conjugated goat anti-mouse IgG+IgM (Invitrogen) diluted 1:10,000 and developed with the Western-Star chemiluminescent substrate. Immunoprecipitation was also performed with 2 μg of MSP5-specific MAb/ml and protein A/G agarose (Thermo Scientific), and the immunoprecipitate (IP) was electrophoresed, transferred to nitrocellulose, probed with antibodies, and developed as described above. Each polyclonal antibody and anti-MSP5 used for immunoprecipitation pulled down the respective target protein.
In addition, immunoprecipitation reactions were performed with rabbit anti-VirB9-1 IgG as described above, and the IPs were electrophoresed in a single large well, transferred to nitrocellulose, and strips were cut and probed with 2 μg of mouse MAb specific for VirB9-1, VirB9-2, VirB10, MSP2, MSP5, or B. bovis MSA1 (23/10.41, IgG2b) (22)/ml. Secondary antibody was AP-conjugated goat anti-mouse IgG+IgM diluted 1:10,000. Blots were developed with Western-Star chemiluminescent substrate for 30 min or longer.
RESULTS
Recombinant T4SS proteins.
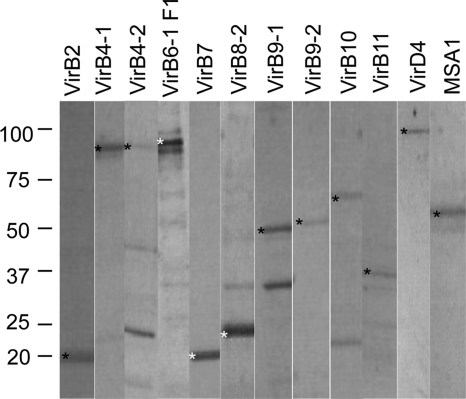
All proteins were expressed with their full-length, with the exception of VirB6-1 F1, for which the first fragment was used because the full-length protein did not express (Fig. 1). Some additional weak bands were observed (i.e., for VirB4-2, VirB8-2, and VirB11). For VirB4-2, these reacted with anti-HIS MAb on immunoblotting, suggesting they are degraded protein fragments (see Fig. S2 in the supplemental material). Visualization of immunoblots of rVirB9-1, rVirB9-2, and rVirB10 probed with anti-FLAG MAb indicated that the purified proteins migrated at 50, 50, and 65 kDa, as well as at 100, 100, and 130 kDa, respectively (Fig. 2A). The corresponding weak bands on Coomassie blue-stained gels (Fig. 2B) were excised for MS/MS analysis, rVirB9-1 and rVirB9-2 were each identified in their respective 50- and 100-kDa bands, and VirB10 was identified in both 65- and 130-kDa bands (Table 1). Because other A. marginale proteins were not identified, this is consistent with the formation of homodimers by rVirB9-1, rVirB9-2, and rVirB10. The same procedure was followed with rVirB7 bands at 20 and 40 kDa (Fig. 2); however, there was no evidence that rVirB7 dimerized. The 20-kDa band was identified as VirB7, and the 40-kDa band was identified as E. coli thioredoxin 1 (Table 1). Although faint bands migrating at ∼140 kDa were observed for VirB9-1 and VirB9-2 on some immunoblots (Fig. 2A), there were no corresponding protein bands visible on the gel, even when 10 time more protein was loaded (Fig. 2B), so these were not analyzed by MS/MS. These may represent protein trimers or multimers with other T4SS proteins (13, 17). In addition to the protein bands that were identified as dimers, products of VirB9-1, VirB9-2, and VirB10 that migrated below the dimeric and monomeric forms were visualized on both the gels and Western blots. Because these bands react with anti-FLAG MAb, which is highly specific for this epitope, these bands likely represent degradation products of the individual proteins resulting in bands smaller than the monomers and dimers, as previously observed for these proteins (33).
Fig 1.
Purity of recombinant T4SS proteins. Recombinant T4SS proteins were loaded at 1 μg/well, separated on 4 to 20% gradient SDS-PAGE gels, and stained with Coomassie blue dye. The proteins were electrophoresed on separate gels, and scanned images were arranged as presented. An asterisk indicates the predicted molecular mass of the recombinant protein, indicated in parentheses as follows: rVirB2 (20 kDa), rVirB4-1 (90 kDa), rVirB4-2 (91 kDa), rVirB6-1 F1 (90 kDa), rVirB7 (20 kDa), rVirB8-2 (23 kDa), rVirB9-1 (46 kDa), rVirB9-2 (46 kDa), rVirB10 (65 kDa), rVirB11 (38 kDa), rVirD4 (93 kDa), and control rMSA1 (58 kDa).
Fig 2.
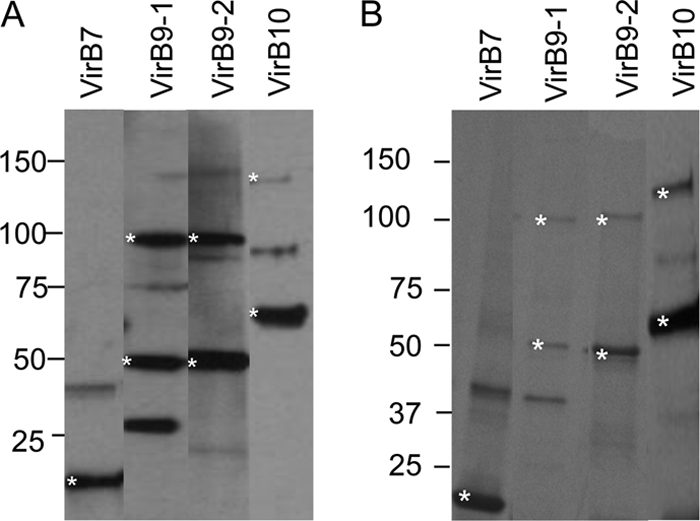
Evidence for dimerization of rVirB9-1, rVirB9-2, and rVirB10. (A) Recombinant proteins (1 μg) were transferred from a 4 to 20% gradient SDS-PAGE gel to a nitrocellulose membrane that was probed with anti-His MAb (rVirB7) or anti-FLAG MAb (rVirB9-1, rVirB9-2, and rVirB10) and developed. Scanned images were rearranged and presented as shown. The results for VirB7 were obtained in a separate experiment. (B) Coomassie blue-stained SDS-PAGE gel corresponding to the Western blot in panel A, with the exception that 10 μg of protein was loaded in each lane to visualize the faint bands. Asterisks denote bands where the A. marginale T4SS protein was identified by MS/MS analysis.
Table 1.
Evidence for dimerization of VirB9-1, VirB9-2, and VirB10
| Protein (mass [kDa])a | Mascot scoreb |
|---|---|
| rVirB7 (20) | 40 |
| rVirB7 (40) | 172c |
| rVirB9-1 (50) | 249 |
| rVirB9-1 (100) | 210 |
| rVirB9-2 (50) | 243 |
| rVirB9-2 (100) | 62 |
| rVirB10 (65) | 99 |
| rVirB10 (130) | 223 |
The molecular masses of protein monomers and dimers are indicated in parentheses.
Using an A. marginale-specific search, Mascot ion scores greater than 19 are significant at P < 0.05.
The Mascot ion score is an E. coli-specific search and identified as thioredoxin 1; no significant hits for A. marginale were detected.
IgG responses to T4SS proteins in OM vaccinees.
All six OM-immunized cattle produced IgG against VirB9-1, VirB9-2, and VirB10, with VirB9-1 inducing the highest titer tested of at least 10,000 (Table 2). Cattle 35113, 35141, 35160, 35280, and 35287 produced strong IgG responses directed at VirB7 with titers of at least 10,000. Cattle 35280 and 35287 also produced IgG against VirB4-2. However, none of the animals produced detectable IgG responses against VirB2, VirB4-1, VirB6-1 F1, VirB8-2, VirB11, or VirD4 compared to preimmunization sera.
Table 2.
IgG responses to recombinant T4SS proteins in cattle immunized with outer membranes
| Proteina | IgG titers from animalb: |
|||||
|---|---|---|---|---|---|---|
| 35113 (11/22) | 35114 (22/24) | 35160 (3/16) | 35280 (16/27) | 35287 (16/22) | 583 (8/23) | |
| VirB2 | <100 | <100 | <100 | <100 | <100 | <100 |
| VirB4-1 | <100 | <100 | <100 | <100 | <100 | <100 |
| VirB4-2 | <100 | <100 | <100 | 1,000 | 3,000 | <100 |
| VirB6-1 F1 | <100 | <100 | <100 | <100 | <100 | <100 |
| VirB7 | ≥10,000 | ≥10,000 | ≥10,000 | ≥10,000 | ≥10,000 | <100 |
| VirB8-2 | <100 | <100 | <100 | <100 | <100 | <100 |
| VirB9-1 | ≥10,000 | ≥10,000 | ≥10,000 | ≥10,000 | ≥10,000 | ≥10,000 |
| VirB9-2 | 3,000 | 1,000 | ≥10,000 | ≥10,000 | 3,000 | ≥10,000 |
| VirB10 | 3,000 | 1,000 | ≥10,000 | 3,000 | 3,000 | ≥10,000 |
| VirB11 | <100 | <100 | <100 | <100 | <100 | <100 |
| VirD4 | <100 | <100 | <100 | <100 | <100 | <100 |
T4SS proteins were expressed in E. coli and purified. Each protein was used at 1 or 2 μg/well and transferred.
Sera were diluted and tested for reactivity to recombinant proteins on immunoblots. A value of <100 indicates that no IgG response was detected at a 1:100 dilution. Positive sera recognizing the antigen at the predicted molecular weight and when preimmune sera were negative were diluted 1:300 to 1:10,000. The titer is defined as the reciprocal of the highest serum dilution giving a positive signal. Animal numbers (DRB3 RFLP types) are indicated in the column subheadings.
T-lymphocyte responses to T4SS proteins in OM vaccinees.
Two-week T-cell lines enriched for CD4+ T cells from the six OM-immunized cattle were tested for antigen specific proliferation to recombinant T4SS proteins (Table 3). VirB9-1, VirB9-2, and VirB10 were highly immunogenic for the majority of the animals. However, animals 35160 and 583 did not respond to VirB9-1, animals 35160 and 35280 did not respond to VirB9-2, and animal 35280 did not respond to VirB10. Animals 35160 and 35287 had significant T-cell responses to VirB2, and animal 35160 had a significant T-cell response to VirD4. Interestingly, 35160 was the only animal that had T-cell responses to VirB6-1, and 35287 was the only animal that responded to VirB4-2. Lastly, there was no detectable T-cell response from any animal to VirB4-1, VirB7, VirB8-2 and VirB11 antigens. These results are representative of three or more experiments.
Table 3.
T-cell responses to recombinant A. marginale T4SS proteins in Holstein cattle with different MHC class II haplotypes
| Antigena | Proliferation of T cells from animalb: |
|||||
|---|---|---|---|---|---|---|
| 35113 (11/22) | 35141 (22/24) | 35160 (3/16) | 35280 (16/27) | 35287 (16/22) | 583 (8/23) | |
| OM | ||||||
| 1 | 736.5 ± 5.7 | 21.9 ± 4.2 | 250.1 ± 13.1 | 50.2 ± 1.8 | 259.4 ± 9.2 | 220.0 ± 13.4 |
| 10 | 941.1 ± 52.8 | 36.7 ± 9.7 | 252.2 ± 9.2 | 44.0 ± 2.1 | 251.1 ± 9.2 | 312.1 ± 2.1 |
| URBC | ||||||
| 1 | 0.2 ± 0.1 | 0.8 ± 0.3 | 0.6 ± 0.1 | 0.3 ± 0.1 | 0.6 ± 0.2 | 0.4 ± 0.3 |
| 10 | 0.9 ± 0.7 | 1.6 ± 0.6 | 0.3 ± 0.1 | 0.8 ± 0.4 | 2.0 ± 2.6 | 0.9 ± 0.9 |
| MSA1 | ||||||
| 1 | 1 ± 1.3 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.4 ± 0.3 | 0.3 ± 0.2 | 0.1 ± 0.0 |
| 10 | 0.3 ± 0.1 | 0.8 ± 0.4 | 1.4 ± 0.2 | 0.4 ± 0.1 | 0.9 ± 0.5 | 0.1 ± 0.0 |
| VirB2 | ||||||
| 1 | 0.4 ± 0.1 | 2.0 ± 0.2 | 191.7 ± 33.6 | 9.4 ± 4.4 | 62.7 ± 17.1 | 3.4 ± 0.5 |
| 10 | 1.5 ± 0.3 | 2.8 ± 0.2 | 46.7 ± 2.2 | 3.3 ± 1.4 | 155.0 ± 2.5 | 7.9 ± 2.0 |
| VirB4-1 | ||||||
| 1 | 1.6 ± 0.7 | 0.5 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.0 | 1.0 ± 0.3 | 1.0 ± 0.2 |
| 10 | 1.0 ± 0.5 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.7 ± 0.2 | 2.6 ± 1.0 |
| VirB4-2 | ||||||
| 1 | 1.3 ± 0.1 | 0.3 ± 0.0 | 0.2 ± 0.2 | 0.2 ± 0.1 | 2.4 ± 1.9 | 1.3 ± 1.1 |
| 10 | 1.1±.1 | 0.1 ± 0.0 | 0.2 ± 0.2 | 0.1 ± 0.0 | 22.8 ± 19.6 | 1.8 ± 0.0 |
| VirB6-1 F1 | ||||||
| 1 | 3.0 ± 0.7 | 2.6 ± 0.7 | 8.2 ± 3.2 | 0.9 ± 0.3 | 0.9 ± 0.3 | 1.5 ± 1.1 |
| 10 | 0.7 ± 0.2 | 2.1 ± 0.4 | 78.4 ± 4.7 | 1.0 ± 0.2 | 1.5 ± 0.8 | 0.8 ± 0.1 |
| VirB7 | ||||||
| 1 | 0.5 ± 0.2 | 0.5 ± 0.1 | 0.7 ± 0.1 | 0.8 ± 0.1 | 1.2 ± 1.0 | 0.7 ± 0.1 |
| 10 | 2.0 ± 0.4 | 1.1 ± 0.3 | 2.2 ± 0.4 | 2.3 ± 2.1 | 1.8 ± 0.7 | 2.0 ± 0.8 |
| VirB8-2 | ||||||
| 1 | 1.3 ± 0.1 | 0.2 ± 0.0 | 0.7 ± 0.9 | 0.0 ± 0.0 | 0.9 ± 0.1 | 1.6 ± 0.5 |
| 10 | 0.9 ± 0.1 | 0.1 ± 0.0 | 0.1 ± 0.1 | 0.0 ± 0.0 | 1.2 ± 0.6 | 1.7 ± 0.6 |
| VirB9-1 | ||||||
| 1 | 385.7 ± 36.5 | 22.5 ± 4.2 | 0.7 ± 0.1 | 46.8 ± 11.2 | 264.7 ± 10.6 | 0.1 ± 0.1 |
| 10 | 779.3 ± 71.4 | 8.0 ± 4.6 | 0.9 ± 0.2 | 103.7 ± 16.4 | 239.8 ± 11.5 | 0.1 ± 0.0 |
| VirB9-2 | ||||||
| 1 | 139.2 ± 11.7 | 35.3 ± 16.0 | 0.8 ± 0.1 | 8.0 ± 2.8 | 32.8 ± 4.1 | 25.9 ± 4.4 |
| 10 | 201.5 ± 27.1 | 32.8 ± 0.7 | 1.7 ± 0.8 | 11.5 ± 2.1 | 109.6 ± 4.3 | 303.7 ± 19.5 |
| VirB10 | ||||||
| 1 | 35.5 ± 3.6 | 3.7 ± 0.5 | 90.6 ± 9.4 | 0.1 ± 0.0 | 96.2 ± 11.6 | 6.7 ± 2.7 |
| 10 | 381.2 ± 39.0 | 5.8 ± 0.3 | 126.2 ± 5.6 | 0.3 ± 0.0 | 162.5 ± 2.3 | 72.9 ± 8.6 |
| VirB11 | ||||||
| 1 | 1.8 ± 0.1 | 0.6 ± 0.1 | 0.1 ± 0.0 | 0.1 ± 0.0 | 1.0 ± 0.4 | 0.0 ± 0.0 |
| 10 | 3.2 ± 1.4 | 0.7 ± 0.2 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.8 ± 0.2 | 0.1 ± 0.0 |
| VirD4 | ||||||
| 1 | 0.4 ± 0.1 | 2.7 ± 0.4 | 30.4 ± 6.2 | 0.3 ± 0.1 | 0.4 ± 0.1 | 2.8 ± 1.2 |
| 10 | 0.5 ± 0.0 | 1.7 ± 0.1 | 99.7 ± 6.4 | 0.5 ± 0.1 | 0.5 ± 0.0 | 6.3 ± 4.0 |
Recombinant T4SS proteins were used at 1 and 10 μg/ml in the T-cell assays, as indicated. A. marginale OM and URBC were positive and negative controls, respectively.
A stimulation index (SI) was calculated as the mean cpm of a 2-week CD4 T-cell line to an individual protein/the mean cpm to medium. Results in boldface indicate significantly greater T-cell responses to a T4SS protein compared to the response to the same concentration of MSA1, where P was <0.05 using the Dunnett's test. Animal numbers (DRB3 RFLP types) are indicated in the column subheadings.
Selection of linked recognition candidates.
Selection of T4SS protein candidates that could undergo linked recognition was based on predicted surface localization, as well as induction of an IgG response, but not a significant CD4 T-cell response, in the same individual. Based on previous results with A. phagocytophilum and A. marginale (2, 18, 42, 43, 61) and bioinformatic predictions for A. marginale (see Fig. S3 and Table S1 in the supplemental material), VirB2, VirB6-1, VirB7, VirB8-2, VirB9-1, VirB9-2, and VirB10 are predicted to be localized on the exterior surface of A. marginale. Therefore, the ability of any of these proteins to induce IgG without a significant CD4 T-cell response would make it a candidate for linked recognition and inclusion in future vaccine trials, as surface exposed proteins could be blocked by neutralizing antibody. These candidates are VirB9-1 for animal 583 (DRB3 RFLP type 8/23), VirB9-1, and VirB9-2 for animal 35160 (3/16), VirB9-2 and VirB10 for animal 35280 (16/27), and VirB7 for all animals except 583 (summarized in Table 4). These proteins were therefore selected to identify protein partners within the T4SS that had T-cell epitopes recognized by these animals.
Table 4.
Summary of IgG and T-cell responses to A. marginale T4SS proteins in six Holstein cattlea
| Antigen | OM-immunized animalb: |
|||||
|---|---|---|---|---|---|---|
| 35113 (11/22) | 35141 (22/24) | 35160 (3/16) | 35280 (16/27) | 35287 (16/22) | 583 (8/23) | |
| VirB2 | ||||||
| IgG | − | − | − | − | − | − |
| T cells | − | − | + | − | + | − |
| VirB4-1 | ||||||
| IgG | − | − | − | − | − | − |
| T cells | − | − | − | − | − | − |
| VirB4-2 | ||||||
| IgG | − | − | − | + | + | − |
| T cells | − | − | − | − | + | − |
| VirB7 | ||||||
| IgG | +* | +* | +* | +* | +* | − |
| T cells | −* | −* | −* | −* | −* | − |
| VirB6-1 F1 | ||||||
| IgG | − | − | − | − | − | − |
| T cells | − | − | + | − | − | − |
| VirB8-2 | ||||||
| IgG | − | − | − | − | − | − |
| T cells | − | − | − | − | − | − |
| VirB9-1 | ||||||
| IgG | + | + | +* | + | + | +* |
| T cells | + | + | −* | + | + | −* |
| VirB9-2 | ||||||
| IgG | − | − | +* | +* | − | + |
| T cells | + | + | −* | −* | + | + |
| VirB10 | ||||||
| IgG | + | + | + | +* | + | + |
| T cells | + | + | + | −* | + | + |
| VirB11 | ||||||
| IgG | − | − | − | − | − | − |
| T cells | − | − | − | − | − | − |
| VirD4 | ||||||
| IgG | − | + | − | − | − | − |
| T cells | − | − | + | − | − | − |
DRB3 RFLP types are indicated in parentheses in the column subheadings. A negative symbol (−) indicates that IgG titers were <100 or that the T-cell response was insignificant. A positive symbol (+) indicates an IgG titer was ≥300 or that the T-cell response was significant.
Candidates for linked recognition.
Detecting the interactions of VirB9-1, VirB9-2, and VirB10 by far-Western blotting.
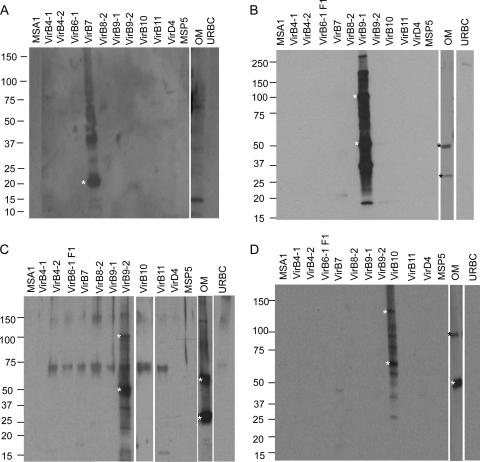
Protein A-purified rabbit IgG against rVirB7, rVirB9-1, rVirB9-2, and rVirB10 was tested for antigen specificity by immunoblotting with native OM and recombinant T4SS proteins (Fig. 3). In repeated assays, anti-VirB7 detected a band in the OM that was greater than the predicted native mass of ∼6 kDa (Fig. 3A). In contrast, IgG against rVirB9-1, rVirB9-2, and rVirB10 recognized the native OM protein at the predicted molecular weight and at a 2-fold-higher molecular weight (Fig. 3B to D). In addition, anti-VirB9-2 detected a fainter, narrow band at ∼140 kDa, which may represent a multimer of the native protein or a multimer of VirB9-2 and another protein(s). When recombinant proteins were used, antibody against rVirB7 reacted with 20- and 40-kDa bands in rVirB7, but not with the other T4SS proteins (Fig. 3A). For rVirB9-1 (Fig. 3B), rVirB9-2 (Fig. 3C), and rVirB10 (Fig. 3D) IgG bound specifically to its target protein and at a band of approximately twice the molecular weight, which we showed are dimers (Fig. 2 and Table 1). Bands larger than the dimer that are present in the immunizing protein detected by the IgGs against VirB9-1 and VirB9-2 may represent multimers of the VirB9 protein with other T4SS proteins, since it is known that the A. tumefaciens core-complex has 14 copies of VirB9 that interact with 14 copies of VirB7 and 14 copies of VirB10 (17). Importantly, the IgGs did not bind to any other T4SS protein, showing that the IgGs were specific for the immunizing protein. An artifactual band often observed in immunoblots with polyclonal rabbit antisera that was determined to be keratin (45) was observed with the anti-VirB9-2 antibody, which detects a diffuse band at ∼68 kDa and another at ∼140 kDa (Fig. 3C). Because of the characteristic sizes and widths of these bands, they are assumed to be contaminating keratin monomers and dimers. These bands were only observed with this antibody and detected in only certain recombinant proteins, but not in native OM. We were not able to eliminate this nonspecific keratin band by lowering the amount of reducing agent in the sample buffer as suggested by others (30).
Fig 3.
Specificity of rabbit polyclonal antibodies against VirB7, VirB9-1, VirB9-2, and VirB10. Purified recombinant T4SS proteins (1 μg) and 10 μg of A. marginale OM (native proteins) were separated on a 4 to 20% gradient SDS-PAGE gel, transferred to a nitrocellulose membrane, and detected by Western blotting with 2 μg of purified, E. coli-adsorbed polyclonal rabbit IgG/ml. (A) anti-VirB7; (B) anti-VirB9-1; (C) anti-VirB9-2; (D) anti-VirB10. Scanned images were rearranged and presented as shown in the figure. The predicted molecular masses for recombinant (r) and native (n) proteins are indicated in parentheses: rVirB7 (20 kDa), nVirB7 (6 kDa), rVirB9-1 (46 kDa), nVirB9-1 (30 kDa), rVirB9-2 (46 kDa), nVirB9-2 (30 kDa), rVirB10 (65 kDa), and nVirB10 (49 kDa). The bands with the predicted sizes of monomeric and dimeric (VirB9-1, VirB9-2, and VirB10) proteins are indicated by asterisks.
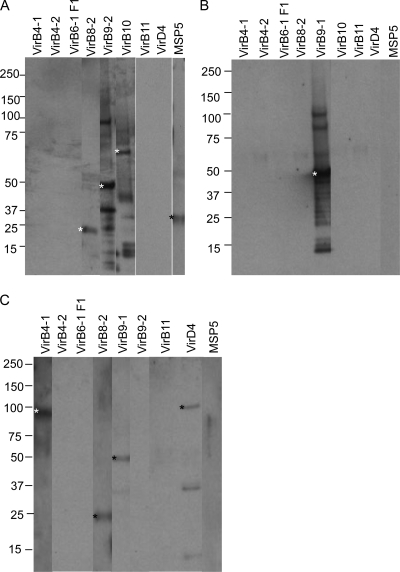
The far-Western blotting strategy was performed as a screening tool to determine which rT4SS proteins bind to rVirB7, rVirB9-1, rVirB9-2, and rVirB10 that could enable them to undergo linked recognition (Fig. 4). The far-Western blots developed with anti-VirB7 failed to detect potential interacting proteins (data not shown), so proteins interacting with VirB7 were not pursued further. However, the results indicate several interactions among T4SS proteins. Recombinant VirB9-1 interacted with rVirB8-2 at the correct molecular mass of 23 kDa; with rVirB9-2 at the predicted mass of 46 kDa, as well as with a predicted dimer and with apparent degradation products; with rVirB10 at the predicted mass of 65 kDa, as well as with apparent degradation products; and with rMSP5 at the predicted mass of 28 kDa (Fig. 4A). Recombinant VirB9-2 interacted with rVirB9-1 at the predicted mass of 46 kDa, as well as with a predicted dimer and apparent degradation products (Fig. 4B). Recombinant VirB10 interacted with rVirB4-1 at the predicted mass of 91 kDa, with rVirB8-2 at the predicted mass of 23 kDa, with rVirB9-1 at the predicted mass of 46 kDa, and with rVirD4 at the predicted mass of 93 kDa and two smaller bands (Fig. 4C). The reactivity of degradation products of these proteins with anti-His MAb can be seen in Fig. S2 in the supplemental material.
Fig 4.
Far-Western blotting to detect interacting recombinant T4SS proteins with specific targets. The indicated purified recombinant T4SS proteins (1 μg) were separated on 4 to 20% gradient SDS-PAGE gels, transferred to a nitrocellulose membrane, and incubated with 10 μg of target protein rVirB9-1 (A), rVirB9-2 (B), and rVirB10 (C). The interactions were detected with E. coli-adsorbed rabbit antisera (1:500) specific for VirB9-1, VirB9-2, and VirB10. Blots were developed with goat anti-rabbit IgG(H+L). Scanned images were rearranged and presented as shown. In panel A, the results for VirB11 and VirD4 were obtained from a separate experiment. An asterisk indicates an interacting protein partner to the target protein with the following predicted molecular masses: rVirB4-1 (90 kDa), rVirB4-2 (91 kDa), rVirB6-1 F1 (90 kDa), rVirB8-2 (23 kDa), rVirB9-1 (46 kDa), rVirB9-2(46 kDa), rVirB10 (65 kDa), rVirB11 (38 kDa), rVirD4 (93 kDa), and MSP5 (28 kDa).
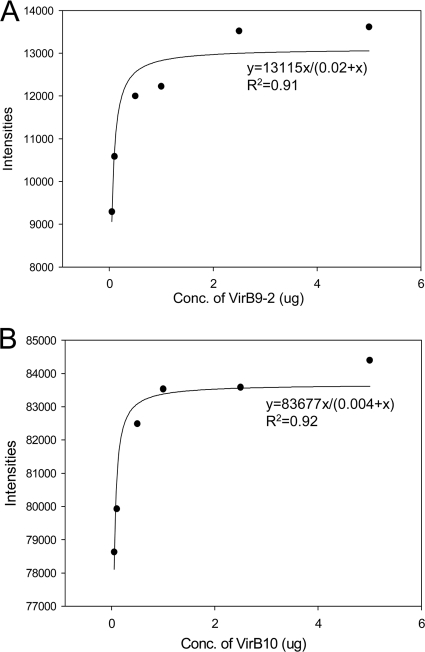
Binding coefficients for VirB9-1 interactions.
The binding coefficient for rVirB9-1 interaction with rVirB9-2 was 0.0173 ± 0.0034 μM and that for rVirB9-1 interaction with rVirB10 was 0.0109 ± 0.0094 μM (Fig. 5). The binding coefficients for the reverse of the interactions were also determined and found to be similar (rVirB9-2 with rVirB9-1, KD = 0.0205 ± 0.0173 μM; and rVirB10 with rVirB9-1, KD = 0.0135 ± 0.0080 μM; see Fig. S4 in the supplemental material), whereas there was no binding detected between VirB9-2 and VirB10 (see Fig. S4 in the supplemental material). The graphed intensities of the spots that yielded the above KD values all had a goodness of fit to the binding curve equation with at least 80% confidence (Fig. 5). These results indicate that rVirB9-1 has stronger binding to rVirB10 than to rVirB9-2.
Fig 5.
Single site saturation binding curves for the interactions of rVirB9-1 with rVirB9-2 and rVirB10. Densitometry analysis was performed to evaluate the interactions of rVirB9-1 with rVirB9-2 (A) and rVirB9-1 with rVirB10 (B) that were applied to dot blots at amounts up to 5 μg, allowed to interact with 10 μg of rVirB9-1, probed with rabbit anti-VirB9-1 antiserum, and developed. Reactions were compared to the density of the reaction of rVirB9-1 to negative control rMSA1. The data are graphically represented as concentration of the protein on the spot versus spot intensity, and the curve was fit to a single site binding curve to yield the binding association constant. The graphs are representative of three independent experiments.
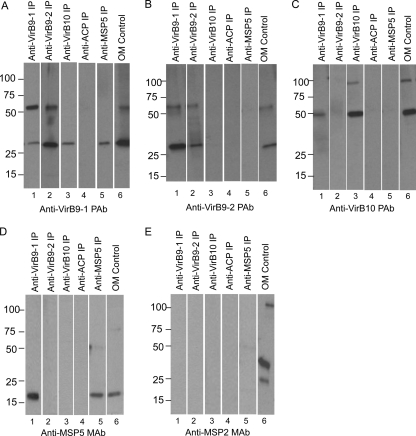
Native VirB9-1, VirB9-2, and VirB10 interactions determined by immunoprecipitation.
Because the anti-VirB7 antibody did not consistently bind to a native protein of the predicted molecular weight, we focused on using antibodies to VirB9-1, VirB9-2, and VirB10 to identify interacting native protein partners using immunoprecipitation of solubilized OM (Fig. 6). The rabbit IgG or mouse MSP5-specific MAb used for immunoprecipitation is shown at the top of each lane; the antibody used for detection is shown under each panel. For anti-VirB9-1 and anti-VirB9-2 immunoprecipitations, bands of the predicted size for VirB9-1 (29.7 kDa) and for VirB9-2 (30.6 kDa) were detected in both the IPs and the OM (Fig. 6A, lanes 1 and 6, and Fig. 6B, lanes 2 and 6, respectively). However, an additional band at ∼60 kDa was also detected with these antibodies. The 60-kDa bands could represent homodimers of these proteins, a heterodimer of VirB9-1 and VirB9-2, or binding to another 30-kDa protein. VirB9-2 (Fig. 6B, lane 1), VirB10 (Fig. 6C, lane 1), and 19-kDa MSP5 (Fig. 6D, lane 1) were also detected in the VirB9-1 IP, but the ∼37-kDa MSP2 was not (Fig. 6E, lane 1). The anti-VirB9-2 IP probed with anti-VirB9-1 IgG also showed ∼30- and ∼60-kDa bands (Fig. 6A, lane 2). VirB10 (Fig. 6C, lane 2), MSP5 (Fig. 6D, lane 2), and MSP2 (Fig. 6E, lane 2) were not detected in the anti-VirB9-2 IP. For the anti-VirB10 IP, a band with the predicted mass for VirB10 (49 kDa) was detected, along with an additional band at ∼100 kDa (Fig. 6C, lanes 3 and 6). This 100-kDa band could be a homodimer of VirB10 or an interaction of VirB10 with another protein(s). VirB9-1 was detected in this IP (Fig. 6A, lane 3), but VirB9-2 (Fig. 6B, lane 3), MSP5 (Fig. 6D, lane 3), and MSP2 (Fig. 6E, lane 3) were not. Immunoprecipitation reactions were also alkylated to prevent reformation of disulfide bonds, electrophoresed, and probed with anti-VirB9-1, anti-VirB9-2, anti-VirB10, and anti-ACP. However, reduction and alkylation had little effect on the interactions or apparent homodimerization (see Fig. S5 in the supplemental material), suggesting that the observed T4SS interactions are not formed with disulfide linkages. The negative control rabbit anti-ACP IgG immunoprecipitation did not pull down any T4SS protein, MSP5, or MSP2 (Fig. 6A to E, lane 4).
Fig 6.
Detection of native protein interactions of VirB9-1, VirB9-2, and VirB10. Solubilized A. marginale OM proteins were immunoprecipitated with purified rabbit IgG against VirB9-1 (lane 1), VirB9-2 (lane 2), VirB10 (lane 3), negative control B. bovis ACP (lane 4), and an MAb specific for MSP5 (lane 5) as indicated for each lane. Solubilized OM were included (lane 6) for a size comparison. Immunoprecipitated pellets and OM were electrophoresed and transferred to nitrocellulose membranes. Detection of the interacting proteins was performed by Western blotting individual strips with purified rabbit IgG against VirB9-1 (A), VirB9-2 (B), and VirB10 (C) and MAbs specific for MSP5 (D) and MSP2 (E), as indicated under each panel. The secondary antibody was Clear Blot-HRP (panels A to D and panel E, lane 5) or goat anti-mouse IgG+IgM (panel E, lanes 1 to 4 and 6). Scanned images were rearranged and presented as shown. The predicted molecular masses for native proteins are indicated in parentheses: VirB9-1 (30 kDa), VirB9-2 (30 kDa), VirB10 (49 kDa), B. bovis ACP (15 kDa), MSP5 (19 kDa), and MSP2 (37 kDa).
To confirm the interaction of native VirB9-1 with MSP5, immunoprecipitation was performed with MAb against MSP5, and it was determined that MSP5 does interact with VirB9-1 (Fig. 6A, lane 5), but not with VirB9-2 or VirB10 (Fig. 6B and C, lane 5, respectively). Furthermore, the highly abundant MSP2 (34) was never identified in the T4SS IPs (Fig. 6E, lanes 1 to 5), supporting the specificity of the detected T4SS interactions. Taken together, our results show that VirB9-1 binds to VirB9-2, VirB10, and MSP5, VirB9-2 only interacts with VirB9-1, VirB10 only interacts with VirB9-1, and native MSP5 interacts only with VirB9-1.
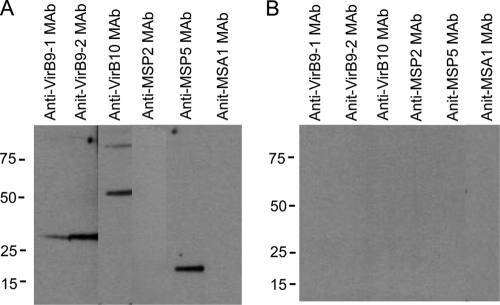
An additional confirmation to detect interactions was performed by following the same immunoprecipitation procedure with rabbit anti-VirB9-1 IgG but using MAbs against VirB9-1, VirB9-2, VirB10, MSP2, MSP5, and MSA1 to probe the Western blot (Fig. 7). The anti-VirB9-1 IP contained 30-kDa VirB9-1, 30-kDa VirB9-2, 49-kDa VirB10, and 19-kDa MSP5 but not negative control MSP2 or B. bovis MSA1 (Fig. 7A). There was no antibody reactivity with IPs using preimmune rabbit IgG (Fig. 7B). This result confirms that VirB9-1 interacts with VirB9-2, VirB10 and MSP5.
Fig 7.
MAb detection of interacting T4SS proteins immunoprecipitated by rabbit anti-VirB9-1 IgG. Solubilized OM proteins were immunoprecipitated with rabbit anti-ViB9-1 (A) or normal rabbit (B) IgG, and the pellets were electrophoresed and transferred to nitrocellulose membranes. Strips were cut and probed with MAbs specific for VirB9-1, VirB9-2, VirB10, MSP2, MSP5, and B. bovis MSA1, as indicated at the top of each lane. The secondary antibody was goat anti-mouse IgG+IgM. Scanned images were rearranged and are presented as shown. The predicted molecular masses for native proteins are indicated in parentheses: VirB9-1 (30 kDa), VirB9-2 (30 kDa), VirB10 (49 kDa), MSP2 (37 kDa), MSP5 (19 kDa), and B. bovis MSA1 (42 kDa).
Comparison of amino acid sequences for VirB9-1, VirB9-2, and VirB10 among A. marginale and A. centrale strains.
Predicted amino acid sequence alignments revealed that VirB9-1, VirB9-2, and VirB10 proteins are highly conserved across A. marginale strains. For VirB9-1 the amino acid identity is 97 to 100% (with the exception of the Mississippi strain for which the VirB9-1 sequence is incomplete), and for VirB9-2 and VirB10 the sequence identity is 100% (see Fig. S6 in the supplemental material). Furthermore, comparison of sequence identities of these proteins to those of the vaccine strain, A. centrale, also revealed a high degree of conservation, which was 98 to 99% identity for VirB9-1 with all A. marginale strains (except Mississippi), 92% for VirB9-2, and 93% for VirB10 for all A. marginale strains (see Fig. S6 in the supplemental material). Because of this high degree of amino acid sequence identity, these T4SS proteins would be good candidates for inclusion in a cross-protective vaccine.
DISCUSSION
This study sought to identify A. marginale T4SS protein candidates for linked recognition that could be incorporated into a vaccine. Linked recognition is a well-known rationale for designing vaccines against pathogens with capsular polysaccharide antigens, which on their own are poorly antigenic. Examples include vaccines for Haemophilus influenzae and Streptococcus pneumoniae, where T-dependent protein antigens are linked to the T-independent polysaccharide to achieve neutralizing antibody responses directed against the polysaccharide (26, 52). Immunization studies using two or more associated proteins are also suggestive that linked recognition enhances protection against bacterial pathogens. For example, protective immunity was induced with bacterial membrane protein complexes or fusion proteins of surface exposed type III secretion system proteins of Yersinia pestis (6, 23). Association of proteins within the context of the membrane was also important for leptospiral outer membrane porin OmpL1 and lipoprotein LipL41 in protection studies (21).
Consistent with previous results (32–34, 56) VirB9-1, VirB9-2, and VirB10 are highly immunogenic and in the present study were the most immunogenic of the 11 A. marginale T4SS proteins examined for cattle with diverse MHC class II haplotypes. We confirmed that cattle with the DRB3 RFLP type 8/23 (animal 583 in the present study and animal 04B91 [33, 56]) have an IgG response, but no T-cell response, to VirB9-1. This suggests that for animals with this haplotype, T-cell help for IgG production is provided through linked recognition by a different, but associated protein. In addition, we have new evidence that linked recognition occurs for cattle with the DRB3 RFLP type 3/16 to produce IgG to VirB9-1 and VirB9-2 (animal 35160), DRB3 RFLP type 16/27 to produce IgG to VirB9-2 and VirB10 (animal 35280), and DRB3 RFLP types 11/22, 22/24, 3/16, 16/27, and 16/22 to produce IgG to VirB7 (animals 35113, 35141, 35160, 35280, and 35287, respectively). All of these proteins are likely to be surface exposed, as predicted by bioinformatics. Furthermore, these proteins were recognized by the immune sera of infected animals (2, 56, 61), and VirB10 was identified in outer membrane complexes following cross-linking with membrane-impermeable cross-linkers (43). Based on the criteria of predicted surface localization and pattern of immune recognition by three animals and haplotypes that had IgG but undetectable T-cell responses to one or two of these proteins, VirB7, VirB9-1, VirB9-2, and VirB10 were designated candidates for linked recognition. We then focused on identifying the interacting protein partners of VirB9 and VirB10 proteins within the T4SS.
Far-Western blotting was used as an initial screening tool to identify potential interacting T4SS proteins. The data suggest that recombinant VirB10 interacts with rVirB4-1, rVirB8-2, VirB9-1, and rVirD4, interactions that have also been observed with homologous proteins of Agrobacterium tumefaciens (3, 5, 15). The interactions of rVirB10 with rVirB4-1 and rVirD4, two putative NTPases, have also been observed for Helicobacter pylori and Rickettsia sibrica (37, 58) and implicate VirB10 in substrate shuttling (11). The VirB4-1 gene is more conserved across the Rickettsiales than VirB4-2, and VirB4-2 has more insertions and deletions, including those in the NTP-binding cleft, relative to other bacteria (19). This is consistent with the observed interaction of VirB10 with VirB4-1 and not with VirB4-2. Recombinant VirB10 and rVirB9-1 both appear to interact with rVirB8-2, indicating that VirB8-2 may also be part of the core complex of the T4SS apparatus (15, 27). However, VirB8-2 was not of particular interest to pursue because it was not antigenic for the OM vaccinees used in the present study. The interaction of rVirB9-1 and rVirB9-2 had not been previously reported, because only bacteria in the family Anaplasmatacae are known to express two full-length VirB9 proteins (19, 20). The interaction of rVirB9-1 with rMSP5 is interesting since none of the other proteins interacted with MSP5. However, it is unknown if MSP5 has T-cell epitopes to stimulate T-cell help to VirB9-1 in 3/16 and 8/23 DRB3 RFLP-typed animals. The interactions of rVirB9 and rVirB10 proteins were confirmed for native proteins in solubilized A. marginale OM using pulldown assays with polyclonal, monospecific antibodies and detection with both polyclonal and monoclonal antibodies. These interactions and the potential formation of homodimers are consistent with what is known about the A. tumefaciens T4SS core complex structure (13, 17). However, this is the first study to report interacting T4SS proteins of the A. marginale, as well as the first to document interactions of the VirB9 and VirB10 proteins in the Anaplasmatacae.
The immunogenicity of VirB9 and VirB10 proteins may be explained by the lack of surface lipopolysaccharide (LPS) leading to increased surface exposure of these proteins that form the outer cap of the T4SS core complex (7, 17, 19, 20). Although Rickettsia spp. synthesize LPS, the Anaplasmataceae do not (20). In agreement with our results and the observation that cattle infected with A. marginale have antibody to VirB9 and VirB10 proteins (2, 56, 61), dogs infected with Ehrlichia canis were also serologically positive for VirB9 (16). Thus, the T4SS VirB9 and VirB10 proteins may generally be good vaccine targets for the Anaplasmatacae.
A combination of cellular and humoral immunity is likely important for the control of anaplasmosis, and vaccine antigens should be highly conserved across multiple strains to ensure cross-protective immunity. Furthermore, inclusion in a vaccine of multiple linked proteins that are naturally associated in the membrane of A. marginale will not only provide more than one immunogenic protein but will offer the opportunity for linked recognition. Immunization with several naturally associated, linked proteins could increase T-cell-dependent IgG responses upon infection in outbred populations that express a broad repertoire of MHC class II molecules. Protecting a large population of genetically heterogeneous individuals requires understanding of MHC class II-restricted epitope presentation. The naturally associated T4SS protein complex that spans the inner and outer membrane provides a model system to examine the importance of membrane protein-protein interactions in stimulating protective immunity. Thus, a protein complex made up of highly conserved VirB9-1, VirB9-2, and VirB10 will be tested as a vaccine in a future study to protect MHC haplotype-defined cattle against A. marginale.
Supplementary Material
ACKNOWLEDGMENTS
The technical assistance of Shelley Whidbee, Beverly Hunter, Bruce Mathison, Nishant Dwivedi, and Emma Karel is appreciated. We also thank Marina Caballero for providing the ACP-specific antibody.
This study was supported by National Institutes of Health/NIAID grant R01 AI053692. K.M. was supported in part by a National Institutes of Health predoctoral fellowship in Protein Biotechnology (T32 GM008336).
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Abbott JR, et al. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J. Immunol. 174: 6702–6715 [DOI] [PubMed] [Google Scholar]
- 2. Araujo FB, et al. 2008. IgG and IgG2 antibodies from cattle naturally infected with Anaplasma marginale recognize the recombinant vaccine candidate antigens VirB9, VirB10, and elongation factor-Tu. Mem. Inst. Oswaldo Cruz 103: 186–190 [DOI] [PubMed] [Google Scholar]
- 3. Atmakuri K, Cascales E, Christie PJ. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bao W, et al. 2009. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuole. J. Bacteriol. 191: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beaupre CE, Bohne J, Dale EM, Binns AN. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhattacharya D, Mecsas J, Hu LT. 2010. Development of a vaccinia virus based reservoir-targeted vaccine against Yersinia pestis. Vaccine 28: 7683–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brayton KA, et al. 2005. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 102: 844–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown WC, Palmer GH, Lewin HA, McGuire TC. 2001. CD4+ T lymphocytes from calves immunized with Anaplasma marginale major surface protein 1 (MSP1), a heteromeric complex of MSP1a and MSP1b, preferentially recognize the MSP1a carboxyl terminus that is conserved among strains. Infect. Immun. 69: 6853–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown WC, Rice-Ficht AC, Estes DM. 1998. Bovine type 1 and type 2 responses. Vet. Immunol. Immunopathol. 63: 45–55 [DOI] [PubMed] [Google Scholar]
- 10. Brown WC, et al. 1998. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66: 5406–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cascales E, Christie PJ. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. U. S. A. 101: 17228–17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan CS, Winstone TM, Turner RJ. 2008. Investigating protein-protein interactions by far-Westerns. Adv. Biochem. Eng. Biotechnol. 110: 195–214 [DOI] [PubMed] [Google Scholar]
- 13. Chandran V, et al. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59: 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das A, Xie YH. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182: 758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Felek S, Huang H, Rikihisa Y. 2003. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect. Immun. 71: 6063–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fronzes R, et al. 2009. Structure of a type IV secretion system core complex. Science 323: 266–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ge Y, Rikihisa Y. 2007. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J. Bacteriol. 189: 7819–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gillespie JJ, et al. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4: e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillespie JJ, et al. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 78: 1809–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haake DA, et al. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 67: 6572–6582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hines SA, McElwain TF, Buening GM, Palmer GH. 1989. Molecular characterization of Babesia bovis merozoite surface proteins bearing epitopes immunodominant in protected cattle. Mol. Biochem. Parasitol. 37: 1–10 [DOI] [PubMed] [Google Scholar]
- 23. Ivanov MI, et al. 2008. Vaccination of mice with a Yop translocon complex elicits antibodies that are protective against infection with F1 Yersinia pestis. Infect. Immun. 76: 5181–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Judd PK, Kumar RB, Das A. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. U. S. A. 102: 11498–11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Judd PK, Kumar RB, Das A. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55: 115–124 [DOI] [PubMed] [Google Scholar]
- 26. Khan AQ, Lees A, Snapper CM. 2004. Differential regulation of IgG anti-capsular polysaccharide and antiprotein responses to intact Streptococcus pneumoniae in the presence of cognate CD4 T cell help. J. Immunol. 172: 532–539 [DOI] [PubMed] [Google Scholar]
- 27. Kumar RB, Xie YH, Das A. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36: 608–617 [DOI] [PubMed] [Google Scholar]
- 28. Lai EM, Chesnokova O, Banta LM, Kado CI. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182: 3705–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai EM, Kado CI. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180: 2711–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee TF, McNellis TW. 2008. Elimination of keratin artifact bands from Western blots by using low concentrations of reducing agents. Anal. Biochem. 382: 141–143 [DOI] [PubMed] [Google Scholar]
- 31. Leroith T, et al. 2005. Sequence variation and immunologic cross-reactivity among Babesia bovis merozoite surface antigen 1 proteins from vaccine strains and vaccine breakthrough isolates. Infect. Immun. 73: 5388–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lopez JE, et al. 2008. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J. Immunol. Methods 332: 129–141 [DOI] [PubMed] [Google Scholar]
- 33. Lopez JE, et al. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect. Immun. 75: 2333–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lopez JE, et al. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73: 8109–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macmillan H, et al. 2006. Analysis of the Anaplasma marginale major surface protein 1 complex protein composition by tandem mass spectrometry. J. Bacteriol. 188: 4983–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macmillan H, Norimine J, Brayton KA, Palmer GH, Brown WC. 2007. Physical linkage of naturally complexed bacterial outer membrane proteins enhances immunogenicity. Infect. Immun. 76: 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malek JA, et al. 2004. Protein interaction mapping on a functional shotgun sequence of Rickettsia sibirica. Nucleic Acids Res. 32: 1059–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGuire TC, Palmer GH, Goff WL, Johnson MI, Davis WC. 1984. Common and isolate-restricted antigens of Anaplasma marginale detected with monoclonal antibodies. Infect. Immun. 45: 697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mitchison NA. 1971. The carrier effect in the secondary response to hapten-protein conjugates. I. Measurement of the effect with transferred cells and objections to the local environment hypothesis. Eur. J. Immunol. 1: 10–17 [DOI] [PubMed] [Google Scholar]
- 40. Mitchison NA. 1971. The carrier effect in the secondary response to hapten-protein conjugates. II. Cellular cooperation. Eur. J. Immunol. 1: 18–27 [DOI] [PubMed] [Google Scholar]
- 41. Mitchison NA. 1971. The carrier effect in the secondary response to hapten-protein conjugates. V. Use of antilymphocyte serum to deplete animals of helper cells. Eur. J. Immunol. 1: 68–75 [DOI] [PubMed] [Google Scholar]
- 42. Niu H, Rikihisa Y, Yamaguchi M, Ohashi N. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell Microbiol. 8: 523–534 [DOI] [PubMed] [Google Scholar]
- 43. Noh SM, et al. 2008. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect. Immun. 76: 2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Norimine J, Brown WC. 2005. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4+ T-lymphocyte responses. Immunogenetics. 57: 750–762 [DOI] [PubMed] [Google Scholar]
- 45. Ochs D. 1983. Protein contaminants of sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 135: 470–474 [DOI] [PubMed] [Google Scholar]
- 46. Palmer GH, Barbet AF, Davis WC, McGuire TC. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231: 1299–1302 [DOI] [PubMed] [Google Scholar]
- 47. Palmer GH, McElwain TF. 1995. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet. Parasitol. 57: 233–253 [DOI] [PubMed] [Google Scholar]
- 48. Palmer GH, McGuire TC. 1984. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J. Immunol. 133: 1010–1015 [PubMed] [Google Scholar]
- 49. Palmer GH, et al. 1988. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect. Immun. 56: 1526–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palmer GH, Rurangirwa FR, Kocan KM, Brown WC. 1999. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol. Today 15: 281–286 [DOI] [PubMed] [Google Scholar]
- 51. Park YH, et al. 2004. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 5: 29–39 [PubMed] [Google Scholar]
- 52. Rosenstein NE, Perkins BA. 2000. Update on Haemophilus influenzae serotype b and meningococcal vaccines. Pediatr. Clin. N. Am. 47: 337–352 [DOI] [PubMed] [Google Scholar]
- 53. Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie PJ. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183: 3642–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmidt-Eisenlohr H, et al. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181: 7485–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharif S, et al. 1998. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with occurrence of disease and milk somatic cell score in Canadian dairy cattle. Anim. Genet. 29: 185–193 [DOI] [PubMed] [Google Scholar]
- 56. Sutten EL, et al. 2010. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect. Immun. 78: 1314–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tebele N, McGuire TC, Palmer GH. 1991. Induction of protective immunity by using Anaplasma marginale initial body membranes. Infect. Immun. 59: 3199–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Terradot L, et al. 2004. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol. Cell Proteomics 3: 809–819 [DOI] [PubMed] [Google Scholar]
- 59. van Eijk MJ, Stewart-Haynes JA, Lewin HA. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23: 483–496 [DOI] [PubMed] [Google Scholar]
- 60. Vidotto MC, McGuire TC, McElwain TF, Palmer GH, Knowles DP., Jr 1994. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect. Immun. 62: 2940–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vidotto MC, Venancio EJ, Vidotto O. 2008. Cloning, sequencing and antigenic characterization of rVirB9 of Anaplasma marginale isolated from Parana State, Brazil. Genet. Mol. Res. 7: 460–466 [DOI] [PubMed] [Google Scholar]
- 62. Wu Y, Li Q, Chen XZ. 2007. Detecting protein-protein interactions by Far Western blotting. Nat. Protoc. 2: 3278–3284 [DOI] [PubMed] [Google Scholar]
- 63. Yokoyama WM. 1994. Production of monoclonal antibodies, p 2.5.2–2.5.17. In Coligan J. E. (ed), Current protocols in immunology, vol 1 Wiley Interscience, Inc, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.