Abstract
The molecular mechanisms that define asymptomatic bacteriuria (ABU) Escherichia coli colonization of the human urinary tract remain to be properly elucidated. Here, we utilize ABU E. coli strain 83972 as a model to dissect the contribution of siderophores to iron acquisition, growth, fitness, and colonization of the urinary tract. We show that E. coli 83972 produces enterobactin, salmochelin, aerobactin, and yersiniabactin and examine the role of these systems using mutants defective in siderophore biosynthesis and uptake. Enterobactin and aerobactin contributed most to total siderophore activity and growth in defined iron-deficient medium. No siderophores were detected in an 83972 quadruple mutant deficient in all four siderophore biosynthesis pathways; this mutant did not grow in defined iron-deficient medium but grew in iron-limited pooled human urine due to iron uptake via the FecA ferric citrate receptor. In a mixed 1:1 growth assay with strain 83972, there was no fitness disadvantage of the 83972 quadruple biosynthetic mutant, demonstrating its capacity to act as a “cheater” and utilize siderophores produced by the wild-type strain for iron uptake. An 83972 enterobactin/salmochelin double receptor mutant was outcompeted by 83972 in human urine and the mouse urinary tract, indicating a role for catecholate receptors in urinary tract colonization.
INTRODUCTION
Urinary tract infections (UTIs) are among the most common infectious diseases of humans, with Escherichia coli responsible for >80% of cases (51). Approximately 50% of women will experience cystitis in their lifetime and asymptomatic bacteriuria (ABU) occurs in 6 to 20% of the population depending on age and gender (21). ABU is an asymptomatic carrier state that resembles commensalism with patients often carrying >105 CFU/ml of urine of a single organism. Many ABU E. coli isolates are phylogenetically related to virulent uropathogenic E. coli (UPEC) strains and some may have evolved from pathogenic strains by virulence attenuation (43, 68).
Iron is essential for bacterial growth and is limited in the urinary tract. Therefore, iron acquisition systems are important colonization factors for ABU E. coli and UPEC. An efficient method for the sequestration of iron is through the production of siderophores, low-molecular-weight Fe3+-chelating compounds, and subsequent uptake via their associated membrane receptors (11, 44). At present four different siderophore systems have been identified in E. coli. Enterobactin is a highly prevalent catecholate siderophore produced by both E. coli K-12 strains and pathogenic strains (12, 32). While enterobactin has a very high stability constant for Fe3+ binding, it is inactivated by host proteins such as serum albumin and siderocalin (neutrophil gelatinase-associated lipocalin) (24, 38). Salmochelin, a glucosylated derivative of enterobactin, is not recognized by siderocalin and thus evades the host immune response (19). Salmochelin biosynthesis and utilization require the iroBCDEN gene cluster as well as the enterobactin biosynthesis and utilization genes. Yersiniabactin is a mixed-type siderophore encoded on the high-pathogenicity island that is widespread in Enterobacteriaceae. The yersiniabactin biosynthesis and uptake system is a virulence determinant in Yersinia species, Klebsiella pneumoniae and several E. coli pathotypes (6, 40, 56). Aerobactin, a hydroxamate siderophore, has a higher Fe3+ binding stability in acidic environments and is maximally produced at low pHs (62).
All four siderophore systems are negatively regulated by ferrous iron and the ferric uptake regulator Fur and are expressed under low-iron conditions (5, 8). E. coli clinical isolates encode and express different combinations of the four siderophores (32, 43). Indeed, studies have shown that the siderophore systems are highly expressed by ABU E. coli and UPEC isolates in vitro in urine and in patients during UTI (3, 27, 52, 54, 58). Transcriptome analysis of E. coli in urine from women with UTIs revealed that iron acquisition systems were the most highly expressed fitness factors common to all patients (27). Siderophore systems are also required for UPEC virulence in the mouse urinary tract (23, 60). Additionally, several iron receptors, including the aerobactin receptor protein IutA, are potential vaccine candidates since they protect mice from infection with UPEC (2). However, the relative contribution of each siderophore system to the growth and survival of E. coli isolates encoding all four systems is not well understood. Furthermore, although some UPEC strains contain the genes encoding siderophore receptors, they fail to express the associated functional siderophore molecule (32).
One of the best-characterized ABU strains is E. coli 83972, a B2 clinical isolate capable of long-term bladder colonization that has been effectively employed as a prophylactic agent for the prevention of UTI in human inoculation studies (4, 33, 42, 59, 65). Bladder infection with E. coli 83972 fails to induce a host inflammatory response and this is associated with the attenuation of virulence determinants including type 1, P and F1C fimbriae (37, 53, 64). With a limited adhesin repertoire, a significant contributing factor to the maintenance of E. coli 83972 in the bladder is its rapid growth rate in urine (54). E. coli 83972 contains genes encoding the enterobactin, salmochelin, aerobactin and yersiniabactin siderophore systems. Transcription of these siderophore genes is highly upregulated during growth of E. coli 83972 in the bladder of deliberately colonized patients, suggesting that siderophores may contribute to urinary tract colonization (29, 52). Differences in iron acquisition have also been proposed as a mechanism by which ABU E. coli outcompetes UPEC in human urine (54). In this study we have defined the specific complement of siderophores produced by E. coli 83972. In addition, we have used E. coli 83972 to extensively study the contribution of enterobactin, salmochelin, aerobactin and yersiniabactin to total siderophore activity and growth in iron-limited environments as well as in vitro and in vivo fitness.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The clinical ABU E. coli isolates were obtained from urine samples of patients at the Princess Alexandra Hospital (Brisbane, Australia) and have been described previously (43). Additional bacterial strains utilized in this study are described in Table 1. E. coli 83972 (OR:K5:H−) is a prototype ABU clinical strain originally isolated from a young Swedish girl who carried it for 3 years without adverse effects (4, 42). E. coli 83972AMP contains the ampicillin resistance gene inserted at the λ-attachment site and was used in competition experiments (17). Strains were routinely grown at 37°C in MM9-glycerol minimal medium as previously described (63) with the addition of 0.3% Casamino Acids, 0.02% thiamine-HCl and 0.2% sodium succinate. Where indicated, 0.2% glucose was used as the carbon source instead of 0.2% glycerol and the pH was adjusted to pH 5.5 or pH 7.0. Human urine was collected from at least three healthy female volunteers with no history of UTI or antibiotic use in the previous 2 months. Pooled urine was filter sterilized and used within 3 days. To ensure iron limitation plasticware was used and 50 μM 2,2′-dipyridyl (DIP) was added to the medium where indicated. Luria-Bertani (LB) agar plates were supplemented with appropriate antibiotics as indicated in the following final concentrations: 50 μg ml−1 kanamycin (Kan), 50 μg ml−1 ampicillin (Amp), and 15 μg ml−1 chloramphenicol (Cam).
Table 1.
Bacterial strains used in this study
| E. coli strain or deletion mutant | Description | Reference or source |
|---|---|---|
| 83972 wild-type | ABU strain | 42 |
| 83972AMP | 83972 attB::bla-rrnBP1-cfp-T0; Ampr | 17 |
| Siderophore biosynthetic gene deletion mutants | ||
| 83972 A | 83972 ΔiucABCD (aerobactin); Kanr | This study |
| 83972 Y | 83972 ΔybtS (yersiniabactin); Kanr | This study |
| 83972 S | 83972 ΔiroB (salmochelin); Kanr | This study |
| 83972 ES | 83972 ΔentB (enterobactin/salmochelin); Kanr | This study |
| 83972 AS | 83972 ΔiucABCD ΔiroB; Kanr | This study |
| 83972 SY | 83972 ΔiroB ΔybtS; Kanr | This study |
| 83972 AY | 83972 ΔiucABCD ΔybtS; Kanr | This study |
| 83972 ASY | 83972 ΔiucABCD ΔiroB ΔybtS; Kanr | This study |
| 83972 EAS | 83972 ΔentB ΔiucABCD; Kanr | This study |
| 83972 ESY | 83972 ΔentB ΔybtS; Kanr | This study |
| 83972 EASY | 83972 ΔentB ΔiucABCD ΔybtS; Kanr | This study |
| Siderophore receptor gene deletion mutants | ||
| 83972 AR | 83972 ΔiutA (aerobactin); Camr | This study |
| 83972 YR | 83972 ΔfyuA (yersiniabactin); Camr | This study |
| 83972 ER | 83972 ΔfepA (enterobactin); Camr | This study |
| 83972 SR | 83972 ΔiroN (salmochelin); Camr | This study |
| 83972 ASR | 83972 ΔiutA ΔiroN; Camr | This study |
| 83972 SYR | 83972 ΔiroN ΔfyuA; Camr | This study |
| 83972 AYR | 83972 ΔiutA ΔfyuA; Camr | This study |
| 83972 EYR | 83972 ΔfepA ΔfyuA; Camr | This study |
| 83972 ESR | 83972 ΔfepA ΔiroN; Camr | This study |
| 83972 EAR | 83972 ΔfepA ΔiutA; Camr | This study |
| 83972 ASYR | 83972 ΔiutA ΔiroN ΔfyuA; Camr | This study |
| 83972 EASR | 83972 ΔfepA ΔiutA ΔiroN; Camr | This study |
| 83972 EAYR | 83972 ΔfepA ΔiutA ΔfyuA; Camr | This study |
| 83972 ESYR | 83972 ΔfepA ΔiroN ΔfyuA; Camr | This study |
| 83972 EASYR | 83972 ΔfepA ΔiutA ΔiroN ΔfyuA; Camr | This study |
| Ferric citrate receptor gene deletion mutants | ||
| 83972 F | 83972 ΔfecA (ferric citrate receptor); Kanr | This study |
| 83972 EASYF | 83972 ΔentB ΔiucABCD ΔybtS ΔfecA; Kanr | This study |
Construction of E. coli 83972 iron acquisition deletion mutants.
E. coli 83972 iron acquisition genes were deleted using the λ Red recombinase gene inactivation method as previously described (14). Plasmids pKD4 and pKD3 were used as the templates for siderophore biosynthetic gene and siderophore receptor gene deletion mutants, respectively, using the primers listed in Table S1 in the supplemental material. Ferric citrate receptor mutants were constructed using pKD4. Plasmid pCP20, expressing the FLP recombinase, was transformed into each deletion mutant to remove the antibiotic resistance cassette prior to subsequent gene deletions. Single, double, triple and quadruple siderophore mutants were constructed. Deletion of the enterobactin biosynthetic gene entB abolishes the synthesis of both enterobactin and salmochelin (ES). All deletion mutants were verified by PCR and DNA sequencing. Mutants with multiple deletions were reconfirmed by PCR once the final deletion had been made; all mutations were correct with no evidence of recombination between λ Red recombinase deletion sites. Mutants utilized in the assays contained the antibiotic resistance cassette used to construct the last mutation; this allowed for selective plating on LB agar plates with antibiotics for the competition assays. Antibiotics were not added to growth medium for any of the assays and therefore did not interfere with growth of the mutants.
Growth and competition assays.
For growth assays, strains grown for 16 h in MM9-glycerol medium were used to inoculate 25 ml of MM9-glycerol medium with 50 μM DIP to an optical density at 600 nm (OD600) of 0.05. Cultures were grown with shaking at 37°C, with OD600 measurements taken every hour. For competition assays, strains grown for 16 h in MM9-glycerol medium were mixed 1:1 to a final OD600 of 0.05 in MM9-glycerol medium with 50 μM DIP. Cultures were grown with shaking at 37°C, with samples plated on LB agar with appropriate antibiotics at 0-, 7-, and 24-h time points. Growth and competition assays in pooled human urine were performed as described above, with and without 50 μM DIP. For competition experiments E. coli 83972AMP was used as the wild-type 83972 strain. E. coli 83972AMP possessed a growth rate identical to that of E. coli 83972 in MM9 medium and pooled human urine and showed no significant difference in fitness when competed against E. coli 83972 in mixed growth assays.
CAS assays.
For liquid chrome azurol S (CAS) assays, strains were grown in MM9-glycerol medium at pH 5.5 or pH 7.0, with shaking at 37°C for 16 h. OD600 measurements of the cultures were taken before the cells were pelleted and the supernatants were collected. Liquid CAS assays were performed using the culture supernatants as previously described (57). Siderophore levels were standardized by the OD600 measurements to account for slight variations in growth with percent siderophore units calculated as previously described (46). For solid CAS assays agar plates based on MM9 medium with 0.2% glycerol or 0.2% glucose at pH 5.5 or pH 7.0 were made as previously described (57). Strains were grown in MM9 medium with 0.2% glycerol or 0.2% glucose at pH 5.5 or pH 7.0 with shaking at 37°C for 17 h. The cultures were standardized to an OD600 of 0.5 before 2 μl of each strain was spotted onto the CAS agar plates. The plates were incubated at 37°C for 17 h. The radius of the halo formed around each colony was measured from the colony edge to the edge of the color change and corresponds with the amount of siderophore activity. The term “siderophore activity” refers to the level of siderophore as measured using either liquid or solid CAS assays.
Extraction and high-performance liquid chromatography (HPLC)/LC-mass spectrometry (MS) analysis of siderophores.
E. coli 83972 strains were grown for 3 h in LB medium, diluted 1:100 into 25 ml of MM9-glycerol medium pH 7.0 and incubated for 17 h with shaking at 37°C. Ferric chloride was added to culture supernatants to a final concentration of 3.75 mM and supernatants were left at room temperature for 15 min before the precipitate was removed. Supernatants were added to a column filled with 1 ml of DEAE-Sepharose (Sigma), and then the column was washed with 3 ml of water. Siderophores were eluted from the DEAE with 3 ml of 7.5 M ammonium formate pH 3.6, desalted using Chrom P solid-phase extraction tubes (250 mg; Supelco), and concentrated to 200 μl as previously described (32).
Analytical HPLC was performed on a Shimadzu LC-20AT liquid chromatograph (flow rate of 1 ml/min) equipped with a Shimadzu SPD-20A UV-visible light (Vis) detector (220 nm), column oven (30°C), and Phenomenex HPLC column (Luna C18; 5-μm particle size; 250 by 4.60 mm). Analytical HPLC analyses were performed with a 60-μl injection volume of the above-prepared siderophore solution, using the following method: gradient of 6 to 40% aqueous CH3CN containing 0.1% formic acid over 20 min, followed by a hold at 40% for the next 10 min. LC-MS analyses were performed on a Waters 2690 liquid chromatograph (flow rate of 0.5 ml/min) equipped with a Waters 2487 dual-wavelength absorbance detector (220 nm), column oven (30°C), and Phenomenex HPLC column (Luna C18; 5-μm particle size; 250 by 4.60 mm) that was coupled to a Waters Quattro Micro API (atmospheric pressure ionization) mass spectrometer. MassLynx software (Waters) was used for data acquisition and processing. LC-MS analyses were performed with a 60-μl injection volume, using the following method: 6 to 40% aqueous CH3CN containing 0.1% formic acid over 40 min, with a hold at 40% for the next 20 min. The cone voltage and desolvation temperature were 50 V and 150°C, respectively.
Mouse model of UTI.
Female C3H/HeJ Mice (8 to 10 weeks old) were purchased from the Animal Resources Centre, Western Australia and housed in sterile cages with ad libitum access to sterile water. The mouse model of UTI with competitive mixed infection was performed as previously described (1). Mice were inoculated with a mixture of 2.5 × 108 CFU of E. coli 83972AMP and 2.5 × 108 CFU of an E. coli 83972 siderophore receptor mutant, both grown for 20 h in LB medium. Urine was collected from each mouse 42 h after inoculation for quantitative colony counts. Mice were euthanized 42 h after inoculation; the bladders and kidneys were excised and processed for quantitative colony counts. The mixed strains were differentiated by their antibiotic resistance with ampicillin resistant E. coli 83972AMP and the 83972 siderophore mutants resistant to chloramphenicol. Data are expressed as the number of CFU per 0.1 g of bladder or kidney tissue or as the number of CFU per ml of urine. Fitness indexes were calculated by dividing the CFU count per ml or per 0.1 g of tissue for each 83972 siderophore mutant by that of 83972AMP. All experiments were carried out in strict accordance with the recommendations in the Animal Care and Protection Act (Queensland, Australia, 2002) and the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (7th ed., 2004). Approval for mouse infection studies was obtained from the University of Queensland Animal Ethics Committee (SCMB/471/09/NHMRC [NF]).
Statistical analysis.
Variation in siderophore activity of ABU E. coli isolates grown under different conditions was tested using a Kruskal-Wallis test with Minitab (version 14) statistical software. Differences in siderophore activity of E. coli 83972 strains were tested using unpaired two-sample t tests. The 630-nm absorbance data, standardized by the OD600 measurements to account for variations in growth, were utilized. Differences in growth rates and cell densities at the 10-h time point between E. coli 83972 strains were tested using unpaired two-sample t tests. For the in vitro competition assays, differences in bacterial CFU/ml of the E. coli 83972 siderophore mutants compared to E. coli 83972AMP were also tested using paired two-sample t tests. For the mouse UTI model, bacterial CFU counts were compared using a nonparametric Wilcoxon matched-pairs signed rank test with GraphPad Prism (version 5) statistical software. For all tests, the level of statistical significance was set at a P value of <0.05.
RESULTS
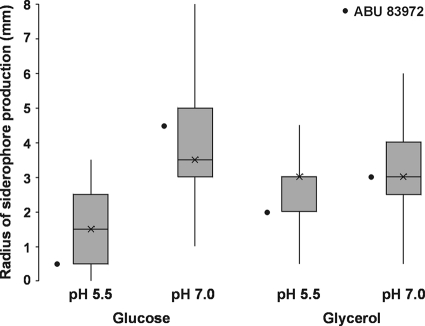
Siderophore activity of ABU E. coli clinical isolates under different environmental conditions does not correspond with genotype.
In order to gain an overview of siderophore biosynthesis by ABU E. coli, we investigated the siderophore activity of a collection of clinical ABU E. coli isolates, as well as of the prototype ABU E. coli strain 83972, grown under different conditions. Chrome azurol S (CAS) agar assays were performed with glucose or glycerol as the carbon source and at pH 5.5 or pH 7.0. Siderophore activity was significantly lower in cultures grown with glucose at pH 5.5 than under the three other conditions (P < 0.001) (Fig. 1). We then investigated whether high siderophore activity (defined as ≥3 mm radius) correlated with the presence of the siderophore receptor gene iutA (aerobactin), iroN (salmochelin), or fyuA (yersiniabactin) as previously determined by PCR screening (43). No correlation between the presence of a specific gene (or any combination of genes) and the level of siderophore activity was apparent for the ABU isolates under any of the four growth conditions. We then focused our attention on ABU strain E. coli 83972 to determine its siderophore content and to evaluate how each siderophore system contributes to total siderophore activity, growth, fitness, and colonization of the urinary tract.
Fig 1.
Total siderophore activity of ABU E. coli isolates (n = 47). Solid CAS assays were performed with glucose or glycerol as the carbon source and at pH 5.5 or pH 7.0. The radius of the halo formed around each colony corresponds with the amount of siderophore activity. The data are presented as box plots with crosses representing the medians. ABU E. coli siderophore activity was significantly lower when cultures were grown with glucose at pH 5.5 than under the other growth conditions (P < 0.001). The siderophore activity of ABU E. coli 83972 is indicated.
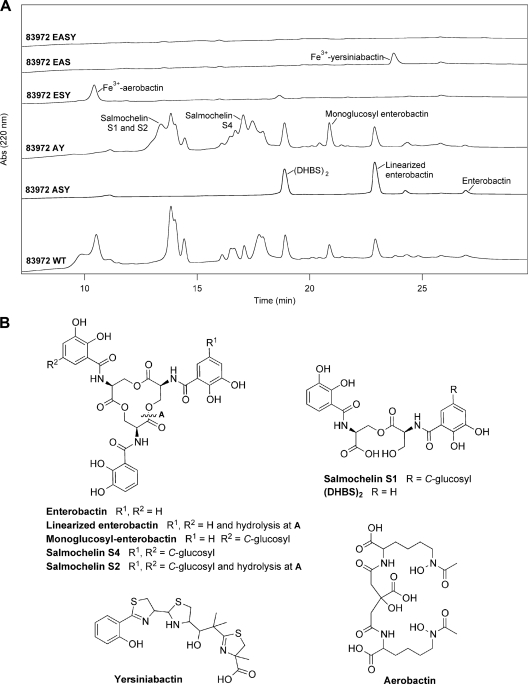
ABU E. coli strain 83972 produces the full repertoire of siderophores encoded by its genome.
To determine if E. coli 83972 is capable of producing enterobactin, aerobactin, salmochelin, and yersiniabactin, siderophore extraction and LC and LC-MS analyses were performed following growth of E. coli 83972 in MM9 medium. In addition, a series of mutants containing deletions in the siderophore biosynthetic genes of E. coli 83972 were made to give single, double, triple, and quadruple siderophore biosynthetic mutants. Throughout the manuscript, these mutants are labeled according to the iron acquisition gene that was deleted: enterobactin (E), aerobactin (A), salmochelin (S), yersiniabactin (Y), and ferric citrate receptor (F). For example, the quadruple enterobactin, aerobactin, salmochelin, and yersiniabactin mutant is referred to as 83972 EASY. In LC and LC-MS analyses, wild-type E. coli 83972 gave peaks that were consistent with the presence of all four siderophores, while siderophore biosynthetic mutants 83972 AY, ASY, ESY, EAS, and EASY produced the expected siderophore profiles (Fig. 2). Strain 83972 ASY, a triple mutant with deletions in all siderophore gene clusters except enterobactin, gave peaks corresponding to enterobactin, linearized enterobactin, and the 2,3-dihydroxybenzoylserine dimer (DHBS)2. Strain 83972 AY, with intact gene clusters for both enterobactin and salmochelin, showed additional peaks for salmochelin S1, S2, and S4 as well as monoglucosyl-enterobactin. As expected, single peaks were seen for the biosynthetic mutants 83972 ESY and 83972 EAS corresponding to Fe3+-aerobactin and Fe3+-yersiniabactin, respectively. Therefore, E. coli 83972 can produce all four siderophores encoded in its genome. The lack of peaks for the quadruple biosynthetic mutant 83972 EASY strongly suggests that this strain does not produce any additional, uncharacterized siderophores.
Fig 2.
Siderophores synthesized by ABU E. coli strain 83972. (A) HPLC analysis showing siderophores synthesized by ABU E. coli strain 83972 wild-type (WT) and 83972 siderophore biosynthetic deletion mutants in MM9 medium at pH 7.0. The identity of each peak was confirmed by LC-MS. (B) Chemical structures of the siderophores enterobactin, salmochelin, yersiniabactin, and aerobactin. (DHBS)2 is the 2,3-dihydroxybenzoylserine dimer. Abs, absorbance.
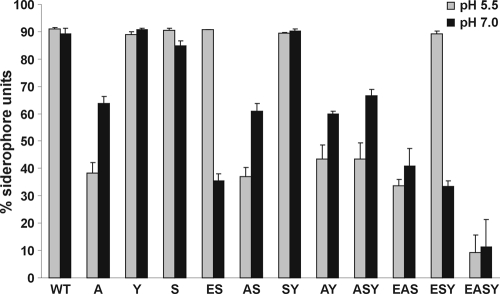
E. coli 83972 relies primarily on aerobactin and enterobactin for iron acquisition in iron-limiting medium.
In order to assess the relative importance of each siderophore to iron acquisition by E. coli 83972 in vitro, total siderophore activity of wild-type 83972 and siderophore biosynthetic mutants was examined via liquid CAS assays. MM9 medium at pH 5.5 and pH 7.0 was utilized since the pH of human urine varies from pH 4.6 to 8, and the optimal pH for iron chelation varies for each siderophore (62). E. coli 83972 displayed high siderophore activity in MM9 medium at both pH 5.5 and pH 7.0 (Fig. 3). In contrast, the quadruple mutant 83972 EASY had negligible siderophore activity, corresponding to abolishment of the four siderophore systems. E. coli 83972 single siderophore mutants showed variation in siderophore levels. Strain 83972 A, which cannot produce aerobactin, showed 53% and 25% decreases in siderophore activity compared to the wild-type strain at pH 5.5 and pH 7.0, respectively (P < 0.05). Strain 83972 ES, deleted for enterobactin (and therefore also salmochelin), showed a 53% decrease in siderophore activity at pH 7.0 compared to the wild-type (P < 0.001). In contrast, strains 83972 Y and 83972 S, with deletions in yersiniabactin and salmochelin synthesis genes, respectively, showed no difference in siderophore activity compared to the wild-type at either pH. Biosynthetic mutants with only one siderophore system left intact—83972 EAS (yersiniabactin only), 83972 ESY (aerobactin only), and 83972 ASY (enterobactin only)—all demonstrated significant siderophore activity compared to the quadruple mutant 83972 EASY at pH 5.5 (P < 0.05). Taken together, this demonstrates that yersiniabactin, aerobactin, and enterobactin are all functional siderophores in E. coli 83972. Since deletion of an enterobactin synthesis gene also abolishes salmochelin synthesis, it was not possible to analyze a strain which produces only salmochelin.
Fig 3.
Liquid CAS assays for ABU E. coli strain 83972 wild-type and siderophore biosynthetic deletion mutants in MM9 medium at pH 5.5 and pH 7.0. Results are the means of biological triplicates plus standard deviations. Bar graph shows percent siderophore units, calculated as [(Ar − As)/Ar] × 100, where Ar is the absorbance of MM9/CAS solution and As is the sample absorbance. The quadruple mutant E. coli 83972 EASY had negligible siderophore activity at both pHs.
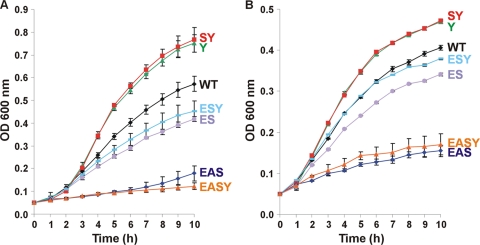
The ability of E. coli 83972 to grow efficiently in iron-limiting medium requires synthesis of enterobactin and aerobactin.
The E. coli 83972 siderophore biosynthetic mutants were utilized to examine how each siderophore system contributes to the ability to grow in iron-limiting medium. Growth curves were established for E. coli 83972 and each siderophore biosynthetic mutant in MM9 medium at pH 7.0, supplemented with 50 μM DIP to ensure iron limitation. Strains 83972 A, 83972 S, 83972 AY, 83972 AS, and 83972 ASY had no change in growth compared to wild-type E. coli 83972 (data not shown). However, all mutants with enterobactin gene deletions (ES) had significantly lower growth rates and cell densities after 10 h of growth than wild-type E. coli 83972 (Fig. 4A). For these ES mutants, two growth patterns were apparent. Strains 83972 ES and 83972 ESY showed moderate decreases in growth rates (35% and 23%, respectively) and cell densities after 10 h of growth (27% and 21%, respectively) compared to wild-type E. coli 83972 (P < 0.05). In contrast, strains 83972 EAS and 83972 EASY were severely compromised compared to wild-type E. coli 83972 for growth rates (88% and 89%, respectively) and cell densities after 10 h of growth (69% and 79%, respectively); these levels were also significantly lower than those of the ES and ESY mutants (P < 0.05). Strains 83972 Y and 83972 SY displayed higher growth rates (63% and 71%, respectively) and higher cell densities after 10 h of growth (31% and 34%, respectively) than wild-type E. coli 83972 (P < 0.05) (Fig. 4A). This increase in growth was not seen with 83972 yersiniabactin deletion strains that were also deleted for enterobactin or aerobactin synthesis genes.
Fig 4.
Growth of ABU E. coli strain 83972 wild type and siderophore biosynthetic deletion mutants in MM9 medium with 50 μM DIP at pH 7.0 (A) and pH 5.5 (B). E. coli 83972 siderophore mutants with the same growth profile as the wild-type strain are not shown. Growth rates were calculated from the exponential phase (2 to 5 h), and cell densities were determined from the 10-h time point. Results are the means of biological triplicates ± standard deviations. Strains 83972 Y, 83972 SY, 83972 ES, 83972 EAS, and 83972 EASY had significantly different growth rates and cell densities after 10 h of growth than the wild-type E. coli 83972 at both pHs (P < 0.05). Strain 83972 ESY had a significantly lower growth rate and final cell density than the wild-type E. coli 83972 at pH 7.0 (P < 0.05).
To determine if the siderophores had different contributions to growth in a more acidic environment, growth assays were also performed in MM9 medium containing 50 μM DIP at pH 5.5. All but one 83972 siderophore biosynthetic mutant followed the same trend as for pH 7.0 (Fig. 4B). The exception was strain 83972 ESY, which showed no significant decrease in growth rate compared to wild-type E. coli 83972 at pH 5.5. E. coli 83972 siderophore biosynthetic mutants had no change in growth compared to wild-type E. coli 83972 when grown in MM9 medium without DIP, suggesting that growth defects were iron dependent (data not shown).
E. coli 83972 siderophore biosynthetic mutants act as “cheaters.”
To determine if the growth deficiencies of the E. coli 83972 siderophore biosynthetic mutants could be rescued by mixed growth with the 83972 wild-type strain, competition assays were performed. E. coli 83972 siderophore mutants were mixed at a 1:1 ratio with an ampicillin-resistant version of 83972 wild-type (83972AMP) and cultured in MM9 medium at pH 7.0 (containing 50 μM DIP). The number of CFU of each strain was enumerated after 7 h of growth. None of the 83972 siderophore mutants, including 83972 Y and 83972 SY that had improved growth in monocultures, was able to outcompete strain 83972AMP, and in all cases the relative fitness was not significantly different from 1 (Fig. 5). Additionally, there was no significant difference in the final cell densities of the mixed cultures compared to those of the monocultures. After 24 h, the proportion of each strain in the mixed culture was similar to that observed after 7 h of growth (data not shown). Therefore, the 83972 siderophore biosynthetic mutants, including the quadruple 83972 EASY mutant, were able to overcome the growth deficiencies observed in single strain growth assays (Fig. 4A), suggesting that they act as cheaters by utilizing siderophores produced by strain 83972AMP for growth.
Fig 5.
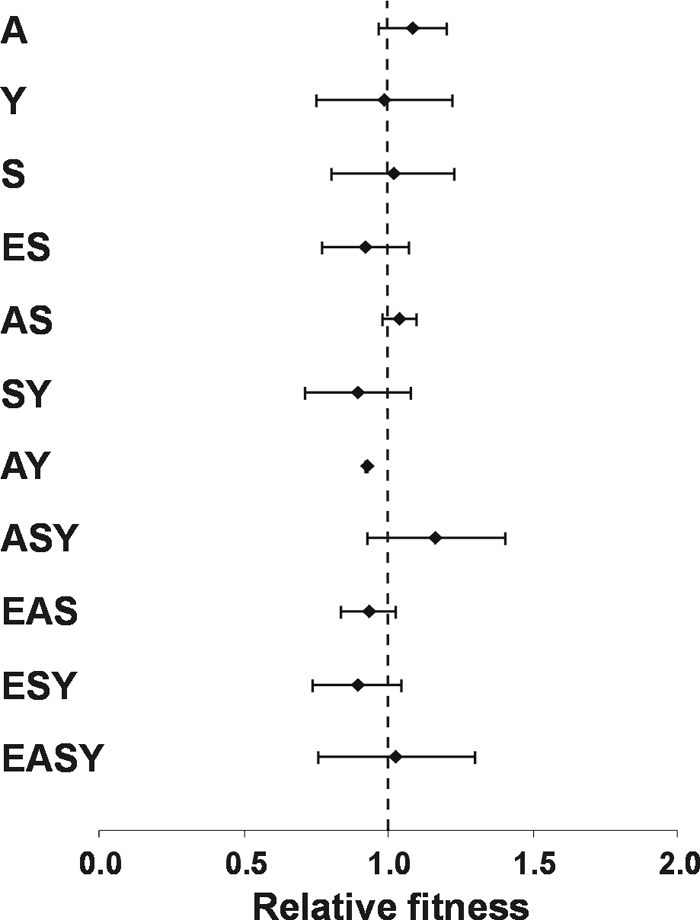
Relative fitness of E. coli 83972 siderophore biosynthetic deletion mutants compared to E. coli 83972AMP in a mixed 1:1 growth experiment in MM9 medium at pH 7.0 containing 50 μM DIP. Fitness indexes were calculated after 7 h of growth by dividing the CFU/ml of each mutant by the CFU/ml of 83972AMP. The dashed line represents a fitness index of 1.0, which indicates no difference in fitness between the two strains. The relative fitness of each E. coli 83972 biosynthetic mutant was not significantly different from 1.
E. coli 83972 siderophore receptor mutants are outcompeted by the wild-type strain during growth in iron-limiting medium.
The ability of siderophore synthesis mutants to act as cheaters necessitated the construction of siderophore receptor deletion mutants in order to dissect the contribution of each siderophore system to the fitness of E. coli 83972. The genes encoding the four siderophore receptors FepA, IroN, IutA, and FyuA were deleted to give single, double, triple, and quadruple siderophore receptor mutants. Unlike the biosynthetic mutants used previously, the receptor mutants synthesize and secrete the siderophores but cannot take up and obtain iron from the siderophores. First, the receptor mutants were assessed for their ability to grow in MM9 medium at pH 7.0 containing 50 μM DIP. The majority of the E. coli 83972 receptor mutants (indicated by R superscripts) had no change in growth rates compared to wild-type E. coli 83972 (i.e., 83972 AR, 83972 YR, 83972 SR, 83972 ER, 83972 ASR, 83972 SYR, 83972 AYR, 83972 EYR, 83972 EAR, 83972 ASYR, and 83972 EAYR) (data not shown). However, all mutants with deletions of both enterobactin and salmochelin receptor genes (ESR) had significantly decreased growth compared to wild-type E. coli 83972 (Fig. 6A). The growth rates of strains 83972 ESR and 83972 ESYR decreased by 90% and 92%, respectively, compared to growth of E. coli 83972, with cell densities after 10 h of growth decreased by 79% and 82%, respectively (P < 0.001). Strains 83972 EASR and 83972 EASYR, both of which contained deletions in the enterobactin, salmochelin, and aerobactin receptor genes, were unable to grow in this iron-limiting medium.
Fig 6.
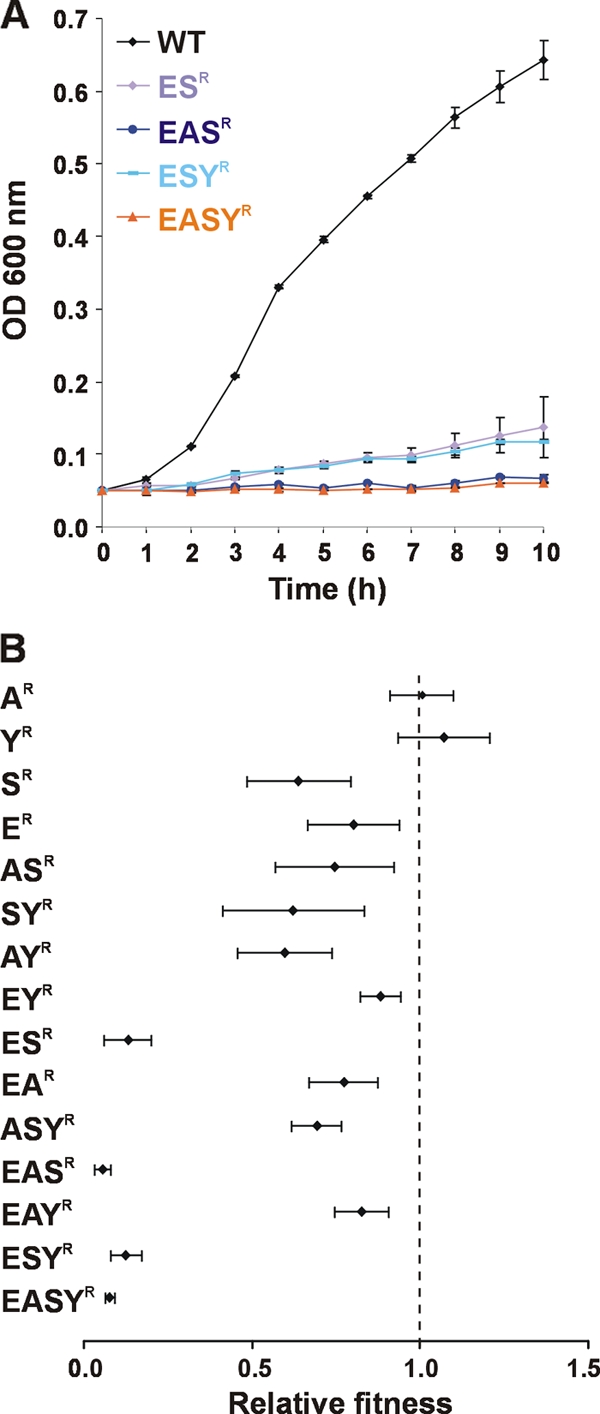
Growth and relative fitness of E. coli 83972 siderophore receptor deletion mutants. (A) Growth in MM9 medium, pH 7.0, containing 50 μM DIP. E. coli 83972 siderophore mutants with the same growth profile as the wild-type strain are not shown. Strains 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR had significantly lower growth rates and lower cell densities after 10 h of growth than wild-type E. coli 83972 (P < 0.001). (B) Relative fitness of 83972 siderophore receptor mutants compared to 83972AMP after a mixed 1:1 growth experiment in MM9 medium at pH 7.0 containing 50 μM DIP. Results are the means of biological triplicates ± standard deviations. The dashed line represents a fitness index of 1.0, which indicates no difference in fitness between the two strains. Strains 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR were severely outcompeted by E. coli 83972AMP (P < 0.05). Strains 83972 SR, 83972 ER, 83972 ASR, 83972 SYR, 83972 AYR, 83972 EYR, 83972 EAR, 83972 ASYR, and 83972 EAYR were also significantly outcompeted by E. coli 83972AMP (P < 0.05).
Growth assays in MM9 medium containing 50 μM DIP at pH 5.5 yielded similar results (data not shown). Again, the only 83972 receptor mutants with statistically significant growth differences compared to wild-type E. coli 83972 were 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR. Compared to wild-type E. coli 83972, the cell densities after 10 h of growth of strains 83972 EASR and 83972 EASYR (79 to 80% reduction) were more severely affected than those of strains 83972 ESR and 83972 ESYR (49 to 51% reduction) (P < 0.05).
We also assessed the E. coli 83972 siderophore receptor mutants in a mixed growth assay by directly competing each mutant against E. coli 83972AMP in MM9 medium containing 50 μM DIP at pH 7.0 for 7 h (Fig. 6B). Strains 83972 AR and 83972 YR showed no difference in fitness compared to E. coli 83972AMP. In contrast, the following mutants were significantly outcompeted by E. coli 83972AMP: 83972 SR, 83972 ER, 83972 ASR, 83972 SYR, 83972 AYR, 83972 EYR, 83972 EAR, 83972 ASYR, and 83972 EAYR (P < 0.05). The greatest fitness defects were seen for the four 83972 mutants lacking the enterobactin and salmochelin receptor genes (ESR, EASR, ESYR, and EASYR); in mixed growth assays with E. coli 83972AMP these strains constituted only 5 to 11% of the final population. This 10-fold decrease in fitness corresponds to the growth deficiency observed for each of the four 83972 ESR mutants in monoculture.
Enterobactin, salmochelin, aerobactin, and ferric citrate uptake have a role in growth of E. coli 83972 in human urine.
The growth of E. coli 83972 wild-type and siderophore deletion mutants was examined in single and mixed cultures using human urine, which is the most physiologically relevant iron-limiting environment for this strain. In contrast to MM9 medium containing 50 μM DIP, all E. coli 83972 siderophore biosynthetic mutants had growth rates and cell densities after 10 h of growth similar to those of wild-type E. coli 83972 when grown in urine (data not shown). The addition of 50 μM DIP to the urine had no effect on the growth of any of the siderophore biosynthetic mutants compared to wild-type E. coli 83972. To investigate whether citrate uptake contributes to iron acquisition by E. coli 83972 during growth in urine, the fecA gene encoding the ferric citrate receptor was deleted in E. coli 83972 (to produce the single mutant 83972 F) and in strain 83972 EASY (to produce the quadruple siderophore synthesis/ferric citrate receptor strain 83972 EASYF). Strain 83972 F exhibited the same growth characteristics as wild-type E. coli 83972 in urine with and without 50 μM DIP (Fig. 7A). However, strain 83972 EASYF had a 63% decrease in growth rate (P < 0.001) and a 49% decrease in final cell density (P = 0.001) compared to wild-type E. coli 83972 in urine containing 50 μM DIP. Mixed growth assays (1:1) were also performed in human urine containing 50 μM DIP to compare the growth of E. coli 83972 F, 83972 EASY, and 83972 EASYF to that of E. coli 83972AMP. Strains E. coli 83972 F, 83972 EASY, and 83972 EASYF showed no significant difference in fitness compared to that of E. coli 83972AMP (data not shown).
Fig 7.
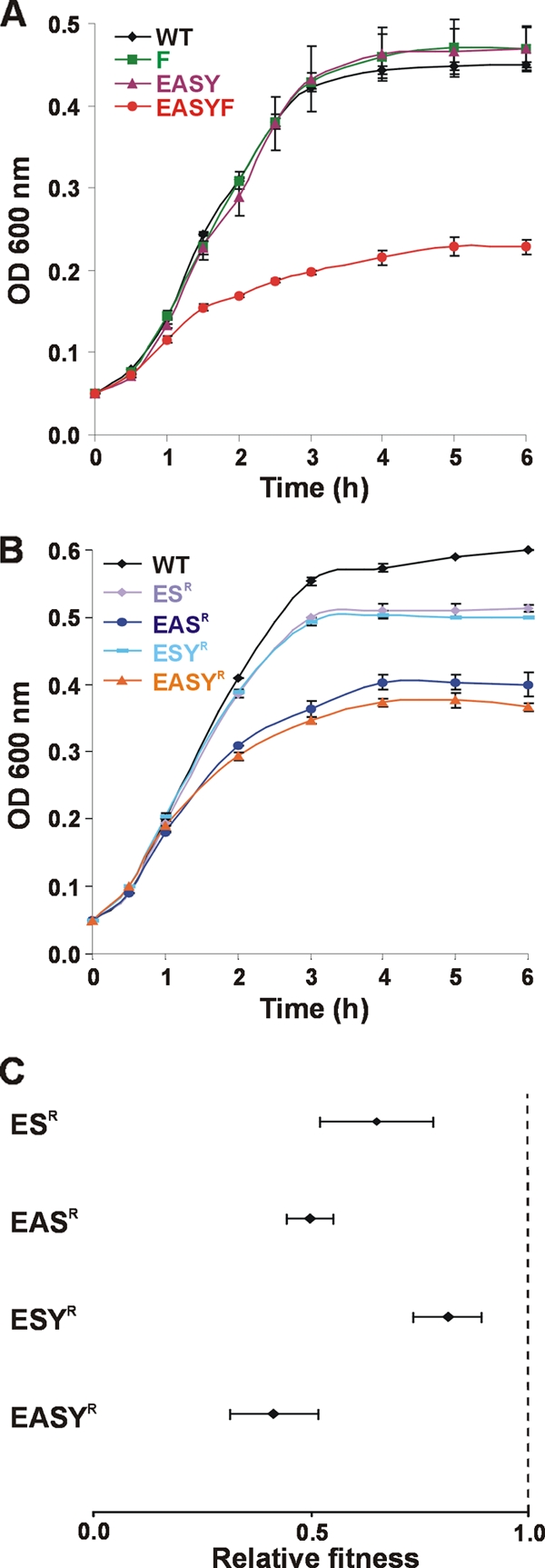
Growth and relative fitness of E. coli 83972 iron acquisition deletion mutants in human urine. (A) Growth of 83972 wild-type, 83972 F, 83972 EASY, and 83972 EASYF in human urine with 50 μM DIP. Strain 83972 EASYF had a significantly lower growth rate (P < 0.001) and final cell density (P = 0.001) than wild-type E. coli 83972. (B) Growth of E. coli 83972 siderophore receptor mutants in human urine (no DIP). E. coli 83972 siderophore receptor mutants with the same growth profile as the wild-type strain are not shown. Strains 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR displayed reduced growth rates and had lower cell densities after 10 h of growth than the wild-type E. coli 83972 (P < 0.05). (C) Relative fitness of 83972 siderophore receptor mutants compared to 83972AMP after a mixed 1:1 growth experiment in human urine (no DIP). Results are the means of biological triplicates ± standard deviations. The dashed line represents a fitness index of 1.0, which indicates no difference in fitness between the two strains. Strains 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR were significantly outcompeted by strain 83972AMP (P < 0.05).
In the case of the siderophore receptor mutants, strains 83972 ESR and 83972 ESYR displayed reduced growth rates (11% and 15%, respectively) and achieved lower cell densities after 10 h of growth (14% and 17%, respectively) than the wild-type E. coli 83972 in human urine (P < 0.05) (Fig. 7B). The growth of strains 83972 EASR and 83972 EASYR was even more compromised, with their growth rates and cell densities after 10 h of growth significantly affected compared to those of wild-type E. coli 83972 (P < 0.05). The difference in growth rates and final cell densities of strains 83972 ESR/83972 ESYR compared to 83972 EASR/83972 EASYR was also significant (P < 0.05). While these trends were similar to those in MM9 medium containing 50 μM DIP, the decrease in growth was not as pronounced, with growth of the receptor mutants not completely abolished in urine. All other 83972 siderophore receptor mutants had no change in growth in urine compared to wild-type E. coli 83972. Identical trends were seen when 50 μM DIP was added to the urine (data not shown).
Mixed assays (1:1) were also performed in urine to compare the growth of each of the 83972 siderophore receptor mutants to E. coli 83972AMP. Strains 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR were significantly outcompeted by strain 83972AMP (P < 0.05) and constituted 29 to 45% of the final population (Fig. 7C). Similar results were seen when 50 μM DIP was added to the urine (data not shown). Thus, all 83972 mutants lacking both enterobactin and salmochelin receptor genes had decreased fitness in human urine.
Enterobactin and salmochelin receptor mutants are outcompeted in the murine UTI model.
To determine if E. coli 83972 siderophore receptor mutants with decreased fitness in iron-limiting medium in vitro also have decreased fitness in vivo, E. coli 83972AMP was competed 1:1 with the 83972 ESR, EASR, ESYR, and EASYR siderophore mutants in the mouse UTI model. E. coli 83972AMP significantly outcompeted mutants 83972 ESR, 83972 EASR, 83972 ESYR, and 83972 EASYR in urine, the bladder, and the kidneys (P < 0.05) (Fig. 8). These four strains have in common mutations that disrupt the enterobactin receptor gene fepA (ER) and the salmochelin receptor gene iroN (SR). There was no significant difference in the competitive index of the double mutant 83972 ESR and the quadruple mutant 83972 EASYR in urine, bladder, and kidneys. These results suggest that the enterobactin and salmochelin receptors FepA and IroN together contribute to colonization of the urinary tract by E. coli 83972.
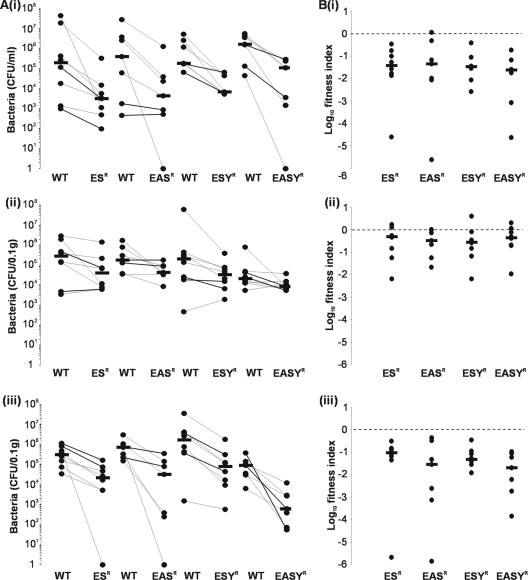
Fig 8.
Fitness of E. coli 83972 siderophore receptor mutants during in vivo competition with E. coli 83972AMP. C3H/HeJ mice were challenged with a 1:1 mixture of 83972AMP and 83972 ESR, 83972 EASR, 83972 ESYR, or 83972 EASYR. (A) Each dot represents total number of CFU recovered from each mouse per ml of urine (i), per 0.1 g of bladder tissue (ii), or per 0.1 g of kidney tissue (iii). Lines connect data points for the same mouse, and horizontal bars represent median values. WT, E. coli 83972AMP. (B) Each symbol represents the log10 fitness index calculated for each individual mouse. Fitness indices were calculated by dividing the mutant CFU/ml of urine (i) or 0.1 g of bladder tissue (ii) or 0.1g of kidney tissue (iii) by that of 83972AMP. Dashed lines represent a log10 fitness index of 0, which indicates no difference in fitness between the two strains. Horizontal bars represent group medians, and each competition group had 7 to 8 mice. All mutants were significantly outcompeted by 83972AMP in all three niches examined (P < 0.05).
DISCUSSION
In this study we utilized the probiotic ABU E. coli strain 83972 to analyze the relative importance of the enterobactin, salmochelin, yersiniabactin, and aerobactin siderophore systems to iron acquisition, growth, and fitness. We show that E. coli 83972 produces all four siderophores during growth under low-iron conditions. To overcome functional redundancy, we sequentially deleted the biosynthetic genes for each of the four systems to obtain all possible combinations of 83972 siderophore biosynthetic mutants. We also sequentially deleted the receptor genes for each of the four siderophore systems to enable competition experiments. This panel of isogenic mutants enabled a detailed investigation of the contribution of each siderophore system to iron acquisition and growth of the prototype ABU E. coli strain 83972.
Environmental factors such as pH and carbon source affect E. coli siderophore activity (62). Thus, we initially analyzed the siderophore activity of a collection of ABU E. coli isolates grown under different conditions, i.e., glucose or glycerol as the carbon source, as well as pH 5.5 and pH 7.0 to represent the pH range of urine. Siderophore activity was significantly lower when strains were grown with glucose at pH 5.5 than under the three other conditions, and we therefore used glycerol defined medium (MM9-glycerol) for detailed in vitro analysis of E. coli 83972 siderophore activity. Overall, the linked enterobactin and salmochelin synthesis and uptake systems were consistently important for siderophore activity, growth, and fitness of E. coli 83972. The other siderophore systems had different contributions depending on the examined phenotype. It is important to remember that deleting the enterobactin biosynthetic gene (entB) results in the loss of both enterobactin and salmochelin siderophores, whereas deleting a salmochelin biosynthetic gene (iroB) abolishes only salmochelin production. With regard to E. coli 83972 total siderophore activity, the loss of enterobactin/salmochelin or aerobactin siderophores significantly decreased siderophore activity, whereas the loss of salmochelin alone or yersiniabactin had no effect. The importance of enterobactin and aerobactin over the other siderophores at pH 7.0 is not simply related to their Fe3+ binding affinities. The binding affinity of yersiniabactin (dissociation constant [Kd] of 10−36 M) is higher than that of aerobactin (Kd = 10−23 M) at neutral to alkaline pHs (47, 50). Enterobactin has the highest Fe3+ binding affinity (Kd = 10−52 M), and the binding affinity of salmochelin is currently unknown (50). The importance of aerobactin but not enterobactin/salmochelin to siderophore activity at pH 5.5 correlates with the higher production of aerobactin and higher stability of Fe3+-aerobactin in acidic environments (62).
The four different siderophores of E. coli 83972 displayed varied contributions to growth in low-iron medium. For example, growth of the enterobactin biosynthetic mutant 83972 ES was decreased while growth of the yersiniabactin biosynthetic mutant 83972 Y increased in comparison to that of E. coli 83972. Inactivation of salmochelin or aerobactin synthesis alone had no effect on growth. However, a role for aerobactin under these growth conditions was demonstrated since strain 83972 EAS had significantly decreased growth compared to strain 83972 ES. The importance of enterobactin for normal growth in vitro has been previously reported for E. coli K-12 as well as Klebsiella pneumonia and Salmonella enterica serovar Typhimurium (39, 40, 66). Additionally, increased growth of a Pseudomonas syringae yersiniabactin mutant has been observed, possibly due to a high metabolic cost of yersiniabactin production (35). In our study, the growth benefit of 83972 Y was not enough to confer an advantage over strain 83972AMP in a mixed competition experiment.
Several of the siderophore receptor mutants demonstrated decreased growth in low-iron medium compared to wild-type E. coli 83972. While the 83972 ER and 83972 SR mutants had no growth defect compared to wild-type E. coli 83972, growth of the double enterobactin/salmochelin receptor mutant 83972 ESR was severely affected. The necessity to delete both catecholate receptors can be explained by their redundancy; IroN is able to transport both ferric enterobactin and ferric salmochelin, and FepA is able to transport ferric enterobactin and, to a lesser extent, ferric salmochelin (31, 49). E. coli 83972 also encodes other catecholate receptors, including Iha (which can transport ferric enterobactin and the enterobactin breakdown product DHBS), Cir (which can transport ferric DHBS and ferric salmochelin), and Fiu (which can transport ferric DHBS) (30, 31, 41). However, the poor growth of E. coli 83972 ESR in low-iron medium suggests that these additional catecholate receptors did not compensate for the loss of FepA and IroN under the conditions employed in these experiments. There were also differences between the growth of the 83972 siderophore biosynthetic and receptor mutants; the receptor mutant 83972 ESR had a significantly decreased growth rate compared to that of the biosynthetic mutant 83972 ES in MM9 medium containing 50 μM DIP at pH 7.0 (P < 0.001). This may be because the secretion of enterobactin and salmochelin by 83972 ESR results in the chelation of iron by molecules that cannot be utilized. This observation has been previously reported for Yersinia pestis yersiniabactin biosynthetic and receptor mutants (18). The difference in growth of strains ESR/ESYR compared to EASR/EASYR indicates a role for aerobactin (but not yersiniabactin) during growth in low-iron medium.
We also examined the fitness of the mutants compared to that of 83972AMP in mixed growth competition experiments. Competition experiments have been utilized to examine siderophore cooperation and cheating characteristics in bacteria (26, 34). The extensively studied pyoverdin siderophore of Pseudomonas aeruginosa has a high metabolic cost of production, which results in cheating in this bacterium (34). P. aeruginosa mutants that cannot produce pyoverdin but can still utilize this siderophore (i.e., cheaters) evolve spontaneously both in vitro and in the lungs of patients with cystic fibrosis (15, 34). In this study, we show that the 83972 biosynthetic mutants, including 83972 EASY, can act as cheaters by utilizing the siderophores produced by 83972AMP to rescue their growth deficiencies. However, since the mutants did not outcompete 83972AMP, there was no fitness advantage gained by avoiding the metabolic cost of production of any of the four siderophores. Overall, this suggests that the additional iron acquisition and flexibility afforded to E. coli 83972 by the production of all four siderophores is worth the cost of producing its range of siderophores.
A series of E. coli 83972 siderophore receptor mutants was constructed and utilized to examine competition and fitness in a scenario where the mutants were unable to cheat from the wild-type E. coli 83972. Siderophore mutants with both enterobactin and salmochelin receptor gene deletions (ESR) exhibited a 10-fold decrease in fitness compared to strain 83972AMP, which correlated with the growth deficiency of these strains in monoculture. While aerobactin and yersiniabactin single receptor mutants had no change in fitness compared to strain 83972AMP, strain 83972 AYR as well as 83972 ER and 83972 SR had a small but significant decrease in fitness which could not be detected in monoculture. Since a previous study found that a yersiniabactin deletion mutant in E. coli Nissle 1917 had decreased fitness in M63 medium, the importance of siderophores may vary between strains and growth conditions (62). The fact that E. coli 83972 has all four siderophore receptors could be advantageous in mixed species infections with bacteria which do not have the full repertoire of receptors. Indeed, E. coli 83972 outcompetes UPEC in human urine and in the mouse UTI model, which is the basis for its use as a prophylactic agent in patients with recurrent UTIs (54).
ABU E. coli 83972 was isolated from the urine of a young girl who carried it for 3 years without UTI symptoms (42). E. coli 83972 grows very well in urine and outcompetes UPEC in human urine and in a murine UTI model (54). Given that urine is the most relevant medium for this strain, we repeated the growth and competition assays in pooled human urine. The 83972 siderophore biosynthetic mutants, including 83972 EASY, did not display a growth defect in urine, as seen in MM9 medium containing 50 μM DIP. It was necessary to further delete the ferric citrate receptor fecA in 83972 EASY and also add 50 μM DIP to see a growth defect in urine. This suggests that pooled urine is not as iron-limiting as MM9 medium and that the citrate content of urine may be enough for iron acquisition in this assay. Of the 83972 receptor mutants, all mutants with both FepA and IroN inactivated (ESR) had decreased growth in urine and decreased fitness compared to wild-type E. coli 83972. These strains were selected to be assessed in the mouse model of UTI.
The importance of the catecholate receptors FepA and IroN to colonization of the mouse UTI model was evident with decreased fitness of the 83972 ESR mutant in urine, the bladder, and the kidneys. Since there was no difference between the competitive indexes of 83972 ESR and 83972 EASYR, the IutA and FyuA receptors may not contribute to E. coli 83972 colonization of this animal model. Previous studies examining the role of iron acquisition systems in vivo have largely focused on the colonization and virulence of pathogenic bacteria. Several studies reported that enterobactin contributes to virulence of S. enterica serovar Typhi and S. Typhimurium (22, 25, 66). However, conflicting studies utilizing S. Typhimurium, K. pneumoniae, or avian-pathogenic E. coli demonstrated that enterobactin is not important for virulence (7, 10, 40, 61). Additionally, it has been shown that the salmochelin system, which is linked to the enterobactin system, contributes to virulence and is more likely to be functional in the host than enterobactin since it is not inactivated by host proteins (10, 13, 19, 45, 55). The enterobactin and salmochelin siderophore systems are intrinsically linked at synthesis, secretion, and receptor levels, making it difficult to completely distinguish the contributions to colonization and virulence by these two catecholate siderophores. Recently, Garcia and coworkers found that aerobactin (IutA), yersiniabactin (FyuA), and heme receptors (ChuA and Hma) but not catecholate receptors were important for virulence of the UPEC strains CFT073 and 536 in a mouse UTI model (23). While a CFT073 ΔfepA Δiha mutant was not attenuated in the mouse model, a CFT073 ΔfepA ΔiroN (ESR) mutant was not examined. Due to the redundancy of the catecholate receptors, it may be necessary to delete most or all of the receptors (i.e., FepA, IroN, Iha, Cir, and Fiu), depending on the strain. Indeed, one study demonstrated that it was necessary to delete FepA, IroN, and Cir receptors in S. Typhimurium before virulence attenuation was achieved (48). Additionally, differences in the importance of the enterobactin and salmochelin systems could be due to the asymptomatic nature of E. coli 83972 compared to UPEC strains. It is feasible that different siderophores are required by E. coli 83972, which has less contact with host proteins since it does not adhere, invade, or elicit an immune response (36). Finally, we note that our UTI model employed the use of TLR4-deficient C3H/HeJ mice since they develop higher levels of bacteriuria and thus enabled differences between E. coli 83972 wild-type and siderophore deletion mutants to be more easily quantified. Thus, it is possible that differences in mouse strain susceptibility may also account for the differences observed between E. coli 83972 and CFT073.
Overall, for E. coli 83972 the linked catecholate enterobactin and salmochelin systems were consistently important for iron acquisition at pH 7.0, growth and fitness in low-iron medium, growth and fitness in human urine, and fitness in the mouse UTI model. This may be partially explained by the high Fe3+ binding affinity of enterobactin. However, we also identified contributions of aerobactin to total siderophore activity at both pH 5.5 and 7.0 and a secondary role in growth where it partially compensated for the loss of enterobactin. While yersiniabactin appears to be the least important siderophore for E. coli 83972, it was produced, functioned effectively as an iron chelator in the absence of the other siderophores, and contributed to fitness of E. coli 83972 in combination with other siderophores. Therefore, aerobactin and yersiniabactin may provide a degree of functional redundancy in certain environments or when the catecholate siderophores are less efficient.
E. coli 83972 maintained all four siderophore systems during its 3-year colonization of the bladder of a young girl, whereas genes for type 1 fimbriae, P fimbriae, F1C fimbriae, and the hemolysin toxin were inactivated (37, 53, 68). Several explanations for the advantage of producing multiple siderophore systems have been proposed: inactivation of some siderophores by host proteins, e.g., enterobactin by serum albumin and siderocalin and aerobactin by tear lipocalin (20, 24, 38); differential suitability for iron acquisition in different environments, e.g., superiority of aerobactin at acidic pHs (62); different preferred host iron sources (9); differential ability of siderophore receptors to utilize exogenous siderophores (e.g., FepA transports corynebactin while IroN transports myxochelin C) (49); and also dual functions of siderophore receptors (e.g., IroN mediates cell invasion and FyuA contributes to biofilm formation [16, 28]). Despite this, there is still the possibility of continued evolution in new hosts where the loss of a specific siderophore system may be advantageous. For example, Zdziarski and colleagues recently reported that an E. coli 83972 isolate during deliberate bladder colonization lost genes for the aerobactin siderophore system, which is immunogenic, while expression of the ferric citrate receptor FecA was upregulated (67). Thus, the major advantages of multiple iron uptake systems in UTI-associated E. coli are likely to reflect the ability to successfully compete for iron against both the host and other bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Australian National Health and Medical Research Council (569676). M.A.S. is supported by an ARC Future Fellowship (FT100100662).
We thank Lydia Teo for expert technical assistance.
Footnotes
Published ahead of print 19 September 2011
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Allsopp LP, et al. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78: 1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alteri CJ, Hagan EC, Sivick KE, Smith SN, Mobley HLT. 2009. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 5: e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alteri CJ, Mobley HLT. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 75: 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson P, et al. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect. Immun. 59: 2915–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bäumler AJ, et al. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180: 1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bearden SW, Fetherston JD, Perry RD. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benjamin WH, Jr, Turnbough CL, Posey BS, Briles DE. 1985. The ability of Salmonella typhimurium to produce the siderophore enterobactin is not a virulence factor in mouse typhoid. Infect. Immun. 50: 392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8: S1409–S1421 [DOI] [PubMed] [Google Scholar]
- 9. Brock JH, Williams PH, Licéaga J, Wooldridge KG. 1991. Relative availability of transferrin-bound iron and cell-derived iron to aerobactin-producing and enterochelin-producing strains of Escherichia coli and to other microorganisms. Infect. Immun. 59: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caza M, Lépine F, Dozois CM. 2011. Secretion, but not overall synthesis, of catecholate siderophores contributes to virulence of extra-intestinal pathogenic Escherichia coli. Mol. Microbiol. 80: 266–282 [DOI] [PubMed] [Google Scholar]
- 11. Chu BC, et al. 2010. Siderophore uptake in bacteria and the battle for iron with the host: a bird's eye view. Biometals 23: 601–611 [DOI] [PubMed] [Google Scholar]
- 12. Cox GB, et al. 1970. Mutations affecting iron transport in Escherichia coli. J. Bacteriol. 104: 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crouch MLV, Castor M, Karlinsey JE, Kalhorn T, Fang FC. 2008. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 67: 971–983 [DOI] [PubMed] [Google Scholar]
- 14. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Vos D, et al. 2001. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch. Microbiol. 175: 384–388 [DOI] [PubMed] [Google Scholar]
- 16. Feldmann F, Sorsa LJ, Hildinger K, Schubert S. 2007. The salmochelin siderophore receptor IroN contributes to invasion of urothelial cells by extraintestinal pathogenic Escherichia coli in vitro. Infect. Immun. 75: 3183–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferrières L, Hancock V, Klemm P. 2007. Biofilm exclusion of uropathogenic bacteria by selected asymptomatic bacteriuria Escherichia coli strains. Microbiology 153: 1711–1719 [DOI] [PubMed] [Google Scholar]
- 18. Fetherston JD, Kirillina O, Bobrov AG, Paulley JT, Perry RD. 2010. The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun. 78: 2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fischbach MA, et al. 2006. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U. S. A. 103: 16502–16507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fluckinger M, Haas H, Merschak P, Glasgow BJ, Redl B. 2004. Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob. Agents Chemother. 48: 3367–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A): S5–S13 [DOI] [PubMed] [Google Scholar]
- 22. Furman M, Fica A, Saxena M, Di Fabio JL, Cabello FC. 1994. Salmonella typhi iron uptake mutants are attenuated in mice. Infect. Immun. 62: 4091–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia EC, Brumbaugh AR, Mobley HLT. 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79: 1225–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goetz DH, et al. 2002. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10: 1033–1043 [DOI] [PubMed] [Google Scholar]
- 25. Gorbacheva, Faundez G, Godfrey HP, Cabello FC. 2001. Restricted growth of ent− and tonB mutants of Salmonella enterica serovar Typhi is human Mono Mac 6 monocytic cells. FEMS Microbiol. Lett. 196: 7–11 [DOI] [PubMed] [Google Scholar]
- 26. Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027 [DOI] [PubMed] [Google Scholar]
- 27. Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6: e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hancock V, Ferrières L, Klemm P. 2008. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 154: 167–175 [DOI] [PubMed] [Google Scholar]
- 29. Hancock V, Seshasayee AS, Ussery DW, Luscombe NM, Klemm P. 2008. Transcriptomics and adaptive genomics of the asymptomatic bacteriuria Escherichia coli strain 83972. Mol. Genet. Genomics 279: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hantke K. 1990. Dihydroxybenzolyserine—a siderophore for Escherichia coli. FEMS Microbiol. Lett. 67: 5–8 [DOI] [PubMed] [Google Scholar]
- 31. Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 100: 3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Henderson JP, et al. 2009. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 5: e1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hull R, et al. 2000. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J. Urol. 163: 872–877 [PubMed] [Google Scholar]
- 34. Jiricny N, et al. 2010. Fitness correlates with the extent of cheating in a bacterium. J. Evol. Biol. 23: 738–747 [DOI] [PubMed] [Google Scholar]
- 35. Jones AM, Lindow SE, Wildermuth MC. 2007. Salicylic acid, yersiniabactin, and pyoverdin production by the model phytopathogen Pseudomonas syringae pv. tomato DC3000: synthesis, regulation, and impact on tomato and Arabidopsis host plants. J. Bacteriol. 189: 6773–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klemm P, Hancock V, Schembri MA. 2007. Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect. Immun. 75: 3688–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klemm P, Roos V, Ulett GC, Svanborg C, Schembri MA. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect. Immun. 74: 781–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konopka K, Neilands JB. 1984. Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 23: 2122–2127 [DOI] [PubMed] [Google Scholar]
- 39. Kwon O, Hudspeth MES, Meganathan R. 1996. Anaerobic biosynthesis of enterobactin in Escherichia coli: regulation of entC gene expression and evidence against its involvement in menaquinone (vitamin K2) biosynthesis. J. Bacteriol. 178: 3252–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lawlor MS, O'Connor C, Miller VL. 2007. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 75: 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Léveillé S, et al. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 74: 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindberg U, et al. 1975. Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic and symptomatic bacteriuria. Acta Paediatr. Scand. 64: 432–436 [DOI] [PubMed] [Google Scholar]
- 43. Mabbett AN, et al. 2009. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 299: 53–63 [DOI] [PubMed] [Google Scholar]
- 44. Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71: 413–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nègre VL, et al. 2004. The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect. Immun. 72: 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Payne SM. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235: 329–344 [DOI] [PubMed] [Google Scholar]
- 47. Perry RD, Balbo PB, Jones HA, Fetherston JD, DeMoll E. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145: 1181–1190 [DOI] [PubMed] [Google Scholar]
- 48. Rabsch W, et al. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 71: 6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rabsch W, Voigt W, Reissbrodt R, Tsolis RM, Bäumler AJ. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 181: 3610–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Raymond KN, Carrano CJ. 1979. Coordination chemistry and microbial iron transport. Acc. Chem. Res. 12: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ronald A. 2003. The etiology of urinary tract infection: traditional and emerging pathogens. Dis. Mon. 49: 71–82 [DOI] [PubMed] [Google Scholar]
- 52. Roos V, Klemm P. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 74: 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roos V, Schembri MA, Ulett GC, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152: 1799–1806 [DOI] [PubMed] [Google Scholar]
- 54. Roos V, Ulett GC, Schembri MA, Klemm P. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Russo TA, et al. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70: 7156–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 70: 5335–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160: 47–56 [DOI] [PubMed] [Google Scholar]
- 58. Snyder JA, et al. 2004. Transcriptome of uropathogenic Eschetichia coli during urinary tract infection. Infect. Immun. 72: 6373–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sundén F, Håkansson L, Ljunggren E, Wullt B. 2006. Bacterial interference—is deliberate colonization with Escherichia coli 83972 an alternative treatment for patients with recurrent urinary tract infection? Int. J. Antimicrob. Agents 28: S26–S29 [DOI] [PubMed] [Google Scholar]
- 60. Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69: 6179–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsolis RM, Bäumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64: 4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K. 2006. Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int. J. Med. Microbiol. 296: 513–520 [DOI] [PubMed] [Google Scholar]
- 63. Wells M, Gösch M, Harms H, van der Meer JR. 2005. Response characteristics of arsenic-sensitive bioreporters expressing the gfp reporter gene. Microchim. Acta 151: 209–216 [Google Scholar]
- 64. Wullt B, et al. 2001. P-fimbriae trigger mucosal responses to Escherichia coli in the human urinary tract. Cell. Microbiol. 3: 255–264 [DOI] [PubMed] [Google Scholar]
- 65. Wullt B, et al. 1998. Urodynamic factors influence the duration of Escherichia coli bacteriuria in deliberately colonized cases. J. Urol. 159: 2057–2062 [DOI] [PubMed] [Google Scholar]
- 66. Yancey RJ, Breeding SAL, Lankford CE. 1979. Enterochelin (enterobactin): virulence factor for Salmonella typhimurium. Infect. Immun. 24: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zdziarski J, et al. 2010. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog. 6: e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. 2008. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect. Immun. 76: 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.