Abstract
Since inactivation of tumor suppressor p53 functions is one of the most common features of human cancer cells, restoring p53 expression and activity is an important focus in cancer therapy. Here we report identification of photoreceptor-specific nuclear receptor (PNR)/NR2E3 as a positive regulator of p53 in a high-throughput genetic screen. In HeLa cells, PNR stimulated p53-responsive promoters in a p53-dependent fashion and induced apoptosis in several cell types. PNR also increased p53 protein stability and specific activity as a transcriptional activator. Our studies of the underlying mechanisms showed that PNR forms complexes with p53 and the acetyltransferase p300, stimulates p53 acetylation, and increases the expression of a subset of p53 target genes. Furthermore, PNR significantly boosted actinomycin D-stimulated p53 acetylation. The unique mechanisms by which PNR stimulates p53 acetylation and functions define this orphan nuclear receptor as a potentially valuable target and tool in p53-associated cancer therapy and offer new insights into the roles of PNR mutation in retinal diseases.
INTRODUCTION
In most cancers, normal p53 functions are abrogated by p53 mutations, transcriptional inhibition, or posttranslational modifications. Since p53 gene transcription is under tight control (35, 36), it is valuable to identify factors that regulate p53 posttranslationally as potential targets for p53-based cancer therapy. MDM2, a major regulator of p53 stability, also blocks the transactivation domain of p53 and enhances p53 nuclear export (12, 13, 20). Nutlins, which are antagonists of MDM2 and promising cancer therapeutic drugs, bind the p53 binding pocket of MDM2, resulting in activation of p53 (47).
One important mechanism for p53 posttranslational regulation is acetylation (2, 3, 10, 21). p53 acetylation at multiple sites directly affects p53 stability, DNA binding, and transactivation. Accordingly, p53 acetylation is commonly targeted by viral proteins to inactivate p53. One example is the inhibition of p53 by human papillomavirus (HPV) oncoprotein E6. HPVs cause over 5% of all human cancers, including essentially all cervical cancers and ∼25% of head and neck cancers as well as other cancers (9, 32). Many HPV-positive (HPV+) cancer cell lines retain a wild-type p53 gene, but E6 abrogates p53 functions both by stimulating p53 ubiquitination and inhibiting p53 acetylation (54). Disrupting E6-mediated inhibition of p53 by knocking down E6 or E6AP significantly restores p53 function and induces cell apoptosis (15).
To identify additional targets for p53-based cancer therapy for HPV+ and potentially other cancer patients, we have now used a high-throughput screen of full-length, mammalian cDNA overexpression plasmids to identify photoreceptor-specific nuclear receptor (PNR/NR2E3) as a gene that enhanced p53 accumulation in HPV+ HeLa cells. PNR/NR2E3, a member of nuclear receptor subfamily 2, is highly expressed in retinal cone and rod cells. With increased characterization, PNR expression has been detected in additional tissues, such as the prostate and uterus (5, 30). Although PNR mutants are implicated as a causative factor for enhanced S-cone syndrome, a cone cell hyperplasia disorder, the mechanism(s) of PNR involvement in the etiology of this disease remains poorly characterized (11). PNR interacts with several transcription factors to inhibit cone opsin expression and enhance rod opsin expression (31). Moreover, PNR binds to and represses the promoter of cyclin D1, which promotes G1/S progression and cell proliferation, implying that wild-type PNR attenuates proliferation of S-cone cells from retinal progenitor cells (42).
In addition to identifying PNR's effects on p53, we show here that PNR stimulates p53 accumulation and functions by enhancing p53 acetylation, a mechanism distinct from the means of regulation of p53 by other nuclear receptors. Since nuclear receptors are proven pharmaceutical targets, PNR, a novel modulator of p53, may serve as a new target for p53-based cancer therapy.
MATERIALS AND METHODS
Plasmids.
The pCMV-SP6-PNR plasmid expressing PNR was constructed by subcloning a full-length wild-type PNR into a pCMV-SP6 expression vector from pcDNA3.1/HisC-PNR (31), kindly provided by S. M. Chen (Washington University). The pCMV-SP6-HA-PNR plasmid expressing N-terminally hemagglutinin (HA)-tagged PNR (see Fig. 7 and 8) was constructed by adding an HA tag coding sequence to the 5′ terminus of PNR with no space. Reporter plasmid p53RE-FLuc, expressing firefly luciferase from a p53-responsive promoter containing two tandem p53-responsive elements, was from Panomics (catalog no. LR0057). A p53RE-FLuc derivative with the p53 binding site inactivated was generated by mutating critical CXXG residues (7) into AXXT with a QuikChange II XL site-directed mutagenesis kit (Agilent catalog no. 200521). The primers used for this mutation were 5′-CGC GTG CTA GCT ACA GAA aAT tTC TAA GaA TtC TGT GCC TTG CCT GGA aTT tCC TGG CaT TtC CTT GGG AGA TCT GGG TAT-3′ and 5′-ATA CCC AGA TCT CCC AAG GaA AtG CCA GGa AAt TCC AGG CAA GGC ACA GaA TtC TTA GAa ATt TTC TGT AGC TAG CAC GCG-3′, where the lowercase letters represent mutated nucleotides. A plasmid expressing human p53 dominant-negative mutant p53C135Y was from Clontech (catalog no. 631922). A pCMV-SP6-Pitx2a plasmid expressing Pitx2a was constructed by subcloning full-length wild-type Pitx2a into a pCMV-Sp6 expression vector from a green fluorescent protein-Pitx2a (GFP-Pitx2a) plasmid (50), kindly provided by Q. Z. Wei (Kansas State University).
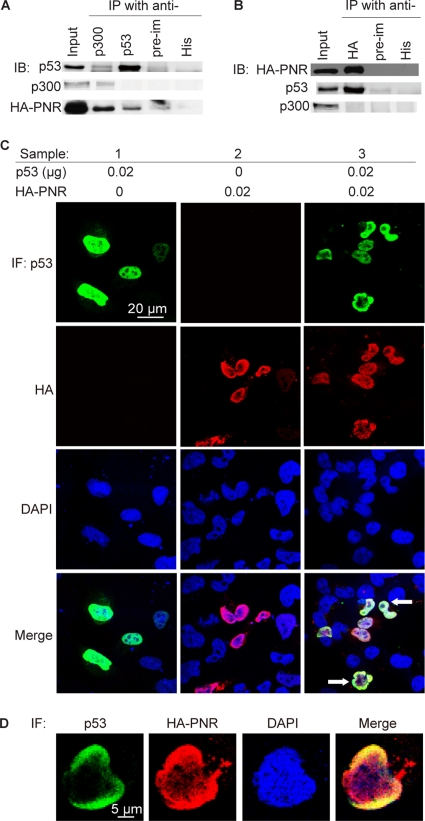
Fig 7.
PNR participates in a complex of p53 and p300 in HeLa cells. (A and B) Two days after cotransfection of HA-tagged PNR, p53, and p300, cells were lysed for immunoprecipitation with the indicated antibodies. Preimmune IgG and anti-His antibody were used as nonspecific binding controls. (A) Anti-p53 and anti-p300 antibodies coimmunoprecipitate (IP) HA-PNR. (B) Anti-HA antibody coimmunoprecipitates p53. (C and D) p53 and PNR colocalize in nuclei. Subcellular localizations of p53 and PNR were visualized by immunofluorescence (IF). Green, p53; red, HA; blue, DAPI. (C) Sample 1, p53 transfection; 2, PNR transfection; 3, cotransfection of PNR and p53. Arrow, colocalization of PNR and p53. (D) Enlarged image of a representative nucleus from a field of cells cotransfected with PNR and p53.
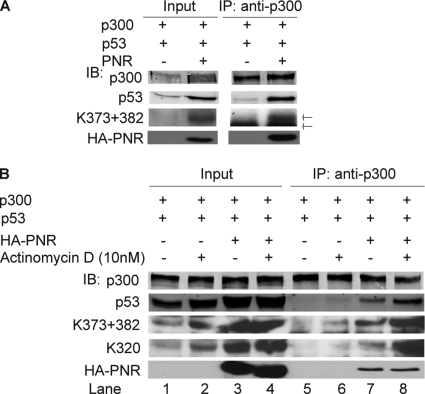
Fig 8.
PNR enhances formation of a complex consisting of p53 and p300 with or without actinomycin D treatment in HeLa cells. (A) PNR enhances p300's binding to total p53 and Ac-p53. Anti-p300 antibody coimmunoprecipitated p53. Both p53 and Ac-p53 (K373 + 382) were measured by immunoblotting. Upper arrow, Ac-p53; lower arrow, rabbit IgG heavy chain. (B) PNR boosts actinomycin D-enhanced association between p53 and p300. One day after cotransfection of the indicated plasmids, cells were treated with 10 nM actinomycin D for 24 h and then processed as described for panel A.
Cell culture.
HeLa cells (ATCC catalog no. CCL-2), p53+/+ and p53−/− HCT116 cells (kindly provided by B. Vogelstein, John Hopkins University), and p53-null H1299 cells (a gift from W. Sugden, University of Wisconsin—Madison) were cultured at 37°C in Dulbecco's modified Eagle medium (DMEM)–10% fetal bovine serum (FBS) (heat inactivated) in a 5% CO2 atmosphere.
Luciferase reporter assays.
Luciferase reporter assays were performed using 96-well plates and reverse transfection (RT). For each well, 0.01 μg of reporter plasmid p53RE-Fluc, 0.005 μg of phRL-SV40 expressing Renilla luciferase, and the indicated amounts of PNR expression plasmid and other plasmids were added to a total of 0.085 μg of DNA in 5 μl of Opti-MEM. TransIT-LT1 (0.17 μl) was mixed with 9 μl of Opti-MEM, mixed with the DNA as described above, transferred to the microplate, and incubated for 30 min at room temperature. A total of 1.2 × 104 cells in 100 μl of medium were then added per well. Two days after transfection, cells were processed using a Dual-Glo luciferase assay (Promega catalog no. E2940) and a Perkin-Elmer luminometer.
Apoptosis assays.
For an annexin V binding assay, 106 cells were seeded per 6-cm-diameter dish and incubated for 18 h at 37°C. Equal amounts of total plasmids were then cotransfected using Lipofectamine 2000 (Invitrogen catalog no. 11668-019) (2:1 Lipofectamine:DNA solution) following the manufacturer's protocol. The medium was changed 6 h after transfection. Two days after transfection, a Vybrant 2 Alexa Fluor 488 apoptosis assay kit (Invitrogen catalog no. V13241) was used to label apoptotic cells according to the manufacturer's instructions. Flow cytometry was used to count the labeled cells, and the data were analyzed using Flowjo software. The percentages of apoptotic cells (see Fig. 3) represent the proportions of Alexa Fluor 488-labeled cells among the transfected cells expressing red fluorescent protein.
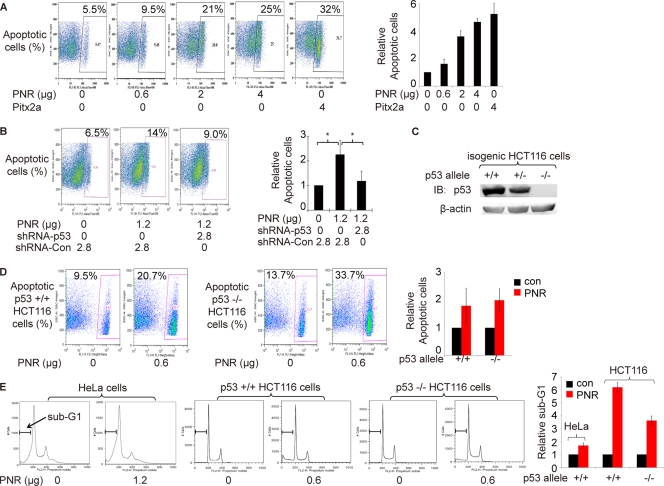
Fig 3.
p53 enhances PNR-induced cell apoptosis. (A) PNR induces HeLa cell apoptosis in a dose-dependent fashion. Two days after transfection, an annexin V binding assay was used to quantitate apoptotic cells by flow cytometry. Pitx2a was used as a positive control (50). The fraction of apoptotic cells in the sample with 0 μg of PNR was normalized to 1. Data representing the results of three repeated experiments are plotted on the right. (B) shRNAs targeting p53 significantly inhibit PNR-induced HeLa cell apoptosis. shRNA-Con, pLKO.1 empty expression vector control. The right plot shows relative levels of apoptosis based on the means of data determined under each set of conditions in 3 independent experiments. The mean for samples with only shRNA-Con was normalized to 1. *, P < 0.05 (one-tailed test). (C) p53 protein levels in three isogenic HCT116 cell lines containing two p53 alleles, one allele, or none. (D and E) PNR induces cell apoptosis in both p53+/+ and p53−/− HCT116 cells. Two days after transfection of PNR expression plasmid, the isogenic HCT116 cells were processed for both an annexin V binding assay and sub-G1 analysis. The data from three repeated experiments is plotted on the right. (D) Annexin V binding assay. PNR mildly induced apoptosis at similar levels in both cell lines. (E) Sub-G1 analysis. PNR induced apoptosis in p53+/+ HCT116 cells at a level ∼2-fold higher than the level seen in p53−/− HCT116 cells. PNR also increased apoptosis at the sub-G1 phase in HeLa cells.
Sub-G1 apoptosis assay.
Cells in 6-cm-diameter dishes were transfected with Lipofectamine 2000 as described above, except that the indicated plasmids were used. Two days after transfection, cells were detached, treated as described elsewhere (http://sciencepark.mdanderson.org/fcores/flow/files/DNA_PI.html), and measured by flow cytometry.
Immunoblotting.
Cells in 6-cm-diameter dishes were transfected with Lipofectamine 2000 as described above, except that the indicated plasmids were cotransfected. Cells were harvested 2 days after transfection. After two washes with cold phosphate-buffered saline (PBS), the cell pellet was lysed with 200 μl of radioimmunoprecipitation assay (RIPA) buffer (Pierce catalog no. 89900) plus protease inhibitors (Roche catalog no. 11873580001) for 15 min on ice. A 50-μl volume of 5× sodium dodecyl sulfate (SDS) sample buffer was added. The sample was sonicated twice for 5 s each time with a probe sonicator at 4°C and heated for 15 min at 99°C with vigorous vortex mixing. After centrifugation at 9,200 × g for 10 min at 4°C, the supernatant was stored at −20°C.
Cell lysates were subjected to electrophoresis on a freshly made 10% SDS-polyacrylamide gel and transferred to nitrocellulose membranes (GE catalog no. RPN303D) in transfer buffer with 10% methanol, except that 5% SDS-polyacrylamide gel electrophoresis (5% SDS-PAGE) was used to resolve p300. The membrane was blocked with Odyssey blocking buffer (LiCor catalog no. 927-40000) for 1 h at room temperature, incubated with primary antibodies for 1 h at room temperature, and washed 5 times for 5 min each time with PBS–0.1% Tween 20. For semiquantitative analysis, the membrane was then incubated with fluorescent secondary antibodies (LiCor goat anti-rabbit antibody conjugated with IRDye 800cw [catalog no. 926-32211] and goat anti-mouse antibody conjugated with IRDye 680 [catalog no. 926-32220]) for 40 min at room temperature, followed by 5 washes performed as described above. The air-dried membranes bearing a dilution curve of each sample were scanned using a LiCor Odyssey imager, and images were analyzed using Odyssey version 3.0 software. For detection of HA-tagged PNR (see Fig. 8), samples were transferred to polyvinylidene difluoride (PVDF) membranes in transfer buffer with 15% methanol, a rat anti-HA antibody conjugated to horseradish peroxidase (HRP; Roche catalog no. 12013819001) was used as the primary antibody, and the secondary antibody was omitted.
For immunoblotting, anti-acetylated p53 antibodies (K373 + 382 [Upstate catalog no. 06-758], K382 [Upstate catalog no. 04-1146], K320 [Upstate catalog no. 06-1283], and K120 [AbCam catalog no. ab78316]) (1:1,000), mouse anti-total p53 antibody (Calbiochem catalog no. OP43) (1:1,000), rabbit anti-PNR antibody (Sigma catalog no. P5373) (1:1,000), rabbit anti-p300 antibody (Santa Cruz catalog no. sc-584) (1:1,000), rabbit anti-β-actin antibody (Santa Cruz catalog no. sc-1616-R) (1:3,000), mouse anti-GFP antibody (Covance catalog no. MMS-118R) (1:6,000), and LiCor fluorescent secondary antibodies (1:10,000) were diluted in Odyssey blocking buffer as indicated and mouse anti-p21 antibody (Santa Cruz catalog no. SC-56335) (1:500), HRP-conjugated rat anti-HA antibody (1:2,000), HRP-conjugated goat anti-mouse secondary antibody (1:10,000), and HRP-conjugated mouse anti-rabbit light chain secondary antibody (1:20,000) were diluted in PBS–0.1% Tween 20–5% nonfat milk as indicated.
Immunoprecipitation.
Cells in 6-cm-diameter dishes were transfected with Lipofectamine 2000 as described above, except that 0.5 μg of p53-expressing plasmid, 1.5 μg of HA-PNR-expressing plasmid, and 1.5 μg of p300-expressing plasmids were cotransfected. Two days after transfection, cells were detached and washed 3 times with cold PBS. Cell pellets were resuspended with 400 μl of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.5 mM PMSF) and incubated for 15 min on ice. A 25-μl volume of 10% NP-40 was added, followed by vigorous vortex mixing for 10 s. The lysate was centrifuged at 2,300 × g for 1 min at 4°C. The supernatant (cytoplasmic extract) was kept on ice. The nuclear pellet was resuspended in 200 μl of buffer B (20 mM HEPES [pH 7.9], 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5 mM PMSF) and sonicated twice for 5 s each time. The resulting nuclear lysate was subjected to vigorous vortex mixing for 15 min and centrifuged at 13,000 × g for 10 min at 4°C. The supernatant (nuclear extract) was combined with the cytoplasmic extract for immunoprecipitation.
Two micrograms of rabbit antibodies against p53 (Santa Cruz catalog no. sc-6243) or p300 (Santa Cruz catalog no. sc-584) or HA (Sigma catalog no. H6908) was mixed with 50 μl of protein A Dynabeads (Invitrogen catalog no. 100-01D). Two micrograms of rabbit anti-His antibody (catalog no. sc-803) and 2 μg of preimmune rabbit IgG (catalog no. sc-2344) from Santa Cruz were used as controls. Immunoprecipitation was performed following the manufacturer's instructions, except that the Dynabeads were washed 6 times for 30 s each time after immunoprecipitation. Dynabeads were then incubated with 100 μl of 1× SDS sample buffer for 20 min at 50°C with vigorous vortex mixing to elute proteins. The beads were centrifuged at 9,200 × g for 1 min at 4°C, and the supernatant was stored at −20°C after 10 min at 99°C.
Immunofluorescence.
Cells were reverse transfected described above in “Luciferase reporter assays,” except that 8-chamber slides were used and all material volumes were doubled for each chamber. Two days after transfection, the cells were washed 3 times with PBS and fixed in 1% paraformaldehyde–PBS for 30 min at room temperature. Cells were washed with PBS 3 times and permeabilized in 0.5% Triton X-100–PBS for 30 min at room temperature. After another 3 washes, the cells were blocked in PBS containing 5% normal horse serum and DAPI (4′,6-diamidino-2-phenylindole) (5 μg/ml) for 5 to 6 h at 4°C. Subsequently, cells were washed 4 times and rabbit anti-p53 antibody (Santa Cruz catalog no. sc-6243) (1:200) and mouse anti-HA antibody (Roche catalog no. 11583816001) (1:100) were added to PBS containing 5% normal horse serum to cover the cells for 8 h at 4°C. The cells were washed 4 times and incubated with goat anti-mouse Alexa Fluor 568-conjugated antibody (Invitrogen catalog no. A11004) (1:1,000) and goat anti-rabbit Alexa Fluor 488-conjugated antibody (Invitrogen catalog no. A31627) (1:1,000) in PBS containing 3% normal horse serum for 1 h at room temperature. After 4 washes, the slide was briefly dried, 40 μl of mounting medium containing DAPI (Vector Laboratories catalog no. H-1200) was added, and the slide was covered with a coverslip for imaging with a confocal microscopy.
Reverse-transcription real-time PCR.
Cells were reverse transfected as described for the reporter assays above, except that 6-cm-diameter dishes were used and all material volumes were increased by 60-fold. Two days after transfection, total RNA was extracted using an RNeasy minikit (Qiagen catalog no. 74106) and treated using DNase for 30 min at 37°C. A 0.1-μg volume of total RNA was added to each reaction mixture.
TaqMan One-Step RT-PCR Master Mix reagents (Applied Biosystems catalog no. 4309169) were used for reverse transcription real-time PCR with a 7900HT real-time PCR system (Applied Biosystems) programmed for 48°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. mRNA levels of β-actin or RPL38 were used as internal controls. The ratio of the TaqMan primer to the probe was fixed at 2:1. The final probe concentration in a 20-μl reaction volume per well in a 96-well plate was approximately 50 to 100 nM. The primer and probe sets (listed here as forward primer, reverse primer, and probe) were as follows: for p53, 5′-TTTCCGTCTGGGCTTCT-3′, 5′-TGGAATCAACCCACAGCT-3′, and 5′–6-carboxyfluorescein (FAM)/TGTGACTTGCACGTACTCCCCTG/IBFQ-3′; for p21, 5′-TTCCTGTGGGCGGATTA-3′, 5′-GAGCAGGCTGAAGGGT-3′, and 5′-FAM/CGTTTGGAGTGGTAGAAATCTGTCATGC/IBFQ-3′; for Puma, 5′-GAGATGGAGCCCAATTAGGTG-3′, 5′-ACATGGTGCAGAGAAAGTCC-3′, and 5′-FAM/AGGGTGTCAGGAGGTGGGAGG/IBFQ-3′; for MDM2, 5′-TGCCAAGCTTCTCTGTGAAAG-3′, 5′-TCCTTTTGATCACTCCCACC-3′, and 5′-FAM/ACCTGAGTCCGATGATTCCTGCTG/IBFQ-3′; for Pirh2, 5′-GGTCAAGAGCGAGGTCAG-3′, 5′-CACAAGCGGCAAGTATAAAGC-3′, and 5′-FAM/ACAGCAAGGTGCCTTTAGGAGACATC/IBFQ-3′; for PNR, 5′-GGGAAGCACTATGGCATCTATG-3′, 5′-CACCTGGCACCTGTAGATG-3′, and 5′-FAM/CGCCGTACGCTCCTCTTGAAGAA/IBFQ-3′; for RPL38, 5′-GCAGATACCTTTACACCCTGG-3′, 5′-CTGGTTCATTTCAGTTCCTTCAC-3′, and 5′-FAM/TGCTTCAGTTTCTCTGCCTTCTCTTTGT/IBFQ-3′; and for β-actin, 5′-TCACCCACACTGTGCCCATCTACGA-3′, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′, and 5′-FAM/ATGCCCTCCCCCATGCCATCCTGCGT/TAMRA-3′.
Protein stability assay.
Cells in 6-cm-diameter dishes were processed as described above for immunoblotting, except that the indicated plasmids were transfected and cells were treated with cycloheximide (Sigma catalog no. C4859) (2 μg/ml) for 0, 15, 30, or 60 min before being harvested for immunoblotting.
RNA interference.
pLKO.1-based short hairpin RNA (shRNA) expression plasmids were cotransfected with the indicated plasmids and transfection reagents in the different assays as described above. Two days after transfection, cells were processed for assays. For each target gene, two shRNA plasmids were validated to efficiently knock down the indicated gene at two different fragments. shRNA expression plasmids targeting p53 (catalog no. RHS4533-NM_000546), PNR (catalog no. RHS4533-NM_014249), p300 (catalog no. RHS4533-NM_001429), GFP (catalog no. RHS4459), and pLKO.1 empty vector plasmid (catalog no. RHS4080) were from Open Biosystems.
Statistical analysis.
P values were calculated using paired Student's t-tests.
RESULTS
PNR enhances p53 accumulation and selectively activates p53-responsive promoters.
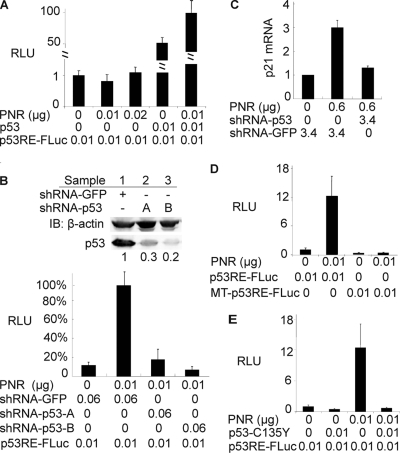
We identified PNR as a modulator of p53 in a screen for genes whose expression increased accumulation of a directly assayable p53-luciferase fusion protein in HPV+ HeLa cells (Fig. 1A). To validate PNR's ability to increase accumulation of a p53-luciferase fusion, we tested PNR's effect on the level of endogenously expressed p53 in HeLa cells. In untransfected HeLa cells, endogenous p53 was only weakly detectable by immunoblotting, presumably due to HPV E6-mediated, proteasome-dependent degradation (Fig. 1B). Transfecting a PNR-expressing plasmid increased the level of endogenous p53 in a dose-dependent fashion by 2- to 3-fold as measured using a quantitative immunoblotting approach (see Materials and Methods).
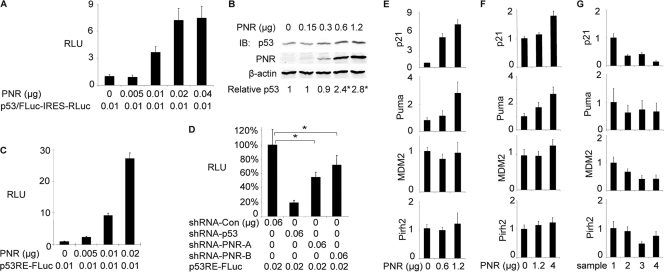
Fig 1.
PNR enhances accumulation of p53 protein in HeLa cells and selectively stimulates p53-responsive promoters in both HeLa cells and HCT116 cells. (A) PNR increases levels of p53 and firefly luciferase fusion protein (p53/FLuc) in a dose-dependent fashion in HeLa cells. A p53/FLuc-IRES-RLuc reporter plasmid expressing the p53/FLuc fusion protein and Renilla luciferase as an internal control was cotransfected with the indicated amounts of PNR expression plasmid into HeLa cells in a 96-well plate. Two days after transfection, the luciferase activities were assayed. RLU, relative luciferase activity; FLuc, firefly luciferase; IRES, internal ribosome entry site; Rluc, Renilla luciferase. (B) PNR enhances accumulation of endogenous p53 in HeLa cells. p53 levels were measured by immunoblotting (IB) 2 days after transfection of PNR expression plasmid into HeLa cells in 6-cm-diameter dishes. The ratio of p53 signal to β-actin signal in the sample with 0 μg of PNR was normalized to 1. *, P < 0.05 (one-tailed test). (C) PNR stimulates p53RE-FLuc, a p53-responsive firefly luciferase reporter plasmid, in a dose-dependent fashion in HeLa cells. Cells were cotransfected with the indicated amounts of a PNR expression plasmid and other plasmids in a 96-well plate. Two days later, the luciferase activities were assayed and normalized to the level in the sample with 0 μg of PNR. (D) Endogenous PNR contributes to p53 response in HeLa cells. Cells in a 96-well plate were cotransfected with 0.02 μg of p53RE-FLuc and the indicated shRNA expression plasmids targeting p53 and PNR. shRNA-Con, pLKO.1 empty lentiviral control. The FLuc activity in the sample with shRNA-Con was normalized to 100%. *, P < 0.05 (two-tailed test). (E, F, and G) PNR selectively stimulates p53 target genes in HeLa cells and p53+/+ HCT116 cells. Cells were transfected with the indicated amounts of the PNR expression plasmid (E and F) or shRNA expression plasmids (G) in 6-cm-diameter dishes. Two days later, mRNA levels of the indicated p53 target genes were measured by qRT-PCR and normalized to β-actin or RPL38 mRNA levels. For each gene, the mRNA level in the sample with 0 μg of PNR (E and F) or shRNA-GFP (G) was normalized to 1. (E) PNR overexpression in HeLa cells. (F) PNR overexpression in HCT116 cells. (G) Knockdown of endogenous PNR in HeLa cells. Sample 1, shRNA-GFP; sample 2, shRNA-PNR-A; sample 3, shRNA-PNR-B; sample 4, shRNA-p53.
Our subsequent studies showed that PNR stimulated expression of the wild-type, unfused luciferase gene from a p53-responsive reporter plasmid in a dose-dependent manner to a level over 20-fold higher than that seen with an empty vector control in HeLa cells (Fig. 1C). PNR also stimulated expression from this p53-responsive reporter in RKO (data not shown), a HPV− but p53+ human colon carcinoma cancer cell line, showing that PNR stimulation of p53 activity is not restricted to HPV+ cells. To determine whether PNR enhances p53 binding to DNA, we used an electrophoretic mobility shift assay with a p53 binding probe and found that nuclear extracts of PNR-transfected HeLa cells exhibited 5-fold-higher p53-specific DNA binding activity than GFP-transfected control cells (data not shown). This implied that PNR increased p53 levels or p53 DNA binding activity or both.
Next, we examined whether PNR could activate endogenous p53 target genes. Four well-characterized p53 target genes, including p21waf1 and Puma, whose functions are related to cell growth inhibition, and MDM2 and Pirh2, which protect cells from excessive p53 activation by negative-feedback regulation (18), were selected for study. As shown by quantitative RT-PCR (qRT-PCR), PNR transfection of HeLa cells resulted in dose-dependent increases in p21 and Puma mRNA levels, although neither MDM2 nor Pirh2 mRNA levels were changed (Fig. 1E). This selective regulation of p53 target genes by PNR also occurred in HCT116, an E6− but p53+/+ human colon cancer cell line (Fig. 1F). Thus, PNR preferentially upregulates a subset of p53 target genes. The nature of this selectivity is further discussed below.
Since the transfected PNR stimulated a subset of p53-responsive genes, we next examined whether endogenous PNR could modulate p53-responsive promoters. Quantitative RT-PCR showed that the low mRNA level of endogenous PNR in HeLa cells was knocked down by ∼80% using shRNA plasmids against PNR (data not shown). As a positive control, an shRNA against p53 inhibited 80% of luciferase activity from a p53-responsive reporter plasmid (Fig. 1D). shRNAs against PNR inhibited approximately 30% to 40% of the luciferase activity of the p53-responsive reporter compared with an empty shRNA expression vector control (Fig. 1D). We also measured changes in mRNA levels for p53 target genes in HeLa cells when endogenous PNR was knocked down by these two shRNA plasmids. As shown in Fig. 1G, the p21 mRNA level was reduced by up to 65%, while lesser changes were detected in the mRNA levels of MDM2, Puma, and Pirh2. To exclude the possibility that PNR stimulation of p53-target genes is HeLa cell specific, endogenous PNR was knocked down in HCT116 cells. Both p53 mRNA and protein levels were significantly decreased, and the p21 mRNA level was also reduced by ∼50% (data not shown). Thus, endogenous PNRs regulate the p53 level and p53-responsive gene expression in HeLa cells and HCT116 cells, regardless of the presence or absence of E6.
PNR stimulation of p53-responsive promoters is p53 dependent.
To test whether PNR stimulates p53-responsive promoters in a p53-dependent manner, we used H1299, a p53-null human lung carcinoma cell line. As expected, exogenous p53 stimulated a p53-responsive luciferase reporter in these p53− H1299 cells (Fig. 2A). Moreover, in keeping with the p53-dependent activity, cotransfected PNR enhanced this p53-mediated stimulation, whereas PNR alone had no effect on the reporter in the absence of transfected p53 (Fig. 2A). To exclude the possibility that unknown factors differentially expressed in H1299 and HeLa cells resulted in nonspecific effects, we used shRNAs to further examine the effects of knocking down endogenous p53 in HeLa cells. Quantitative immunoblotting using shRNA plasmids against p53 showed that the low protein level of endogenous p53 in HeLa cells was knocked down by approximately 70% to 80% (Fig. 2B, upper panel). Although p53-dependent luciferase expression was ∼10-fold higher with exogenous PNR (0.01 μg of PNR plasmid) (Fig. 1C and 2B [lower panel]), knocking down p53 inhibited reporter expression by a similar (∼5- to 10-fold) fraction in either the presence (Fig. 2B, lower panel) or absence (Fig. 1D) of exogenous PNR. This proportional response implies that the dramatic PNR stimulation of the p53-responsive reporter was directly dependent on the presence of p53. Consistent with this, shRNAs against p53 also abolished the upregulation of endogenous p21 transcription in HeLa cells by exogenous PNR (Fig. 2C) and, in the absence of exogenous PNR, suppressed the mRNA levels of p21 and MDM2, with lesser effects on two other p53 target genes, PUMA and Pirh2 (Fig. 1G). Together, these results indicated that PNR alone does not activate p53-responsive promoters; rather, it stimulates p53 target genes in a p53-dependent fashion.
Fig 2.
PNR-mediated stimulation of p53-responsive promoters requires p53. (A) In p53-null H1299 cells, PNR alone fails to stimulate p53RE-FLuc whereas PNR in the presence of exogenous p53 stimulates p53RE-FLuc. The RLU in the sample with 0 μg of p53 and PNR was normalized to 1. (B) shRNAs targeting p53 abolish PNR-mediated stimulation of p53RE-FLuc in HeLa cells. shRNA-GFP-expressed shRNA targeting GFP was used as a control. The RLU in the sample with both shRNA-GFP and PNR was normalized to 100%. Upper panel: immunoblot showing β-actin loading control and shRNA-mediated reductions in p53 protein accumulation. The numbers below each band represent the level of p53 protein normalized to β-actin and to the p53 level in sample 1. Lower panel: reporter assay. (C) shRNAs targeting p53 significantly inhibit PNR-mediated increases of p21 mRNA levels in HeLa cells. The p21 mRNA level was measured by qRT-PCR normalized to the sample with only shRNA-GFP transfected. (D) Mutation of p53-responsive elements abrogates PNR-mediated stimulation of MT-p53-RE-FLuc in HeLa cells. The RLU in the sample with only p53RE-FLuc was normalized to 1. (E) Dominant-negative mutant p53-C135Y abrogates the PNR-mediated stimulation of p53RE-FLuc in HeLa cells. The RLU in the sample with only p53RE-FLuc was normalized to 1.
To validate further the idea that PNR specifically activates p53-responsive promoters, we mutated key CXXG elements to AXXT in the two p53-responsive elements in the luciferase reporter plasmid, thus suppressing p53 binding (7). As expected, transfecting PNR did not stimulate expression from this mutated reporter plasmid in HeLa cells, demonstrating that PNR stimulation requires the known p53-responsive elements (Fig. 2D). Finally, we examined p53C135Y, a p53 dominant-negative mutant that sequesters wild-type p53 into a DNA binding-deficient heterotetramer (6). This mutant was cotransfected with PNR and the p53-responsive reporter into HeLa cells that express endogenous, wild-type p53. PNR's ability to stimulate p53 transactivation was abrogated by the cotransfected p53C135Y (Fig. 2E). Together, our data implied that de novo p53 binding to its responsive DNA elements is required for PNR-mediated stimulation of p53 transactivation.
PNR stimulates apoptosis in multiple cell lines.
Since p53 activation results in tumor inhibition via induction of apoptosis or cell cycle arrest (8), we tested whether transfecting PNR potentiates p53-mediated growth inhibition in HeLa cells. To elucidate whether PNR enhances apoptosis, we monitored the cell surface exposure of phosphatidylserine, a marker of cells in early-stage apoptosis (17). Two days after transfection, we used flow cytometry to measure the percentage of HeLa cells labeled by the phosphatidylserine binding protein annexin V. As a positive control, we used Pitx2a, which, in HPV+ HeLa cells, reactivates p53 and induces apoptosis by binding to the HPV E6 oncoprotein (50). As expected, Pitx2a induced apoptosis by ∼5- to 6-fold compared with an empty vector control (Fig. 3A). Similarly, PNR induced apoptosis in a dose-dependent fashion by up to ∼4- to 5-fold. When p53 was knocked down by shRNA, the PNR-mediated induction of cell apoptosis was drastically attenuated, implying that p53 may contribute to PNR-induced HeLa cell apoptosis (Fig. 3B). In addition to HPV+ HeLa cells, we found that PNR transfection similarly induced apoptosis in HPV− but p53+ RKO cells (data not shown).
In addition to its effects on p53, PNR can inhibit cell proliferation by repressing transcription of tumor-supporting genes, including cyclin D1 and TBX2 (40). To test the degree to which PNR stimulation of apoptosis was independent of or dependent on p53, we examined the effect of PNR on apoptosis in two isogenic HCT116 cell lines that contained either two wild-type p53 alleles or none (4) (Fig. 3C). To measure apoptosis frequencies in these cells, we used fluorescence-activated cell sorter (FACS) analysis to assay annexin V binding (Fig. 3D) and also the fraction of cells with reduced (“sub-G1”) DNA content (Fig. 3E) that were associated with the characteristic partial loss of DNA fragmented in apoptosis (16, 44). For HeLa cells, the sub-G1 DNA content assay confirmed that PNR stimulated the fraction of apoptotic cells (Fig. 3E), which is consistent with the previously described annexin V binding results (Fig. 3A). For the two isogenic HCT116 cell lines, PNR increased the fraction of apoptotic cells ∼2-fold, as shown by the annexin V binding assay (Fig. 3D), while ∼6-fold and ∼3-fold increases in p53+/+ and p53−/− HCT116 cell apoptosis levels were measured by the sub-G1 DNA content assay (Fig. 3E). Thus, PNR can induce apoptosis in the absence of p53, but (under at least some circumstances) p53 also appears to enhance the PNR-induced cell apoptosis.
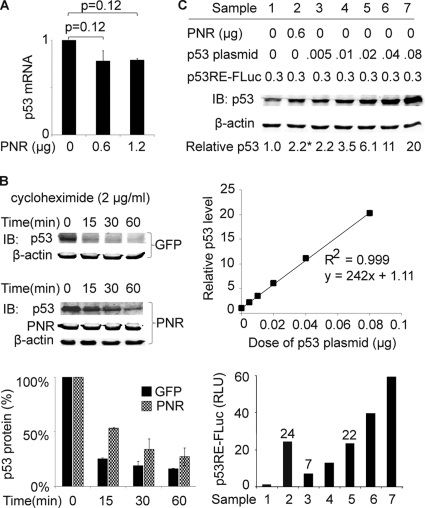
PNR modulates p53 at a posttranslational level.
PNR increased the accumulation of p53 protein (Fig. 1A and B). In contrast, using qRT-PCR, we found that expression of exogenous PNR did not increase the level of p53 mRNA (Fig. 4A). Next, we tested whether PNR increased p53 protein stability. HeLa cells were treated with cycloheximide (2 μg/ml) to block de novo protein synthesis. After 15, 30, and 60 min of incubation with cycloheximide, cells were harvested and p53 protein levels were measured by immunoblotting. Compared with the GFP transfection control results, transfecting PNR increased the half-life of p53 from 7 min or less to 15 min (Fig. 4B).
Fig 4.
PNR modulates p53 posttranslationally in HeLa cells. (A) PNR does not stimulate p53 transcription. Two days after transfection, p53 mRNA levels were measured by qRT-PCR and normalized to that of the sample with 0 μg of PNR. (B) PNR stabilizes p53 protein. Levels of endogenous p53 remaining after treatment with cycloheximide (2 μg/ml) for the indicated times were measured by immunoblotting, and the level in the 0-min sample was normalized to 1. Upper panel: GFP control transfection (1.2 μg). Middle panel: PNR transfection (1.2 μg). Lower panel: plot of remaining p53 protein levels versus treatment time. (C) PNR stimulates the specific activity of p53 as a transcriptional factor. Increasing amounts of the p53 expression plasmid were cotransfected with a constant amount of p53RE-FLuc to provide standard curves of p53 protein levels versus reporter activity. For both p53 protein measurement by immunoblotting (upper panel) and p53 transcriptional activity measurement by reporter assay (lower panel), the results determined under the conditions represented by sample 1 were normalized to 1. The middle panel shows the relative intensities of p53 immunoblotting signal (y axis) versus doses of p53 plasmid transfected (x axis). Sample numbers correspond to the conditions shown in the upper panel (sample 1, empty vector control; sample 2, PNR; samples 3 to 7, p53 standard curve). *, P < 0.05 (one-tailed test).
The results presented above showed that PNR stimulates both transactivation and accumulation of p53. It remained unclear whether the stimulation of p53 transactivation exclusively resulted from enhanced p53 accumulation. Increasing amounts of a p53-expressing plasmid were cotransfected with a constant amount of the p53-responsive luciferase reporter into HeLa cells, and p53 protein levels were quantitatively measured by immunoblotting (Fig. 4C, upper panel). The results shown in the plot in Fig. 4C (middle panel) revealed a linear relationship between the amount of p53-expressing plasmid transfected and the p53 protein signal intensity in the immunoblot, confirming that p53 protein can be quantitated by immunoblotting in this tested range. Simultaneously, we measured the luciferase activity expressed from the p53-responsive reporter (Fig. 4C, lower panel) in the same cells. We found that 0.6 μg of PNR plasmid increased the level of p53 protein by 2-fold (Fig. 4C, upper panel, lanes 1 and 2) but increased p53-dependent transcriptional activity by 24-fold (Fig. 4C, lower panel, lanes 1 and 2) compared with an empty vector control. For comparison, 0.02 μg of a p53-expressing plasmid increased the p53 transactivation signal of the reporter plasmid by a similar 22-fold while increasing the p53 protein level by 6-fold (Fig. 4C, upper panel, lanes 1 and 5). These results suggest that the high level of transactivation by p53 in the presence of PNR may not be attributable solely to increased p53 protein accumulation. In addition, the results suggest that PNR stimulation increased the specific transcriptional activity of p53 by about 3-fold under these conditions. Thus, PNR may affect p53 transactivation by modulating p53 at two posttranslational levels: protein stability and specific transcriptional activity per molecule of p53.
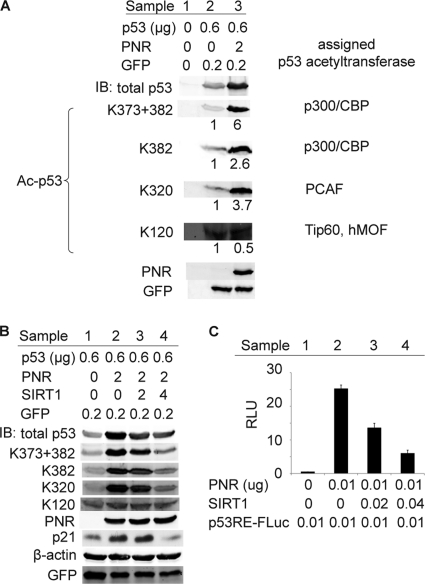
PNR acts largely by stimulating p53 acetylation.
Acetylation is a major mode of p53 posttranslational regulation that enhances p53 protein stability, DNA binding, transcriptional activity, and apoptosis induction (22, 24, 45), which resemble the phenotypes that we observed for PNR in HeLa cells. Thus, we examined whether PNR could regulate p53 acetylation by immunoblotting analysis performed with an antibody specifically recognizing p53 acetylation at K373 and K382. As shown in Fig. 5A, PNR significantly increased p53 acetylation and, consistent with previous data (Fig. 1B and 4B), increased the level of total p53 protein. After normalizing acetylated p53 to total p53 by the use of quantitative immunoblotting, PNR stimulated p53 acetylation at K373 + 382 by ∼6-fold under these experimental conditions. Site-specific p53 acetylation at K382 and other lysines, including acetylation by acetyltransferase PCAF at K320 and by acetyltransferases Tip60/hMOF at K120 (18), was examined by immunoblotting using appropriate specific antibodies. Notably, PNR stimulated p53 acetylation at K320 and K382 but not at K120 (Fig. 5A). Thus, PNR preferentially stimulates acetylation of p53 at certain lysines, possibly through selective interactions with relevant site-specific acetyltransferases.
Fig 5.
PNR-stimulated p53 acetylation correlates with p53 transactivation and accumulation in HeLa cells. (A) PNR stimulates p53 acetylation. Total p53 and acetylated p53 (Ac-p53) levels were measured by immunoblotting with anti-p53 and anti-Ac-p53 antibodies, respectively. Acetylations by the indicated acetyltransferases, which were measured with their specific anti-Ac-p53 antibodies, can occur at multiple lysines of p53. The numbers below each band represent the levels of Ac-p53 per unit of total p53 normalized to that in sample 2. GFP was used as a transfection control. (B and C) Coexpressing p53 deacetylase SIRT1 inhibits PNR-mediated stimulation of p53 accumulation and transactivation. (B) SIRT1 inhibits PNR-mediated p53 acetylation and accumulation in a dose-dependent fashion. Ac-p53, total p53, PNR, and GFP were measured as described for panel A. Endogenous p21 and β-actin protein levels were also measured. SIRT1 abolished the PNR-mediated increase in p21 protein accumulation. (C) SIRT1 inhibited PNR-mediated stimulation of p53RE-FLuc expression in a dose-dependent fashion in a 96-well plate.
Next, we tested whether reversing PNR-mediated p53 acetylation was sufficient to inhibit PNR-mediated p53 transactivation and accumulation. SIRT1 is a well-characterized p53 deacetylase (25, 48). Coexpressing SIRT1 with PNR in HeLa cells significantly inhibited PNR-mediated p53 acetylation at multiple sites other than K120 in a dose-dependent manner (Fig. 5B). SIRT1 also inhibited PNR stimulation of total p53 accumulation, implying that PNR's enhancement of p53 accumulation results from stimulation of p53 acetylation. Consistent with our results showing p21 mRNA levels (Fig. 1E), PNR significantly enhanced endogenous p21 protein levels (Fig. 5B, samples 1 and 2). However, coexpressing SIRT1 also abolished this enhancement (Fig. 5B, samples 3 and 4). Simultaneously, coexpressing SIRT1 with PNR reversed the PNR-mediated stimulation of p53 transactivation in a dose-dependent fashion (Fig. 5C), implying that PNR stimulates p53 transactivation primarily through stimulating p53 acetylation. From these results, we concluded that PNR-mediated stimulation of p53 acetylation is responsible for the PNR-mediated stimulation of p53 transactivation and accumulation.
p53 acetylation is essential for activating a subset of p53 target genes but dispensable for activating some other p53 target genes (18). Our tests of PNR's effects on four selected p53 target genes matched the previously identified effects of p53 acetylation by upregulating p21 and Puma mRNAs but inducing no change in MDM2 and Pirh2 mRNA levels (Fig. 1E and F). These gene-specific differential effects notably extend the correlation between the effects of PNR and those of p53 acetylation in HeLa cells.
PNR forms complexes with and enhances interaction between p300 and p53.
To dissect the mechanism of PNR-induced p53 acetylation, we focused on the interaction between p53 and p300, a major p53 acetyltransferase. shRNAs against p300 mildly decreased the level of endogenous p300 protein by less than 40% while inhibiting PNR-stimulated p53 transactivation by 60% to 70% (Fig. 6A). Simultaneously, knocking down p300 also significantly repressed the PNR-mediated stimulation of p53 acetylation and accumulation (Fig. 6B). These data suggested that p300 is involved in the PNR-mediated stimulation of p53 acetylation.
Fig 6.
p300 is involved in the PNR-mediated stimulation of p53 in HeLa cells. (A) Two different shRNA plasmids targeting p300 inhibited the PNR-mediated stimulation of p53RE-FLuc expression. The RLU in the PNR+shRNA-Con sample was normalized to 100%. (B) shRNA targeting p300 inhibits the PNR-mediated stimulation of p53 acetylation and accumulation. The indicated proteins were measured by immunoblotting. The numbers below the p300 band represent the levels of p300 protein normalized to the p300 level in sample 1. shRNA-Con, pLKO.1 empty lentiviral control.
The “LXXLL” motif is a common binding site for nuclear receptors like PNR (27). We found one LXXLL motif in p53 (amino acids [aa] 22 to 26) at the N terminus and two in p300 (aa 81 to 85 at the N terminus and aa 2051 to 2055 at the C terminus). Thus, we tested whether PNR, p300, and p53 could form complexes with each other. First, we cotransfected HA-tagged PNR, p300, and p53 into HeLa cells. Anti-p300 and anti-p53 antibodies successfully precipitated p300 and p53, respectively (Fig. 7A). HA-PNR was detected by an anti-HA antibody in the immunoprecipitates and by the anti-p300 and anti-p53 antibodies, while the control IgGs failed to coimmunoprecipitate HA-PNR. Simultaneously, an anti-HA antibody successfully immunoprecipitated HA-PNR while the control IgGs did not (Fig. 7B). p53 was specifically detected in the immunoprecipitate by the anti-HA antibody. We did not detect a clear band of p300 in the immunoprecipitate product by the use of the anti-HA antibody, although the reverse coimmunoprecipitation of PNR by the anti-p300 antibody strongly suggested that p300 may interact with PNR. In addition, we found by immunofluorescence analysis using HeLa cells that transfected PNR and p53 both located inside nuclei and partially overlapped (Fig. 7C and D). Thus, in combination, these results showed that, in HeLa cells, PNR and p53 form a complex in which p300 is likely incorporated.
In general, increased interaction between p53 and its p300 acetyltransferase would be expected to be associated with increased p53 acetylation. We therefore immunoprecipitated p300 from the whole-cell lysate with or without PNR transfection and then used quantitative immunoblotting to measure p53 protein levels in these immunoprecipitates. As expected, more p53 was detected in the whole-cell lysate with PNR transfection (Fig. 8A). Similarly, more p53 was detected in anti-p300 immunoprecipitates from PNR-transfected cells than in those from control cells. After normalizing p53 levels in each immunoprecipitate to that in the corresponding whole-cell lysate, we found that PNR stimulation increased p300-p53 association by ∼2- to 3-fold under these conditions. Simultaneously, we found that PNR significantly increased acetylated p53 levels in the anti-p300 immunoprecipitates (Fig. 8A). These results implied that PNR enhances p53 acetylation by promoting the intermolecular interaction between p53 and p300.
PNR boosts actinomycin D-stimulated association between p53 and p300.
Wild-type p53 is usually in a quiescent state in cells until activated by various DNA damage stresses. Actinomycin D is an antineoplastic, genotoxic agent that forms a stable complex with DNA, stimulates p53 acetylation at K305 and K382, and induces cell apoptosis (49). We tested whether actinomycin D treatment of HeLa cells could enhance the association between p53 and p300. One day after cotransfection of the indicated plasmids (Fig. 8B), HeLa cells were treated with 10 nM actinomycin D for another 24 h before being lysed for immunoprecipitation with anti-p300 antibody as described above. As shown in Fig. 8B, lanes 1 and 2, actinomycin D somewhat increased accumulation in the cell lysate of both total p53 and p53 acetylated at K320 and K373 + 382. Although the signal was relatively weak in the absence of added PNR, close inspection showed that the coimmunoprecipitation of total and acetylated p53 with p300 also was enhanced (Fig. 8B, lanes 5 and 6). Expressing exogenous PNR boosted all of these signals, further revealing that actinomycin D stimulated both the overall accumulation (Fig. 8B, lanes 2 and 4) and the coimmunoprecipitation with p300 (Fig. 8B, lanes 6 and 8) of total and acetylated p53. Thus, PNR synergized with actinomycin D to jointly stimulate p53 acetylation and interaction with p300.
DISCUSSION
p53, which plays crucial roles in regulating cell cycle arrest, apoptosis, and senescence, is regulated by a variety of posttranslational modifications, including ubiquitination, phosphorylation, methylation, and acetylation (45). p53 acetylation at multiple sites in its DNA binding domain and C-terminal regulatory domain modulates the stability of p53, DNA binding, and coactivator interactions and is essential for p53-activated transcription of a subset of p53 target genes, including p21 and other genes associated with cell cycle arrest and apoptosis (25, 41, 45). Acetylation is also crucial for the transcription-independent preapoptotic functions of p53 (52). Thus, stimulation of p53 acetylation is a potentially valuable target for restoration of normal p53 functions in cell growth arrest and apoptosis in cancer cells.
Here, we reported the unexpected finding that orphan PNR serves as a positive regulator of p53 acetylation and activity. Building on our identification of PNR as a p53 activator in a high-throughput genetic screen, we demonstrated that PNR is specifically engaged in enhancing acetylation of p53 and thus potentiating acetylated p53 functions such as apoptosis (Fig. 3). Expressing PNR enhanced formation of a complex between p53 and the acetyltransferase p300 (Fig. 7), resulting in increased p53 acetylation (Fig. 5A). Coordinately, PNR stimulated p53 transcriptional activity and stability (Fig. 1C and 4B). In agreement with the finding that p53 acetylation is required for transcriptional regulation of a subset of genes (18, 45), we further found that PNR selectively upregulated expression of acetylated p53-dependent genes p21 and Puma but not of acetylation-independent p53 target genes MDM2 and Pirh2 (Fig. 1E and F). Together, our results demonstrate that PNR regulates p53 functions by stimulating p53 acetylation.
Although prior work has documented other regulatory interactions between nuclear receptors and p53, results showing regulation of p53 acetylation by a nonacetyltransferase nuclear receptor are unprecedented. In contrast, estrogen receptor α, another nuclear receptor, interacts with p53, binds with p53 to p53 response elements in the promoters of p53 target genes, and inhibits p53-mediated transactivation and transrepression (23, 37). The orphan nuclear receptor Coup-TF II stimulates transcription of p53 and p53-responsive reporter genes (14) and is downregulated in concert with the p53 target gene p21 in multiple breast cancer cell lines (29). Coup-TF I, a cousin of Coup-TFII, stimulates transcription of MDM2, an E3 ligase essential for p53 degradation (33). In contrast, p53 regulates transcription of nuclear receptors HNF4A and TR2 (26, 28). Accordingly, our finding that PNR potentiates p53 acetylation and related acetylated p53 functions provides a novel link between orphan nuclear receptors and p53. Notably, our preliminary results further show that other members of nuclear receptor subfamily 2, which includes PNR, also stimulate p53 activity through acetylation (unpublished data). Thus, regulation of p53 functions by enhancement of p53 acetylation may be a general paradigm shared by nuclear receptor subfamily 2 members. This finding has therapeutic implications, since several PNR subfamily orphan receptors have broad tissue distribution and are aberrantly expressed in cancers (19, 30, 34, 43, 53).
One attractive set of targets for PNR-based therapeutic approaches includes the cancers caused by human papillomaviruses (HPVs). Most such HPV-linked cancers retain a wild-type p53 gene, but p53 accumulation and functions are downregulated posttranslationally by multiple actions of HPV oncogene E6 (38, 39, 46). Thus, approaches that stimulate p53 stability and function by the use of PNR or related nuclear receptors, either alone or in conjunction with E6 inhibitors, could be undertaken to control HPV-associated malignancies. Moreover, since we have found that PNR stimulation of p53 activity is not limited to HPV+ cells (Fig. 2A and unpublished data), PNR and related nuclear receptors should be valuable against other HPV− cancers that retain functional p53 genes. Furthermore, the benefits of PNR-based cancer therapies may extend beyond activating p53, since, in addition to affecting p53+ cells, we have found that PNR also induces apoptosis of p53−/− HCT116 cells (Fig. 3D and E). Similarly, prior work has shown that PNR has intrinsic tumor suppression activities that operate by binding and repressing the promoters of cyclin D1 (42) and another cell cycle regulator, TBX2 (1, 42).
PNR is an orphan nuclear receptor whose natural ligand(s) remains to be identified. However, compounds with a 2-phenylbenzimidazole core have been found to be potent agonists of PNR (51). Thus, natural or artificial ligands for PNR, and perhaps for other subfamily 2 nuclear receptors, may stimulate p53 acetylation and induce cell apoptosis and thus have therapeutic potential to be developed into anticancer drugs. In addition, the novel functional link between p53 and PNR also makes it intriguing to explore the possible roles of p53 in retinal diseases linked to PNR mutations, including enhanced s-cone syndrome, Leber's congenital amaurosis (LCA), retinitis pigmentosa, macular degeneration, etc. (11, 40).
ACKNOWLEDGMENTS
This work was supported by the NIH through grants CA-22443 and CA-125387. P.A. is an investigator of the Howard Hughes Medical Institute.
We thank Bill Sugden, Norman Drinkwater, Paul Friesen, Elaine Alarid, Shannon Kenny, Robert Kalejta, Linhui Hao, James Bruce, and Shouhong Guang for valuable advice and Shiming Chen, Qize Wei, Ann Palmenberg, Arthur Polans, William See, Saraswati Sukumar, and Bert Vogelstein for generously sharing reagents and materials. We also thank Kathleen Schell (University of Wisconsin Carbone Cancer Center Flow Cytometry Facility) and Lance Rodenkirch (University of Wisconsin Keck Laboratory for Biological Imaging) for helpful suggestions.
Footnotes
Published ahead of print 24 October 2011
REFERENCES
- 1. Abrahams A, Parker MI, Prince S. 2010. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life 62:92–102 [DOI] [PubMed] [Google Scholar]
- 2. Banin S, et al. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674–1677 [DOI] [PubMed] [Google Scholar]
- 3. Brooks CL, Gu W. 2003. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15:164–171 [DOI] [PubMed] [Google Scholar]
- 4. Bunz F, et al. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497–1501 [DOI] [PubMed] [Google Scholar]
- 5. Choi MY, et al. 2006. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development 133:4119–4129 [DOI] [PubMed] [Google Scholar]
- 6. Clontech I. 2008. Protocol PT3348-5, version PR842524. http://www.clontech.com/xxclt_ibcGetAttachment.jsp?cItemId=17908
- 7. el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45–49 [DOI] [PubMed] [Google Scholar]
- 8. Fojo T. 2002. p53 as a therapeutic target: unresolved issues on the road to cancer therapy targeting mutant p53. Drug Resist Updat. 5:209–216 [DOI] [PubMed] [Google Scholar]
- 9. Gillison ML, Lowy DR. 2004. A causal role for human papillomavirus in head and neck cancer. Lancet 363:1488–1489 [DOI] [PubMed] [Google Scholar]
- 10. Gu W, Roeder RG. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595–606 [DOI] [PubMed] [Google Scholar]
- 11. Haider NB, et al. 2000. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet. 24:127–131 [DOI] [PubMed] [Google Scholar]
- 12. Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299 [DOI] [PubMed] [Google Scholar]
- 13. Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25–27 [DOI] [PubMed] [Google Scholar]
- 14. Huang Q, et al. 2004. Identification of p53 regulators by genome-wide functional analysis. Proc. Natl. Acad. Sci. U. S. A. 101:3456–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang M, Milner J. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041–6048 [DOI] [PubMed] [Google Scholar]
- 16. Kajstura M, Halicka HD, Pryjma J, Darzynkiewicz Z. 2007. Discontinuous fragmentation of nuclear DNA during apoptosis revealed by discrete “sub-G1” peaks on DNA content histograms. Cytometry A 71:125–131 [DOI] [PubMed] [Google Scholar]
- 17. Koopman G, et al. 1994. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84:1415–1420 [PubMed] [Google Scholar]
- 18. Kruse JP, Gu W. 2009. Modes of p53 regulation. Cell 137:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee YF, Lee HJ, Chang C. 2002. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J. Steroid Biochem. Mol. Biol. 81:291–308 [DOI] [PubMed] [Google Scholar]
- 20. Li M, et al. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302:1972–1975 [DOI] [PubMed] [Google Scholar]
- 21. Li M, et al. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648–653 [DOI] [PubMed] [Google Scholar]
- 22. Li M, Luo J, Brooks CL, Gu W. 2002. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277:50607–50611 [DOI] [PubMed] [Google Scholar]
- 23. Liu W, et al. 2006. Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J. Biol. Chem. 281:9837–9840 [DOI] [PubMed] [Google Scholar]
- 24. Luo J, et al. 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 101:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo J, et al. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137–148 [DOI] [PubMed] [Google Scholar]
- 26. Maeda Y, et al. 2006. Tumour suppressor p53 down-regulates the expression of the human hepatocyte nuclear factor 4alpha (HNF4alpha) gene. Biochem. J. 400:303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McInerney EM, et al. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mu X, Liu Y, Collins LL, Kim E, Chang C. 2000. The p53/retinoblastoma-mediated repression of testicular orphan receptor-2 in the rhesus monkey with cryptorchidism. J. Biol. Chem. 275:23877–23883 [DOI] [PubMed] [Google Scholar]
- 29. Nakshatri H, et al. 2000. The orphan receptor COUP-TFII regulates G2/M progression of breast cancer cells by modulating the expression/activity of p21(WAF1/CIP1), cyclin D1, and cdk2. Biochem. Biophys. Res. Commun. 270:1144–1153 [DOI] [PubMed] [Google Scholar]
- 30. Nishimura M, Naito S, Yokoi T. 2004. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug Metab. Pharmacokinet. 19:135–149 [DOI] [PubMed] [Google Scholar]
- 31. Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. 2005. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 14:747–764 [DOI] [PubMed] [Google Scholar]
- 32. Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. 2009. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 5:e1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qi JS, Yuan Y, Desai-Yajnik V, Samuels HH. 1999. Regulation of the mdm2 oncogene by thyroid hormone receptor. Mol. Cell. Biol. 19:864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qin J, Chen X, Yu-Lee LY, Tsai MJ, Tsai SY. 2010. Nuclear receptor COUP-TFII controls pancreatic islet tumor angiogenesis by regulating vascular endothelial growth factor/vascular endothelial growth factor receptor-2 signaling. Cancer Res. 70:8812–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raman V, et al. 2000. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 405:974–978 [DOI] [PubMed] [Google Scholar]
- 36. Reisman D, Loging WT. 1998. Transcriptional regulation of the p53 tumor suppressor gene. Semin. Cancer Biol. 8:317–324 [DOI] [PubMed] [Google Scholar]
- 37. Sayeed A, et al. 2007. Estrogen receptor alpha inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res. 67:7746–7755 [DOI] [PubMed] [Google Scholar]
- 38. Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495–505 [DOI] [PubMed] [Google Scholar]
- 39. Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136 [DOI] [PubMed] [Google Scholar]
- 40. Swaroop A, Kim D, Forrest D. 2010. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11:563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sykes SM, et al. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24:841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takezawa S, et al. 2007. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 26:764–774 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Tanaka T, De Luca LM. 2009. Therapeutic potential of “rexinoids” in cancer prevention and treatment. Cancer Res. 69:4945–4947 [DOI] [PubMed] [Google Scholar]
- 44. Tang Y, Luo J, Zhang W, Gu W. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24:827–839 [DOI] [PubMed] [Google Scholar]
- 45. Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. 2008. Acetylation is indispensable for p53 activation. Cell 133:612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas MC, Chiang CM. 2005. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell 17:251–264 [DOI] [PubMed] [Google Scholar]
- 47. Vassilev LT, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848 [DOI] [PubMed] [Google Scholar]
- 48. Vaziri H, et al. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159 [DOI] [PubMed] [Google Scholar]
- 49. Wang YH, Tsay YG, Tan BC, Lo WY, Lee SC. 2003. Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J. Biol. Chem. 278:25568–25576 [DOI] [PubMed] [Google Scholar]
- 50. Wei Q. 2005. Pitx2a binds to human papillomavirus type 18 E6 protein and inhibits E6-mediated P53 degradation in HeLa cells. J. Biol. Chem. 280:37790–37797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolkenberg SE, et al. 2006. Identification of potent agonists of photoreceptor-specific nuclear receptor (NR2E3) and preparation of a radioligand. Bioorg. Med. Chem. Lett. 16:5001–5004 [DOI] [PubMed] [Google Scholar]
- 52. Yamaguchi H, et al. 2009. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J. Biol. Chem. 284:11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu X, Mertz JE. 2003. Distinct modes of regulation of transcription of hepatitis B virus by the nuclear receptors HNF4alpha and COUP-TF1. J. Virol. 77:2489–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zimmermann H, Degenkolbe R, Bernard HU, O'Connor MJ. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]