Abstract
Antibiotic-induced changes in the intestinal microbiota predispose mammalian hosts to infection with antibiotic-resistant pathogens. Clostridium difficile is a Gram-positive intestinal pathogen that causes colitis and diarrhea in patients following antibiotic treatment. Clindamycin predisposes patients to C. difficile colitis. Here, we have used Roche-454 16S rRNA gene pyrosequencing to longitudinally characterize the intestinal microbiota of mice following clindamycin treatment in the presence or absence of C. difficile infection. We show that a single dose of clindamycin markedly reduces the diversity of the intestinal microbiota for at least 28 days, with an enduring loss of ca. 90% of normal microbial taxa from the cecum. Loss of microbial complexity results in dramatic sequential expansion and contraction of a subset of bacterial taxa that are minor contributors to the microbial consortium prior to antibiotic treatment. Inoculation of clindamycin-treated mice with C. difficile (VPI 10463) spores results in rapid development of diarrhea and colitis, with a 4- to 5-day period of profound weight loss and an associated 40 to 50% mortality rate. Recovering mice resolve diarrhea and regain weight but remain highly infected with toxin-producing vegetative C. difficile bacteria and, in comparison to the acute stage of infection, have persistent, albeit ameliorated cecal and colonic inflammation. The microbiota of “recovered” mice remains highly restricted, and mice remain susceptible to C. difficile infection at least 10 days following clindamycin, suggesting that resolution of diarrhea and weight gain may result from the activation of mucosal immune defenses.
INTRODUCTION
Clostridium difficile is a Gram-positive, anaerobic, spore-forming bacterium that primarily causes infections in hospitalized patients and residents of long-term health care facilities (29, 31). C. difficile is the most common cause of nosocomial diarrhea in the United States and causes a spectrum of intestinal disease, ranging from mild diarrhea to potentially fatal pseudomembranous colitis and toxic megacolon. The incidence and mortality of C. difficile infection are increasing, and associated costs in the United States are estimated to exceed $3.2 billion annually (22, 24, 30, 33).
C. difficile spores persist and remain viable for long periods of time in the environment, including health care institutions, facilitating transmission of this pathogen to patients. C. difficile spores, upon ingestion by vulnerable patients, germinate, produce toxin, and induce intestinal epithelial damage, inflammation, and mucosal pathology. C. difficile colitis most commonly follows the administration of antibiotics for prophylaxis against (e.g., to prevent surgery-associated infections) or treatment of bacterial infections (31). Although most antibiotics can increase the risk of developing C. difficile colitis, clindamycin, fluoroquinolones, or cephalosporins are most highly associated with subsequent development of C. difficile infection (2). A prevailing hypothesis is that antibiotic therapy kills native intestinal microbial populations that normally compete with or otherwise antagonize invading pathogens (34, 40) such as C. difficile. However, it remains unclear whether specific microbial species or families provide colonization resistance against C. difficile infection. Conversely, the in vivo stimuli that induce C. difficile spore germination and infection, beyond the relatively undefined changes to the commensal microbiota that accompany antibiotic administration, remain poorly understood.
Our understanding of complex microbial populations inhabiting mammalian mucosal surfaces and the gastrointestinal tract has increased dramatically in recent years thanks to the availability of deep DNA pyrosequencing platforms. Second-generation 16S rRNA gene sequencing techniques, utilizing massively parallel 454 pyrosequencing, have been used across a variety of experimental conditions to obtain more detailed and comprehensive information about microbial population dynamics (5, 26, 27, 32, 41). This approach has helped clarify the impact of numerous exogenous factors on the commensal microbiota, including the effect of diet (38), inflammation (35), and antibiotic therapy (6). Recent studies have also demonstrated that antibiotic administration to mice can enable hospital acquired pathogens such as vancomycin-resistant Enterococcus to establish microbial domination of the gastrointestinal tract, a phenomenon that also occurs in some patients following allogeneic hematopoietic stem cell transplantation (39). Furthermore, characterization of patients with recurrent C. difficile infections by 16S rRNA gene sequencing has suggested that relapse of colitis is associated with reduced intestinal microbial diversity (4).
Clindamycin, a lincosamide antibiotic with broad-spectrum activity against Gram-positive and obligate anaerobic bacteria, is excreted in bile and becomes highly concentrated in feces. Treatment with clindamycin is one of the most important risk factors for the development of C. difficile colitis (37). Using 16S DNA fingerprinting techniques, Jansson and coworkers have shown that clindamycin produces transient decreases in total intestinal bacteria and long-lasting deficits in Bacteroides species diversity (18). Clindamycin treatment of mice previously infected with C. difficile spores resulted in a transient loss of commensal microbiota diversity and in vivo germination and growth of C. difficile and toxin production, with accompanying colitis. Concurrent clindamycin treatment and active C. difficile infection resulted in a transient increase in the frequencies of proteobacteria and enterococci in feces (25). The losses and gains in bacterial taxa following clindamycin therapy, however, and the potential impact of these changes on susceptibility to C. difficile infection remain incompletely defined.
Here, we used a mouse model to investigate the effects of clindamycin with or without C. difficile infection on the intestinal microbiota. Clindamycin administration followed by C. difficile inoculation resulted in an early phase of infection characterized by diarrhea, weight loss, and high mortality over the course of 5 days. Surviving mice entered a chronic phase of infection during which diarrhea and weight loss resolved, but C. difficile colonization and toxin production persisted. While a single dose of clindamycin did not result in lasting decreases in intestinal microbial density, 16S rRNA gene pyrosequencing revealed a dramatic loss in microbiota diversity with many bacterial species present prior to clindamycin treatment becoming undetectable. Intestinal bacterial diversity remained depressed and susceptibility to C. difficile persisted for at least 10 days after clindamycin treatment. Our results suggest that the broad decreases in intestinal microbial diversity, or perhaps the loss of specific bacterial species, following clindamycin treatment render mice susceptible to C. difficile infection and provide a starting point for the discovery of microbial species or products that prevent infection by this pathogen.
MATERIALS AND METHODS
Mice.
All experiments were performed with C57BL/6J female mice, 6 to 8 weeks old, purchased from Jackson Laboratories and housed in sterile cages with irradiated food and acidified water. Mouse handling and weekly cage changes were performed by investigators wearing sterile gowns, masks, and gloves in a sterile biosafety hood. For the 28-day time course experiments, mice from three separately housed colonies were kept in the same facility, and one mouse from each of the three colonies was analyzed per time point to yield triplicate measurements. For experiments involving clindamycin treatment, animals received a single dose of clindamycin (200 μg) by intraperitoneal injection on day −1. For experiments involving C. difficile infection, mice were administered 103 CFU of C. difficile strain VPI 10463 spores by oral gavage. For experiments where clindamycin was not administered prior to inoculation with C. difficile, a dose of 105 CFU of spores was used. All animals were maintained in a specific-pathogen-free facility at Memorial Sloan-Kettering Cancer Center Animal Resource Center. Experiments were performed in compliance with Memorial Sloan-Kettering Cancer Center institutional guidelines and approved by the institution's IACUC.
C. difficile culture.
C. difficile strain VPI 10463 (ATCC 43255) was purchased from the American Type Culture Collection (ATCC). For isolation of spores, C. difficile was cultured in BHIS medium at 37°C in an anaerobic chamber (Coy Labs) and patched onto BHIS agar. Ten days later, bacteria were recovered from plates and spores were obtained as previously described (28). Briefly, spores were washed in ice-cold phosphate-buffered saline (PBS) multiple times and resuspended in 20% HistoDenz (Sigma-Aldrich), which was layered onto 50% HistoDenz, and centrifuged at 15,000 × g at 4°C for 15 min. After isolation of pelleted spores, the samples were washed five times in ice-cold PBS. The absence of live vegetative bacteria was confirmed by microscopic examination and the inability to grow in the absence of taurocholate.
Quantitative culture of C. difficile spores and vegetative forms.
Stool pellets or intestinal contents from ileum, cecum, or colon were suspended in deoxygenated PBS in an anaerobic chamber. Tenfold dilutions of the suspension were plated on BHIS plates containing taurocholate, d-cycloserine, and cefoxitin for specific selection of C. difficile. For enumeration of spore forms, the suspension was wet heated at 60°C for 20 min to kill vegetative forms and then plated on BHIS plates containing taurocholate, d-cycloserine, and cefoxitin. Plates were placed in a 37°C incubator within the anaerobic chamber overnight.
C. difficile toxin A and toxin B determination.
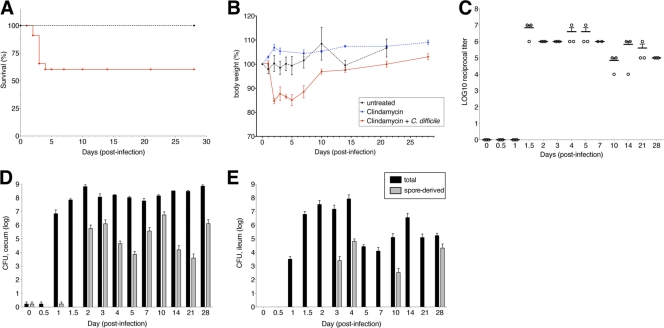
The presence of C. difficile toxins was determined using a cell-based cytotoxicity assay as previously described (17). Briefly, human embryonic lung fibroblast WI-38 cells (ATCC) were plated in a 96-well plate overnight for the formation of a monolayer. Intestinal or fecal contents were resuspended in sterile PBS and spun at 10, 000 × g for 10 min. Tenfold dilutions of the supernatant were added to WI-38 cells for overnight incubation, after which the effect of intestinal content supernatants on cell rounding was assessed. The presence of C. difficile toxins A and B was confirmed by neutralization by antitoxin antisera (Techlab, Blacksburg, VA). The data are expressed as the log10 reciprocal value of the first dilution at which cell rounding was not observed.
Histologic analysis.
Cecum and colon tissues were removed from mice, fixed overnight at 4°C in freshly made ice-cold 4% paraformaldehyde. Tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin. The severity of enteritis was quantified by using a grading system that includes evaluation (on a scale of 0 to 3 for each parameter) of edema of the mucosa, inflammatory cell infiltration, epithelial cell loss, and goblet cell loss (21). Histologic analysis and scoring of these parameters was performed blindly by a pathologist.
Sample collection and DNA extraction.
Immediately after mice were euthanized, the contents of the cecum and the distal 2 cm of the small intestine (ileum), excluding the last 1 cm proximal to the cecum, were recovered by manual extrusion. The samples were snap-frozen in liquid nitrogen prior to storage at −80°C. DNA was extracted from samples using a Power Soil DNA isolation kit (MoBio Laboratories).
V1-V3 16S rRNA gene amplification and 454 pyrosequencing.
For each sample, three replicate 25-μl PCRs were performed, each containing 20 ng of purified DNA, 0.2 mM concentrations of deoxynucleoside triphosphates, 1.5 mM MgCl2, 1.25 U of Platinum Taq DNA polymerase, 2.5 μl of 10× PCR buffer, and 0.2 μM concentrations of each primer designed to amplify the V1-V3 region, as described in the Human Microbiome Project Provisional 16S 454 Protocol (http://www.hmpdacc.org/tools_protocols.php): (i) the modified primer 8F (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGAGTTTGATCCTGGCTCAG-3′), composed of 454 primer B (underlined) and the universal bacterial primer 8F (in italics), and (ii) the modified primer 534R (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNCATTACCGCGGCTGCTGG-3′), composed of 454 primer A, a unique 6- or 7-base barcode, and the broad-range bacterial primer 338R (in italics). The cycling conditions used were: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min. The three replicate PCR products were pooled and subsequently purified using a Qiaquick PCR purification kit (Qiagen). The purified PCR products were sequenced on a 454 GS FLX Titanium pyrosequencing platform according to the Roche 454 recommended procedures.
Sequence analysis.
Sequences were converted to standard FASTA format using vendor 454 software. Sequences were not included for analysis if they (i) were shorter than 200 bp or longer than 440 bp, (ii) contained undetermined bases, (iii) had a 454 sequence quality average below 25, (iv) had no exact match to the forward primer or a barcode, or (v) did not align with the V1-V3 region. Sequences were grouped into OTUs (operational taxonomic units) using MOTHUR version 1.19.0 and were aligned using the SILVA reference alignment as a template and the Needleman-Wunsch algorithm with default scoring options. Sequence distances were calculated with MOTHUR as described previously (39). Relative abundances of specific bacterial taxa present in samples were calculated by dividing the total number of counts of all of the OTUs classified as a particular taxon in a sample by the total number of OTU counts in that respective sample. To determine the proportion of C. difficile in a sample, the pyrosequencing reads were subjected to BLAST analysis against a reference 16S C. difficile strain VPI-10463 sequence (GenBank accession no. AF072473.1). BLAST hits with 100% identity and 100% coverage were considered to be C. difficile.
Quantification of intestinal microbiota density by qPCR.
16S rRNA gene copies were quantified by quantitative PCR (qPCR) on DNA extracted from cecal and ileal content samples using 0.2 μM concentrations of the universal bacterial primer 8F (5′-AGAGTTTGATCCTGGCTCAG) and the broad-range bacterial primer 338R (5′-TGCTGCCTCCCGTAGGAGT-3′) and the DyNAmo SYBR green qPCR kit (Finnzymes). Standard curves were generated by serial dilution of the PCR blunt vector (Invitrogen) containing one copy of the 16S rRNA gene derived from a member of the Porphyromonadaceae family. The cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min.
Statistics.
Statistical analyses of histopathology scores and C. difficile intestinal CFU were performed by Mann-Whitney test using the GraphPad Prism software package (version 5.0). P values of <0.05 were considered to be significant.
Sequencing data analysis.
All 16S DNA sequence analysis and data plotting was carried out in Matlab (Mathworks, Natick, MA) using the statistics and bioinformatics toolboxes and scripts written in-house. Principal component analysis (PCA) was carried out on the abundances of OTUs defined at a 97% sequence similarity. Abundance maps and phylogenetic tree representations used the top 80 OTUs, which account for 94.3% of the sequences analyzed. Bar plot representations show top 15 microbial groups at the genus level. The abundance maps (Fig. 5), the phylogenetic trees (Fig. 6C), the time series (Fig. 8), and trajectories in principal component space (Fig. 9B and C) were constructed using compounded data from the replicate samples.
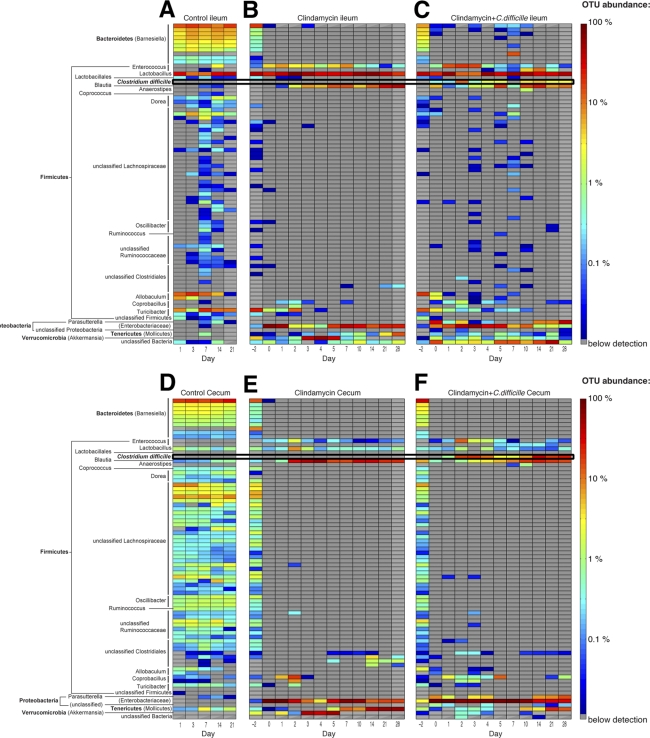
Fig 5.
Clindamycin treatment depletes a broad range of microbial populations. Heat map showing the relative abundance of phylotypes (OTUs defined with 97% similarity) in the ileum (A, B, and C) and cecum (D, E, and F) over time following inoculation with 103 C. difficile spores (A and D), clindamycin administration (B and E), or clindamycin administration plus C. difficile inoculation (C and F).
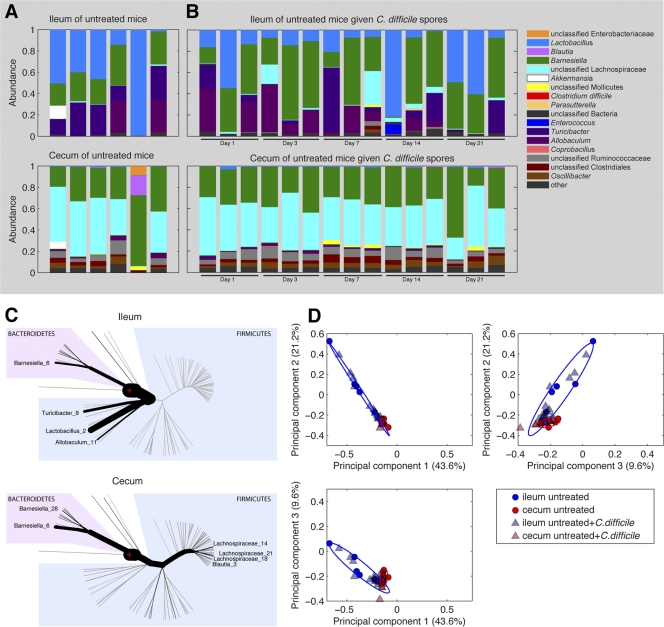
Fig 6.
Characteristics of the microbiota of the ileum and cecum of untreated mice. Phylogenetic classification of the 16S rRNA gene of the most predominant bacterial taxa in the ilea and ceca of untreated mice. (A) Each bar represents the microbiota of an individual mouse. (B) The relative abundances of major bacterial OTUs of the ilea and ceca of untreated mice administered 105 CFU of C. difficile spores on day 0 over the subsequent 21-day period are shown. (C) The relative abundance of major bacterial OTUs of untreated mice are given with reference to a phylogenetic tree. The thickness of branches corresponds to the relative abundance of the respective phylotypes. The number following phylotype name distinguishes different OTUs within the same genus. (D) PCA of ileal and cecal microbiota samples. Each point represents an individual mouse.
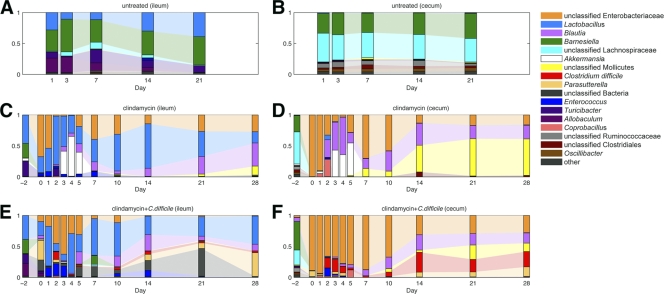
Fig 8.
Clindamycin and C. difficile infection alter intestinal microbiota composition. Mice were administered clindamycin (200 μg) intraperitoneally on day −1 and/or inoculated with C. difficile spores on day 0, and samples of cecal and ileal content were obtained from a representative mouse from three separately housed colonies on the days indicated. Each bar represents the pooled microbiota of three separately housed mice on the indicated day. The most predominant bacterial phylotypes are color coded as indicated.
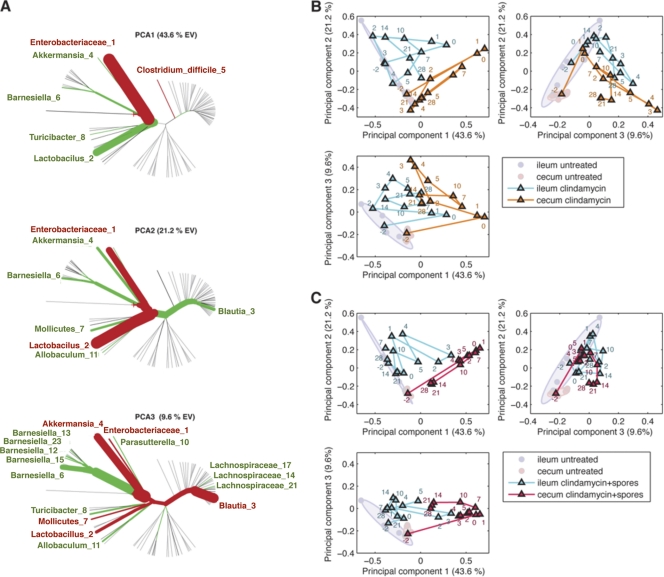
Fig 9.
PCA of the effect of clindamycin and C. difficile infection on the intestinal microbiota composition. (A) Principal components generated to explain maximal variance of samples from all treatment arms. The thickness of phylogenetic tree branches indicates the relative weight of that phylotype on the principal component. The color of branches indicates the sign of the phylotype (red, positive; green, negative). (B and C) Longitudinal principal coordinate analysis of cecal and ileal microbiota from mice treated with clindamycin (B) or clindamycin plus C. difficile spores (C). Untreated mice are represented as circles. Clindamycin was administered on day −1, and C. difficile spores were given on day 0. Each point represents the average of three separately housed mice. Number labels indicate the day (postinfection) on which the samples were harvested.
RESULTS
A single dose of clindamycin renders mice susceptible to C. difficile colitis.
While clindamycin treatment of C. difficile infected mice induces a supershedder state (25), it remains unclear whether a single dose of clindamycin is sufficient to render mice susceptible to subsequent C. difficile infection. Therefore, we treated mice with 200 μg of clindamycin or PBS by intraperitoneal injection on day −1 and subsequently challenged them with 103 C. difficile spores by oral gavage on day 0. Clindamycin-treated mice infected with C. difficile initially developed diarrhea, weight loss, and 40% of mice died within 5 days of infection (Fig. 1A and B). Surviving mice regained weight and recovered from diarrhea beyond the fifth day of infection. Mice receiving either clindamycin without subsequent C. difficile spores or C. difficile spores without preceding clindamycin survived and did not develop diarrhea or weight loss.
Fig 1.
Clindamycin predisposes mice to C. difficile infection and associated disease. Mice (n = 10 per group) received 105 C. difficile spores (strain VPI 10463) without clindamycin (untreated), a single dose of clindamycin (200 μg) intraperitoneally, or a single dose of clindamycin, followed by 103 CFU of C. difficile spores 24 h later. Survival (A) and weight loss (B) were monitored over the subsequent 4-week period. (C) The cytotoxicity of cecal content derived from clindamycin-treated, C. difficile-infected mice was quantified using a cell-based assay. Total (black) and spore (gray) CFU in the cecum (D) and ileum (E) were quantified by culture. Each bar represents the average for three mice. Panels A and B and panels D and E, respectively, share keys.
Persistent burden of C. difficile and toxin production.
We next investigated whether C. difficile growth and toxin production persisted in mice beyond the initial phase of infection. We treated with clindamycin on day −1 and infected mice with C. difficile spores 1 day later (designated day 0). Animals were euthanized either before clindamycin treatment (day −2) or on days 0, 1, 2, 3, 4, 5, 7, 10, 14, and 28, and the contents of the cecum and ileum were obtained for quantitative culture and C. difficile toxin determination. The density of C. difficile in the cecum and ileum increased 1 day after infection with C. difficile in clindamycin-treated mice, peaking at 109 CFU/g at 2 days postinfection (Fig. 1D and E). Both vegetative and spore forms of the bacterium persisted through day 28 (Fig. 1D and E), well beyond the initial phase of infection associated with diarrhea, weight loss, and mortality. Similarly, toxin levels peaked between days 1 and 2 and remained high through day 28 after infection (Fig. 1C).
Intestinal damage following clindamycin-associated C. difficile infection.
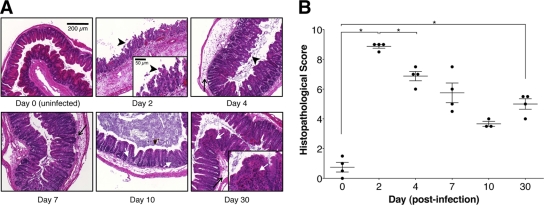
In light of the persistence of C. difficile colonization and toxin production following the early symptomatic phase of infection, we next evaluated the effect of C. difficile infection on intestinal tissues. Mice received clindamycin (200 μg) by intraperitoneal injection on day −1 and 103 C. difficile spores by oral gavage on day 0. Mice were sacrificed before infection and on days 2, 4, 7, 10, and 30 postinfection. Histological analysis of the ceca of mice sacrificed 2 days postinfection revealed pathology characterized by edema, infiltration of inflammatory cells, and epithelial cell damage, compared to the ceca of uninfected mice (Fig. 2A). Intestinal pathology was most pronounced during the first days following infection, but intestinal epithelial loss and inflammatory cell infiltration persisted for at least 30 days postinfection compared to uninfected ceca (Fig. 2). A similar degree and progression of tissue damage was also observed in the colon (data not shown).
Fig 2.
C. difficile infection following clindamycin treatment results in persistent intestinal inflammation. Mice were treated with clindamycin (200 μg) intraperitoneally on day −1 and infected with 103 C. difficile spores on day 0. (A and B) Uninfected mice and mice infected 2, 4, 7, 10, and 30 days postinfection were sacrificed, and their ceca were isolated and fixed, and tissue sections were stained with hematoxylin and eosin. Cecum sections were scored for edema (thin arrows), inflammatory cell infiltration (white arrows), and epithelial cell loss (arrowheads). Scale bar, 200 μm (inset, 50 μm). *, P < 0.05 (Mann-Whitney test).
Single-dose clindamycin treatment does not reduce overall 16S rRNA gene copy numbers in ileum and cecum.
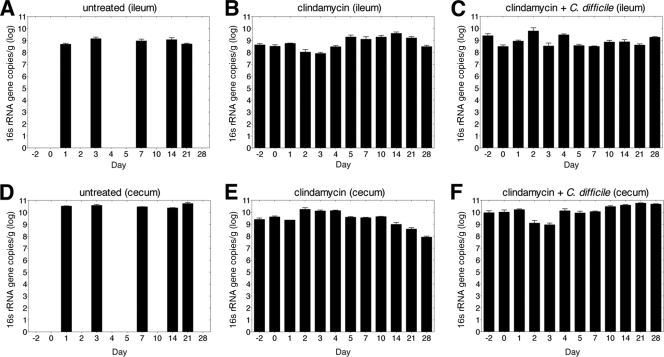
We next investigated how clindamycin and C. difficile infection impact the intestinal microbiota. First, we determined the effect of antibiotic on microbial density in the cecum and ileum by quantitative PCR (qPCR) of the total amount of bacterial 16S rRNA encoding genes. The density of 16S rRNA gene sequences was stable over the course of 21 days in the ileum and cecum of mice that did not receive clindamycin (Fig. 3A and D). 16S rRNA gene copy numbers were not altered by single-dose clindamycin administration (Fig. 3B, C, E, and F). In other follow-up experiments, we detected up to 10-fold decreases in the 16S rRNA gene copies 24 h after clindamycin administration (data not shown). Although qPCR of the bacterial 16S rRNA gene has been used to estimate bacterial density, interpretation of these results is complicated by the fact that 16S rRNA genes can be present in variable copy numbers in different bacterial species. Thus, quantitative shifts in total bacterial 16S rRNA genes may result from changes in bacterial density or changes in the frequency of bacteria with more or fewer 16S rRNA gene copies per genome.
Fig 3.
Single-dose clindamycin treatment does not reduce 16S rRNA gene copy numbers in the ileum and cecum. The quantity of 16S rRNA gene copies was determined by qPCR of ileal (A, B, and C) and cecal (D, E, and F) samples from mice that received clindamycin (200 μg) intraperitoneally on day −1 and/or inoculation of 103 C. difficile spores on day 0. Samples were obtained on days (postinfection) indicated. Each bar represents the average of three mice.
Clindamycin induces profound, long-lasting decreases in enteric microbial diversity.
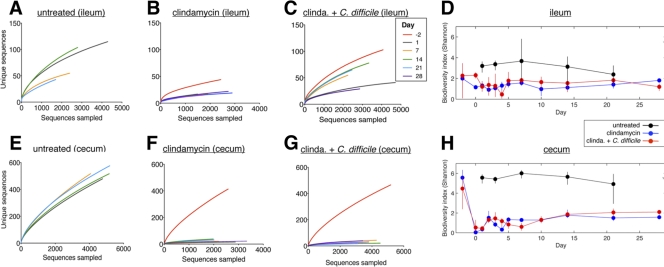
Because antibiotic-induced changes in the commensal microbiota have been implicated in the development of C. difficile colitis, we determined the impact of a single dose of clindamycin with or without C. difficile infection on the composition of the intestinal microbiota over the course of 28 days. We obtained 16S rRNA gene sequences from the ileum and cecum of mice from three separately housed colonies per time point using the 454/Roche pyrosequencing platform (39). The number of sequences obtained was 542,253, with an average of 3,116 sequences per sample. The overall microbiota richness (i.e., the number of unique phylotypes present in a given sample) was assessed by constructing rarefaction curves. Richness of the intestinal microbiota of untreated mice was similar to that of mice that received C. difficile spores without antibiotics, where the steep slope of these curves reflected microbial communities so rich that they had not been sampled completely (Fig. 4E). Consistent with prior studies (13), the richness of samples obtained from the cecum (Fig. 4E) was greater than samples obtained from the ileum (Fig. 4A). Administration of clindamycin on day −1 resulted in sustained depression of rarefaction curves, indicating profound long-lasting decreases in microbiota richness (Fig. 4F and G).
Fig 4.
Clindamycin induces profound, long-lasting decreases in intestinal microbial diversity. Rarefaction analyses of microbial communities in the ilea (A, B, and C) and ceca (E, F, and G) of mice that received C. difficile spores (A and E), clindamycin (B and F), or clindamycin and spores (C and G). Clindamycin was administered on day −1, and C. difficile spores were administered on day 0. The Shannon biodiversity index values of microbial communities in the ileum (D) and cecum (H) were also calculated at the time points indicated. Untreated, C. difficile-inoculated mice were only measured on days 1, 7, 14, and 21. Panels A, B, C, E, F, and G and panels D and H, respectively, share keys.
Overall biodiversity (i.e., phylogenetic richness and evenness) of the intestinal microbiota was evaluated by calculating the Shannon index of each sample (8). The Shannon index values of the intestinal microbiota in untreated mice were approximately 2 and 5 in the ileum and cecum, indicating relatively high microbial biodiversity, with the cecum having higher values, as expected (13). The effect of clindamycin was particularly pronounced in the cecum, where the index dropped significantly in both C. difficile-infected (P < 0.007) and uninfected cohorts (P < 0.009) from approximately 5 to <1, indicating a profound loss of microbial diversity. Cecal biodiversity rebounded modestly but remained persistently low (≤2) over the subsequent 4 weeks. The biodiversity in ilea of clindamycin treated mice also dropped but not as markedly (P > 0.05). In contrast, the biodiversity of untreated mice receiving C. difficile was never significantly different from that of untreated mice both in the cecum (P > 0.2) and in the ileum (P > 0.1) (Fig. 4D and H). Infection with C. difficile did not alter the biodiversity of the intestinal microbiota in mice treated with clindamycin or PBS alone.
To more precisely illustrate the broad losses of intestinal microbial diversity following clindamycin treatment, we plotted the relative frequencies of the 80 most abundant microbial populations over time in a heat map (Fig. 5). After clindamycin administration, ca. 87% of all OTUs initially present in the cecum were depleted and remained below the limit of detection for the following 4 weeks. The loss of microbial populations encompassed a broad taxonomic range and included both major and minor contributors to the initial microbial consortium (Fig. 5).
C. difficile inoculation does not alter intestinal microbiota composition in untreated mice.
The previous results demonstrated that clindamycin treatment resulted in a rapid loss of microbial species and, hence, decreased microbiota richness and diversity. We next turned our attention to the composition of the residual microbiota following clindamycin administration. Previous studies have shown that clindamycin induces long-term changes in intestinal Bacteroides communities (18), and concurrent C. difficile infection and clindamycin treatment have been shown to transiently alter the composition and diversity of the intestinal microbiota (25). However, the temporal microbial shifts induced by clindamycin treatment and those attributable to concurrent C. difficile infection remain undefined.
Consistent with previous studies, our analysis shows that the microbiota of the ileum and cecum in untreated mice are distinct (Fig. 6A). In untreated mice many bacterial populations were found in both compartments, but the ileum was primarily populated with Lactobacillus and Turicibacter species, while the major residents of the cecum consisted of Barnesiella and Lachnospiraceae species (Fig. 6A and C). The microbiota composition of untreated mice that received C. difficile spores was similar to that of untreated mice over 21-day period monitored. We used PCA to generate orthogonal combinations of OTUs that explained the maximum variance between samples. Using this approach we were able to delineate regions in PCA space that are typical of ileal and cecal microbiota from untreated mice in PCA plots (Fig. 6D). In addition, the microbiota of untreated mice that received C. difficile spores remained within the same PCA space as untreated mice not receiving C. difficile for the 21-day period monitored (Fig. 6D).
Administration of clindamycin induced dramatic and rapid changes in intestinal microbiota composition.
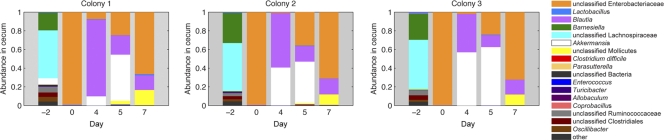
One day after a single dose of clindamycin, the cecal microbiota underwent profound losses of Lachnospiraceae and Barnesiella populations and became dominated by Enterobacteriaceae species (Fig. 7). Analysis of ileal samples revealed a similar trend, with increased Enterobacteriaceae proportions and contractions of previously dominant populations, which were modest for Lactobacillus but substantial for Turicibacter. These initial alterations in microbiota composition post-clindamycin treatment were reproducible across three independently housed mouse colonies (Fig. 7). The intestinal microbiota of mice treated with C. difficile spores alone resembled those of untreated mice, and no shifts in intestinal microbiota composition over the 3 week period monitored were detected (Fig. 8A and B). Treatment with clindamycin, however, resulted in marked expansion of the Enterobacteriaceae species, and this initial period of dominance was followed by multiple shifts in microbiota composition over the next 4 weeks (Fig. 8C and D). From days 2 through 7, the relative abundance of Akkermansia species increased in the cecum, peaking at day 4 but contracting by day 7. The relative proportion of Enterobacteriaceae decreased during this period, but re-emerged as the dominant population by day 7. Subsequently, unclassified Mollicutes species increased to become the dominant population from day 14 through the final sample collection on day 28. During this period, the relative abundance of Enterobacteriaceae species decreased (Fig. 8D). These shifts in the cecal microbiota were largely mirrored in the ileum, although increases in Mollicutes from days 14 to 28 were relatively small compared to those in the cecum. In addition, Lactobacillus remained a major component of the ileal microbiota throughout the time course (Fig. 8C). Although expansions and contractions of microbial taxa were generally similar among mice harvested at the same time point, there were occasional differences between individual mice (see Fig. S1 in the supplemental material). The reason for these aberrations is currently unknown but likely reflects environmental effects or subtle differences in the initial microbiota composition of different mice.
Fig 7.
Clindamycin treatment alters the composition of the microbiota. Mice from three separately housed colonies were administered clindamycin (200 μg) on day −1, and samples of cecal and ileal content were obtained for analysis on the days indicated. Each bar represents the microbiota of an individual mouse.
C. difficile infection alters microbiota composition in clindamycin-treated mice.
Infection with C. difficile at day 0 (1 day following clindamycin administration) resulted in similar shifts in the intestinal microbiota (Fig. 8E and F). Consistent with culture-based quantification, C. difficile was detected in the cecum by 16S rRNA gene sequencing on day 1 following infection. C. difficile relative abundance increased over the next 4 days, concomitant with decreases in Enterobacteriaceae species. In contrast to mice treated with clindamycin alone, Akkermansia did not increase in treated mice infected with C. difficile during this initial day 1 to 5 period. During days 4 to 7 postinfection, C. difficile abundance decreased moderately, concurrent with increases in Enterobacteriaceae species, but quickly rebounded and remained a major component of the cecal microbiota from day 14 through the end of sampling on day 28. Similar to mice treated with clindamycin alone, Mollicutes species abundance also increased during this period and Enterobacteriaceae species abundance decreased. However, this increase in Mollicutes abundance from day 10 to 28 was far less than that observed in uninfected mice (Fig. 8F). Changes in ileum microbiota of clindamycin treated C. difficile-infected mice largely reflected those in the cecum, with some exceptions. Compared to the cecal microbiota, C. difficile was less abundant, late increases in Mollicutes species were not apparent, and Lactobacillus remained a major component of the microbiota in the ileum throughout the infection (Fig. 8E).
In order to better compare global changes in the cecal and ileal microbiota over time, we used PCA to cluster the sampled bacterial communities along three orthogonal axes of maximal variance, which together explained ca. 77% of the variance observed among all samples (Fig. 9A). Cecal and ileal microbiota samples from untreated mice clustered separately but remained distinct from samples obtained from clindamycin-treated mice (Fig. 9B and C). However, clustering of ileal samples at time points after clindamycin treatment closely paralleled the trajectories of cecal samples, indicating that changes in the ileum microbiota mirrored changes in the cecum. Samples obtained from mice on day 0 (1 day after clindamycin administration) clustered on principal component axes 1, 2, and 3 opposite of untreated samples, indicating that the treated mice contain markedly different cecal microbiota compositions (Fig. 9C). While the PCA coordinates of samples progressively changed at times following clindamycin administration, the coordinates of treated samples never approximated those of untreated samples. This further supported that aggregate changes in the intestinal microbiota that occur over a 28-day period following clindamycin therapy do not produce a microbiota comparable to the initial untreated state.
Brief clindamycin treatment confers long-term susceptibility to C. difficile infection.
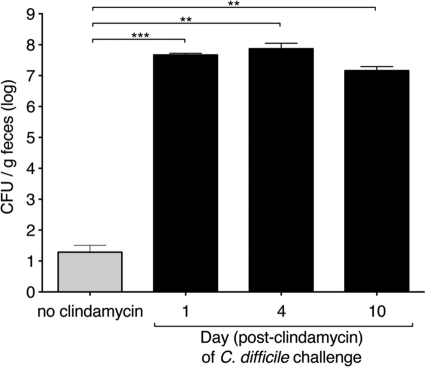
Our results indicate that clindamycin has short-term effects on microbial density and much more prolonged effects on the microbiota composition of the ileum and cecum. In order to determine whether the longer-term clindamycin-induced changes on microbial diversity and composition correlate with susceptibility to C. difficile infection, we challenged mice with oral C. difficile inoculation at 1, 4, or 10 days following a single intraperitoneal injection of clindamycin. At all time points post-clindamycin administration, mice challenged with C. difficile spores exhibited diarrhea and weight loss and had >106 CFU C. difficile per g of feces (Fig. 10). Mice infected at all time points post-clindamycin treatment exhibited weight loss and mortality rates comparable to mice infected 1 day post-clindamycin treatment (see Fig. 1A and B). Thus, despite recovery of microbial density, mice remain susceptible to C. difficile infection at least 10 days post-clindamycin administration. This result suggests that persistent reductions in microbial complexity following clindamycin treatment may account for sustained susceptibility to C. difficile-induced colitis.
Fig 10.
Single-dose clindamycin treatment confers long-term susceptibility to C. difficile infection. Mice received 105 spores of C. difficile (without clindamycin) or 200 μg of clindamycin intraperitoneally 1, 4, or 10 days before inoculation with 103 spores of C. difficile. One day following inoculation, C. difficile CFU in the fecal content were quantified. Figure 10 represents pooled results of two independent experiments. Each bar represents the average of six mice. ***, P < 0.001; **, P < 0.003.
DISCUSSION
The intestinal commensal flora confers resistance to a wide range of intestinal pathogens, extending from Salmonella (11) to hospital acquired pathogens such as vancomycin-resistant Enterococcus (9, 39) and C. difficile (4, 25). Over the past half-century, various approaches have contributed to our understanding of the role native intestinal microbial populations play in conferring colonization resistance to invasive enteric pathogens, such as Shigella (12, 15). While illuminating, such early culture-based studies were limited by the inability to study uncultured organisms that are estimated to constitute ca. 75% of the intestinal microbiota (10, 36). The sampling bias of culture-based approaches were eventually circumvented through the use of molecular methods, such as terminal restriction fragment length polymorphism and denaturing gradient gel electrophoresis, which utilize polymorphisms in highly conserved regions of bacterial 16S rRNA gene to identify previously undetected commensal species (19). Here, we apply the high-throughput molecular approach involving massively parallel pyrosequencing (38) to perform unbiased and extensive longitudinal analyses of native intestinal microbial populations following clindamycin therapy, with or without associated C. difficile infection.
We show that a single dose of clindamycin induces a profound and lasting loss of microbial diversity, eliminating a broad range of bacterial populations that remain undetectable for at least 4 weeks. The extended duration of the impact of clindamycin is consistent with human studies, which demonstrate that Bacteroides species in the fecal microbiota are diminished 2 years following clindamycin therapy (18). Our findings in laboratory mice indicate that the effect of clindamycin on the microbiota composition is more extensive, diminishing or eliminating ca. 90% of bacteria taxa from the microbiota of the cecum. In contrast, ciprofloxacin treatment of healthy human volunteers impacts approximately one-third of bacterial taxa, many of which recover after a 4-week antibiotic-free period (7), and mice treated with a amoxicillin-metronidazole-bismuth cocktail recovered dominant phylotype proportions 2 weeks following antibiotic cessation (1). Thus, compared to other antibiotic treatments that have been studied using similar deep-sequencing approaches, clindamycin depletes a broader range of intestinal bacteria.
Studies of the effect of antibiotics on the human microbiota have been largely limited to the analysis of fecal samples. Although the microbiota of the cecum, colon, and feces are quite similar (13, 39), microbial populations in the small intestine are distinct and thus are likely to undergo different changes following antibiotic administration. Mouse models provide opportunities for a much more detailed study of the intestinal microbiota under controlled replicate conditions that circumvent interindividual variability that frequently besets the study of microbiota dynamics in humans (1, 4, 7). Indeed, in our study we found that despite the complex temporal dynamics of the residual microbiota following clindamycin treatment, the major shifts in the composition of the microbiota were similar in uniformly treated, but separately housed, replicate mouse colonies. These shifts included a succession of dominance by the taxonomically diverse phylotypes Enterobacteriaceae, Akkermansia, and Mollicutes, each of which were initially present in untreated mice in relatively low numbers. Consistent with our findings, Enterobacteriaceae and Akkermansia have been identified in the normal flora of mice and humans (1, 6), and rapid expansion of Enterobacteriaceae phylotypes has been shown to occur in the intestine following a variety of antibiotic treatments (1, 25, 39). Thus, the depth and high temporal resolution of our survey revealed that dramatic shifts in residual microbiota composition, albeit complex, were similar in separately housed groups of mice. That said, it is likely that the microbial shifts we have documented in our studies are highly dependent on the starting microbiota of the experimental mouse population. All mice used in our study were obtained from a single vendor and were cohoused prior to the initiation of the experiment. Given recent studies demonstrating that mice obtained from different vendors harbor distinct intestinal microbial populations (16), it is likely that the effects of antibiotic treatment will be variable in different studies. Indeed, we believe that differences in the starting microbiota of experimental mice is a likely explanation for differences between our results and those published by Lawley et al. (25). Those studies demonstrated that the effect of clindamycin on microbial diversity and the composition of the microbiota were transient, with recovery occurring over the course of 7 to 10 days following the cessation of antibiotic treatment (25).
We found that mice remained susceptible to C. difficile colonization and disease at least 10 days following administration of a single dose of clindamycin, despite a series of marked shifts in the composition of the residual microbiota. This result suggests that microbial populations important for resistance to C. difficile infection are among those eliminated rapidly and lastingly following clindamycin administration. It is possible that clindamycin-sensitive bacteria are specifically responsible for suppressing C. difficile colonization. Alternatively, clindamycin-resistant bacteria may be responsible for C. difficile colonization resistance but depend upon clindamycin-sensitive bacteria for vital functions (for example, nutrient metabolism), since even minimal microbiomes may exist in interdependent consortia, as evidenced by failure of monocolonization with specific microbial species (14). In addition, the mechanism by which normal flora confer C. difficile colonization resistance may be direct or indirect, since work from our group has shown that microbial products augment innate immune barrier function (3, 23) and confer resistance to C. difficile infection (17). Given the broad impact of clindamycin on taxonomical representation in the intestinal microbiota, the range of phylotypes that may be critical for resistance to C. difficile infection remains vast. Nevertheless, our study suggests new avenues to identify precisely how clindamycin-induced changes in the intestinal microbiota predispose to C. difficile colonization and disease.
C. difficile infection of clindamycin-treated mice induced additional effects on the intestinal microbiota. Compared to mice treated with clindamycin alone, C. difficile-infected mice sustained little expansion of Akkermansia and mitigated the expansion of Enterobacteriaceae and Mollicutes phylotypes. Although it is possible that C. difficile directly impairs expansion of other microbial groups by suppression or competition for ecological niches, it is also possible that C. difficile infection indirectly modulates intestinal microbiota composition by augmenting mucosal inflammation. Indeed, intestinal inflammation induced by enteric pathogens has been shown to affect colonic bacteria density (28), suppress populations of indigenous intestinal bacteria (20), and otherwise alter microbiota composition in ways that favor colonization and expansion of exogenous microbial populations (28), including pathogenic bacteria (35). Our results indicate C. difficile infection induces intestinal inflammation that persists for at least 30 days. While the magnitude of the inflammatory processes is diminished at later time points, the sustained inflammatory tone may affect measurable changes in the residual microbiota at these time points.
Successful treatment of recurrent C. difficile infection in humans with fecal transplantation demonstrates the therapeutic effectiveness of reconstituting a complex microbiota but also underscores our crude understanding of which microbial populations are critical for colonization resistance to C. difficile infection. The murine model of sustained vulnerability to C. difficile infection following clindamycin treatment that we have described here provides an opportunity to identify microbial species or consortia that are capable of preventing the germination of C. difficile spores and/or growth and toxin production by vegetative C. difficile bacteria in the gastrointestinal tract.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (grant 1RO1AI42135 to E.G.P.), the Lucille Castori Center for Microbes, Inflammation, and Cancer, and the Tow Foundation.
Footnotes
Published ahead of print 17 October 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Antonopoulos DA, et al. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77: 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bishara J, Peled N, Pitlik S, Samra Z. 2008. Mortality of patients with antibiotic-associated diarrhoea: the impact of Clostridium difficile. J. Hosp. Infect. 68: 308–314 [DOI] [PubMed] [Google Scholar]
- 3. Brandl K, et al. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455: 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang JY, et al. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197: 435–438 [DOI] [PubMed] [Google Scholar]
- 5. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37: D141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A. 108 (Suppl 1): 4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dinsdale EA, et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452: 629–632 [DOI] [PubMed] [Google Scholar]
- 9. Donskey CJ, Hanrahan JA, Hutton RA, Rice LB. 2000. Effect of parenteral antibiotic administration on the establishment of colonization with vancomycin-resistant Enterococcus faecium in the mouse gastrointestinal tract. J. Infect. Dis. 181: 1830–1833 [DOI] [PubMed] [Google Scholar]
- 10. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Endt K, et al. 2010. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6: e1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freter R. 1962. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. II. The inhibitory mechanism. J. Infect. Dis. 110: 38–46 [DOI] [PubMed] [Google Scholar]
- 13. Garner CD, et al. 2009. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect. Immun. 77: 2691–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geuking MB, et al. 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34: 794–806. [DOI] [PubMed] [Google Scholar]
- 15. Hentges DJ, Freter R. 1962. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. I. Correlation between various tests. J. Infect. Dis. 110: 30–37 [DOI] [PubMed] [Google Scholar]
- 16. Ivanov II, et al. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4: 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarchum I, Liu M, Lipuma L, Pamer EG. 2011. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect. Immun. 79: 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jernberg C, Löfmark S, Edlund C, Jansson JK. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1: 56–66 [DOI] [PubMed] [Google Scholar]
- 19. Jernberg C, Löfmark S, Edlund C, Jansson JK. 2010. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology (Reading, Engl.) 156: 3216–3223 [DOI] [PubMed] [Google Scholar]
- 20. Keeney KM, Finlay BB. 2011. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr. Opin. Microbiol. 14: 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kelly CP, et al. 1994. Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J. Clin. Invest. 93: 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelly CP, LaMont JT. 1998. Clostridium difficile infection. Annu. Rev. Med. 49: 375–390 [DOI] [PubMed] [Google Scholar]
- 23. Kinnebrew MA, et al. 2010. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 201: 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kyne L, Hamel MB, Polavaram R, Kelly CP. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34: 346–353 [DOI] [PubMed] [Google Scholar]
- 25. Lawley TD, et al. 2009. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77: 3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71: 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lupp C, et al. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2: 119–129 [DOI] [PubMed] [Google Scholar]
- 29. McDonald LC, Owings M, Jernigan DB. 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerging Infect. Dis. 12: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Brien JA, Lahue BJ, Caro JJ, Davidson DM. 2007. The emerging infectious challenge of Clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect. Control Hosp. Epidemiol. 28: 1219–1227 [DOI] [PubMed] [Google Scholar]
- 31. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7: 526–536 [DOI] [PubMed] [Google Scholar]
- 32. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U. S. A. 103: 12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sohn S, et al. 2005. Varying rates of Clostridium difficile-associated diarrhea at prevention epicenter hospitals. Infect. Control Hosp. Epidemiol. 26: 676–679 [DOI] [PubMed] [Google Scholar]
- 34. Stecher B, Hardt W-D. 2008. The role of microbiota in infectious disease. Trends Microbiol. 16: 107–114 [DOI] [PubMed] [Google Scholar]
- 35. Stecher B, et al. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5: 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suau A, et al. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65: 4799–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sullivan A, Edlund C, Nord CE. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1: 101–114 [DOI] [PubMed] [Google Scholar]
- 38. Turnbaugh PJ, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ubeda C, et al. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120: 4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vollaard EJ, Clasener HA. 1994. Colonization resistance. Antimicrob. Agents Chemother. 38: 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.