Abstract
The Ku heterodimer (Ku70/Ku80) is a main component of the nonhomologous end-joining (NHEJ) pathway that repairs DNA double-strand breaks (DSBs). Ku binds the broken DNA end and recruits other proteins to facilitate the processing and ligation of the broken end. While Ku interacts with many proteins involved in DNA damage/repair-related functions, few interactions have been mapped to the N-terminal von Willebrand A (vWA) domain, a predicted protein interaction domain. The mutagenesis of Ku70 vWA domain S155/D156 unexpectedly increased cell survival following ionizing radiation (IR) treatment. DNA repair appeared unaffected, but defects in the activation of apoptosis and alterations in the DNA damage signaling response were identified. In particular, Ku70 S155A/D156A affected the IR-induced transcriptional response of several activating transcription factor 2 (ATF2)-regulated genes involved in apoptosis regulation. ATF2 phosphorylation and recruitment to DNA damage-induced foci was increased in Ku70-deficient cells, suggesting that Ku represses ATF2 activation. Ku70 S155A/D156A substitutions further enhanced this repression. S155A substitution alone was sufficient to confer enhanced survival, whereas alteration to a phosphomimetic residue (S155D) reversed this effect, suggesting that S155 is a phosphorylation site. Thus, these findings infer that Ku links signals from the DNA repair machinery to DNA damage signaling regulators that control apoptotic pathways.
INTRODUCTION
One of the most dangerous forms of DNA damage is the DNA double-strand break (DSB), which can lead to aberrant genomic rearrangement if not repaired properly (25, 26). In eukaryotic cells, DSBs trigger signaling pathways that induce cell cycle checkpoints and alter gene transcription, allowing DNA integrity to be reestablished through the action of repair complexes (9, 30, 68, 71).
The DNA damage response (DDR) pathway is initiated by a phosphorylation cascade that triggers chromatin modifications which enhance the accessibility of the broken DNA to repair factors and promote the subsequent accumulation of DDR factors into foci at the site of damage (57, 68). The Mre11-Rad50-NBS1 (MRN) complex immediately binds the DSB independently of other factors (32), functioning to recruit the serine/threonine (S/T) phosphoinositide-3-kinase (PI3K) family member ATM (ataxia telangiectasia mutated), an essential regulator of the DNA damage response that is responsible for many phosphorylation events at the site of DNA damage (36, 37). An important signal amplification step involves the ATM phosphorylation of the histone variant H2AX to create a platform to which other DDR proteins are able to bind (17). ATM activates signaling cascades that trigger the activation of cell cycle checkpoints, leading to cell cycle arrest through the phosphorylation of several substrates, including p53, MDC1, BRCA1, Chk1, and Chk2. ATM also contributes to the establishment of apoptotic pathways (36).
Two main pathways function to repair DSBs, homologous recombination (HR), which uses a homologous chromosome or sister chromatid as the template to repair the broken DNA, and nonhomologous end joining (NHEJ), which simply religates the two broken ends together (25). In mammals, NHEJ is the predominant DSB repair pathway, functioning throughout the cell cycle, and is exclusive to the G1 and S phases (41, 47). NHEJ also mediates the rejoining of programmed breaks generated in V(D)J recombination during B- and T-cell maturation (41, 47). NHEJ can be subdivided into two subpathways, the core or classical NHEJ pathway (C-NHEJ), which represents the main end-joining activity in the cell, and alternative NHEJ activities (A-NHEJ) consisting of microhomology-mediated repair that function as backup pathway(s) to join DSBs (25, 41, 52).
The C-NHEJ complex in higher eukaryotic cells consists of DNA-dependent protein kinase (DNA-PK), which is composed of the Ku heterodimer and DNA-PK catalytic subunit (DNA-PKcs), Artemis, a DNA processing enzyme, a DNA ligase complex, XRCC4/DNA ligase IV, and a recently identified factor called Cernunnos-XLF (41, 47, 65). Other accessory factors, including polynucleotide kinase (PNK) and DNA polymerases μ and λ, have been implicated in some aspects of C-NHEJ (41, 47).
Ku is the DNA-binding component of the C-NHEJ repair machinery. Upon recognition and binding to the broken DNA end, Ku recruits DNA-PKcs to form the active protein kinase complex DNA-PK (41, 47). DNA-PKcs is a large (p450) S/T kinase that is a member of the PI3K group that includes ATM, ATM-related (ATR), and mammalian target of rapamycin (mTOR) (1, 27, 49). The importance of DNA-PK in maintaining genomic integrity is underscored by the profound immunodeficiency, radiosensitivity, and prevalence of tumors in mice lacking any of the three subunits (18, 39, 53, 67). However, DNA-PKcs knockout mice display milder defects than Ku−/− mice, suggesting that Ku has additional functions that are independent of those of DNA-PKcs (42, 67). Besides DNA end recognition, Ku appears to protect broken DNA from aberrant nucleolytic processing (15). Ku also has been shown to bind to telomeres and to function in telomere maintenance, notably by anchoring telomeres to the nuclear periphery, contributing to telomeric silencing and preventing telomere shortening (15, 58).
In addition to its main function in DNA repair, several reports have suggested that DNA-PK also is involved in signaling to regulate specific aspects of the DDR. DNA-PK participates in replication protein A2 (RP-A2) and nuclear factor κB (NF-κB) phosphorylation in response to DNA damage and also contributes to the modification of histone H2AX (reviewed in references 12 and 27). Roles for Ku in signaling to the apoptotic machinery also have been documented (27).
Aside from a recently identified end-processing activity (59), much of Ku's function appears to be mediated by protein-protein interactions with other factors. A number of proteins interact with Ku, including C-NHEJ core proteins and factors implicated in the DDR and in telomere maintenance, transcription, and replication (12, 63). Within the mammalian C-NHEJ complex, the interaction of Ku with XRCC4/DNA ligase IV is required to recruit the complex to DNA and to stimulate the ligase activity (13, 43). Yeast Ku also interacts with factors of the RSC complex that mediates ATP-dependent chromatin remodelling in yeast (66). Interaction with DNA repair and damage-response factors Mre11, Werner, and PARP also have been documented (12, 63).
Ku is a heterodimer of two proteins, Ku70 and Ku80, that form a complex that is conserved throughout evolution both structurally and functionally (4, 15, 63, 72). Ku homologs are found in organisms ranging from bacteria to humans (4). The two subunits of Ku in eukaryotes feature three structurally similar domains, an amino-terminal α/β domain, a central β-barrel domain, and an α-helical carboxy-terminal arm, that come together in the heterodimer to form a quasisymmetrical ring structure that envelopes up to two helical turns of DNA ends, as seen in the crystal structure (72). The Ku70 carboxy-terminal domain sequence shows similarities with SAP domains that are involved in DNA binding (3, 72), whereas the Ku80 C-terminal domain forms a globular structure with similarity to protein domains involved in protein-protein interactions (24, 77). The amino-terminal domains of Ku70 and Ku80 (α/β domains [72]) share similarity with von Willebrand factor A (vWA), a domain that mediates protein-protein interactions (4, 72, 74). The vWA domains of Ku fall into the ancient conserved vWA proteins, a group of evolutionarily conserved intracellular proteins (74). However, while Ku interacts with many proteins, few have been mapped to the vWA domains (12, 15, 47, 63). Site-directed mutagenesis of the Saccharomyces cerevisiae YKU80 and YKU70 genes identified that α-helices on the surface of the vWA domain confer different Ku functions (56). The Yku80 α-helix 5 is critical for telomeric functions, while Yku70 α-helix 5 is required for C-NHEJ. This likely results from differences in the orientation of the two vWA domains, with the Ku70 vWA domain facing outwards in close proximity to the DNA end and the Ku80 vWA domain facing inwards, thus facilitating telomeric functions (56, 72).
In mammals, very little is known about the precise function of the Ku70/Ku80 N-terminal vWA domains. In this study, we introduced point mutations in various regions of the Ku70 vWA domain with the intent of identifying structural determinants that direct Ku function in response to DNA damage (Fig. 1). The mutation of Ku70 vWA α-helix 5 residues (D192A/D195R) resulted in a sharp decrease in survival. These substitutions, previously shown to confer a DNA repair defect in yeast (56), markedly impaired the DNA repair function of Ku in mouse embryonic fibroblasts (MEFs), suggesting that the function of these residues is conserved between yeast and mammals. Unexpectedly, the mutagenesis of residues adjacent to α-helix 4 (S155A/D156A) resulted in increased survival following ionizing radiation (IR) treatment. C-NHEJ appeared unaffected, but a marked decrease in the activation of apoptosis and alterations in the DNA damage signaling response, as well as in the transcriptional profile of gene expression following DNA damage, were identified. In particular, this mutation affected an activating transcription factor 2 (ATF2)-dependent transcriptional pathway that modulates several genes implicated in the activation of apoptosis. The D192A/D195R survival defect was rescued by introducing the S155A/D156A substitution, inferring that separate regions of the Ku70 vWA domain confer two different Ku functions in response to DNA damage. Further, S155 was identified as the critical residue regulating cell survival. Thus, importantly, the defects resulting from these mutations suggest that the N-terminal vWA domain of Ku70 is implicated in the activation of apoptotic pathways by linking signals of DNA repair completion (or lack thereof) to the signaling machinery that controls the activation of cell death pathways.
Fig 1.
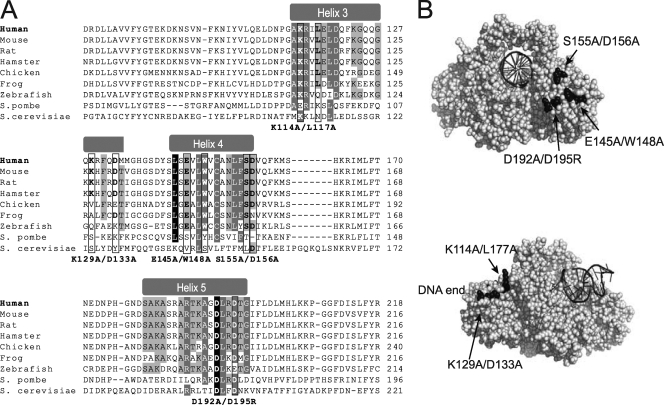
(A) Sequence alignment of the Ku70 N-terminal vWA domain (α-helices 3 to 5) from a selection of Ku70 eukaryotic homologs. The position of the vWA domain α-helices is indicated at the top. Conservation between species of the residues within α-helices 3 to 5 is highlighted according to percent identity (PID) (light gray, >40%; dark gray, >60%; black, >80% PID). The residues mutated in this study are boxed, and the substitutions introduced are indicated below the alignments. (B) Space-filling representations of the human Ku dimer structure bound to DNA (72) (Protein Data Bank no. IJEY; depicted using PyMol). The upper image shows the front view of the Ku dimer (facing the DNA end). The Ku70 vWA domain E145/W148, S155/D156, and D192/D195 residues are highlighted in black, and their position is indicated. DNA is represented as a black helix. The lower image shows the side view of the Ku dimer (DNA end to the left). The position of the Ku70 vWa domain K114/L117 and K129/D133 residues is shown.
MATERIALS AND METHODS
Plasmid expression constructs.
Ku70 human cDNA was cloned from the BamHI site in pEGFP Ku70 (6) into the HpaI site of pMSCVpuro retroviral vector (Clontech). Ku70 point mutations were introduced by site-directed mutagenesis using Pfu polymerase (Stratagene) with primers bearing the targeted point mutations (primers are listed in Table S3 in the supplemental material). All mutations were confirmed by sequencing. pGL3-Promoter and pRL-SV40 plasmids were from Promega.
Cell culture and treatments.
Ku70−/− MEFs were obtained from S. Matsuyama (Case Western, Cleveland, OH) (19). All cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2.
pMSCV vectors containing wild-type and mutant Ku70, as well as the empty pMSCVpuro vector, were transfected via calcium phosphate into the Phoenix Ampho retroviral packaging cell line. The medium-containing virus was collected 48 h later and used to infect Ku70−/− MEFs. Twenty-four hours postinfection, the media were replaced with 2.5 μg/ml puromycin containing medium to select and maintain cells infected with the pMSCV vector. Cells were maintained as a pool for all subsequent experiments.
For irradiation experiments, cells were plated the night before at 50 to 70% confluence. Irradiations were performed with a Faxitron RX-650 at a dose rate of 1.42 Gy/min.
Extracts and Western blot analyses.
Whole-cell extracts were prepared as described previously (64). Nuclear extracts were prepared as described previously (2). For Western blot analysis, extracts were resolved by SDS-PAGE (either 8 or 10%), transferred onto a polyvinylidene difluoride (PVDF) membrane, and hybridized with antibodies to Ku70 (N3H10; Neomarkers), Ku80 (M-20; Santa Cruz), β-actin (I-19; Santa Cruz), GADD153/CHOP (F-168; Santa Cruz), ATF3 (C-19; Santa Cruz), PCNA (clone PC-10; Millipore), 69/71 phospho-ATF2 (Cell Signaling), and ATF2 (N-96; Santa Cruz).
Clonogenic survival assays.
Cells were plated in triplicate at a single-cell density, irradiated 6 h later with various doses of irradiation (IR), and then incubated for 7 days. The plates were washed with phosphate-buffered saline (PBS) and stained with 0.5% crystal violet in 20% methanol. Colonies were counted and survival was assessed by calculating the ratio of colony number on the irradiated plates to the number of the unirradiated controls.
Caspase assays.
Cell extracts were prepared in lysis buffer (1 mM KCl, 10 mM HEPES [pH 7.4], 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 μg/ml leupeptin, 2 μg/ml aprotinin, and 10% glycerol). Caspase activity was measured in caspase assay buffer containing 25 mM HEPES, pH 7.4, 10 mM DTT, 10% sucrose, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, and 10 μM caspase-3 substrate containing N-acetyl-Asp-Glu-Val-Asp-(7-amino-4-trifluoromethyl-coumarin) (DEVD-AFC; Biomol International). The fluorescence produced by DEVD-AFC cleavage was measured on a SpectraMax M5 fluorimeter (excitation, 400 nm; emission, 505 nm) during a 2-h interval. Caspase activity was calculated as the ratio of the fluorescence output in treated samples to that of corresponding untreated controls.
Pulsed-field gel electrophoresis (PFGE).
One day prior to irradiation, 3.5 million cells were seeded onto a 10-cm plate. Following 40 Gy of IR or mock treatment, cells were harvested into agarose plugs using the Bio-Rad contour-clamped homogeneous electric field (CHEF) genomic DNA plug kit. Agarose plugs were run on a 0.8% gel using the Bio-Rad CHEF-DR II system for 48 h (switch time of 200 to 500 s, 120° angle, 3 V/cm). The gels were stained with ethidium bromide, images were captured using a Bio-Rad ChemiDoc and ImageLab software, and staining was quantified using ImageJ. The fraction of activity released (FAR) corresponding to unrepaired DNA was determined by calculating the ratio of the DNA migrating below the plug to the total DNA loaded (DNA remaining in the plug and fraction entering the gel).
Plasmid repair luciferase assays.
The pGL3-Promoter luciferase reporter plasmid was digested with BglII, which cuts between the promoter and the luciferase coding region. PMSCV-infected MEFs were transfected with 750 ng of linearized PGL3-Promoter and 5 ng of pRL-SV40 in 12-well plates using Fugene 6 (Roche) by following the manufacturer's instructions. Forty-eight hours posttransfection, the cells were harvested in 0.3 ml 1× passive lysis buffer (Promega), and luciferase assays were performed with 30 μl of extract with the Promega dual-luciferase reporter assay system (50 μl of both LAR II and Stop & Glo reagents) using an Orion II luminometer (Titertek-Berthold).
Immunofluorescence.
One day prior to irradiation, cells were seeded at 60 to 80% confluence on 10-mm glass coverslips. At the given time points postirradiation, cells were washed in cold PBS and fixed in 3% paraformaldehyde. Cells were permeabilized in 0.5% Triton-X and blocked in 5% fetal bovine serum (FBS), followed by incubation with the primary antibody to phosphoserine 139 H2AX (20E3; Cell Signaling), ATF2 (C-19; Santa Cruz), or 69/71 phospho-ATF2 (Cell Signaling). Slides then were incubated with an anti-rabbit Alexa 497 secondary antibody (Invitrogen). Coverslips were mounted onto glass slides using ProLong Gold containing 4′,6′-diamidino-2-phenylindole (DAPI) (Invitrogen). Cell pictures were taken with an Olympus BX51 microscope at ×40 magnification and Image-Pro Plus software (Media Cybernetics, Inc.). For γ-H2AX and phospho-ATF2, pixel density was measured with ImageJ software and used as a measure of focus content per picture. DAPI nucleic staining was used for cell counting, and the pixel density was averaged per cell for approximately 500 cells. For the quantification of ATF2 foci, all pictures were set to an equal contrast threshold on ImageJ, and cells were scored positive if they contained at least one focus.
Reverse transcriptase PCR (RT-PCR).
Total RNA was isolated using the Qiagen RNeasy RNA extraction kit. RNA (2 μg) was reverse transcribed with the Superscript II cDNA kit (Invitrogen). Quantitative PCR was performed using Bio-Rad MyiQ single-color real-time PCR detection and the Bio-Rad IQ SYBR green mix. Primers are listed in Table S3 in the supplemental material. The relative quantification of specific gene expression was determined by the ΔΔCT method, with the target gene threshold cycle (CT) values normalized to that of the beta-2-microglobulin control. The change of gene expression in irradiated samples was calculated relative to that of the unirradiated controls.
Sequence alignments.
Sequences were obtained from the NCBI database and aligned using MUSCLE software (16). Percent identity calculations were performed using Jalview software.
Statistical analyses.
Differences between two groups were compared using an unpaired two-tailed t test, and analysis of variance (ANOVA) was used for comparisons of multiple groups. Results were considered significant when P < 0.05.
RESULTS
Identification of Ku70 mutations that impair survival in response to IR.
To investigate the contribution of the Ku70 vWA domain in Ku70's function in the response to DSBs, several point mutations were produced in the human Ku70 vWA domain. These mutations targeted residues located on the solvent-exposed surface of the protein and showing various degrees of conservation across Ku70 homologs (Fig. 1). For instance, residues in Ku70 α-helix 5, previously involved in DNA repair in yeast, are fairly well conserved (Fig. 1A) (56). In contrast, α-helix 3 is much less conserved (Fig. 1A). We produced five different Ku70 mutations: two mutations in α-helix 3, one in α-helix 4, one in α-helix 5, and one in a loop region bordering α-helix 4 (S155A/D156A) (Fig. 1B). At each location, we altered two amino acids to maximize the likelihood of disrupting a protein-interacting surface. Wild-type human Ku70 as well as the various mutant human Ku70 cDNAs were stably introduced into Ku70−/− immortalized MEFs via a murine stem cell virus construct. To determine whether the mutations interfered with Ku's function in response to IR, we measured the radiosensitivity of MEFs expressing wild-type or mutant Ku70 at various doses of IR (between 2 and 10 Gy) using a clonogenic assay. Cells lacking Ku70 are severely deficient in DSB repair and therefore have very low survival rates following treatment with IR (20). Consistently with previous reports (20, 21), the reexpression of human Ku70 in the Ku70−/− MEFs restored wild-type MEF survival in response to IR (Fig. 2A). Western blot analyses indicated that the level of Ku70 restored was similar to that in wild-type cells (data not shown). Also, we verified that the expression of Ku80, which is reduced to undetectable levels in the absence of the Ku70 subunit, was reestablished upon the expression of the human Ku70 construct (Fig. 2D and 3D).
Fig 2.
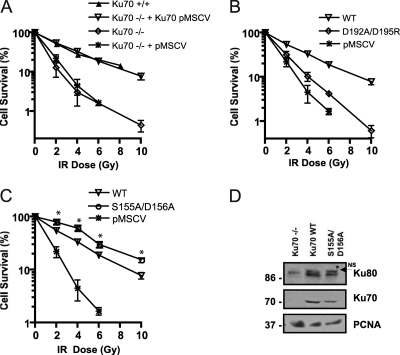
Analysis of cell survival properties of Ku70−/− MEFs expressing Ku70 bearing substitutions in the N-terminal vWA domain. (A) Reexpression of human wild-type Ku70 via retroviral infection restores wild-type survival levels of MEFs following ionizing radiation. The clonogenic survival of MEFs (wild type [Ku70+/+], Ku70 deficient [Ku70−/−], Ku70−/− expressing empty vector [pMSCV], or Ku70 cDNA [Ku70 pMSCV]) was tested at the IR doses indicated. Results are the means from three separate experiments performed in triplicate, with error bars representing the standard deviations (SD). Error bars are included for all data points but may not be visible when they are smaller than symbol size. (B) Ku70−/− MEFs expressing Ku70 mutant bearing substitutions D192A/D195R exhibit radiation sensitivity. A clonogenic assay was done as described for panel A with Ku70−/− MEFs expressing wild-type Ku70 (WT), Ku70 with substitutions (D192A/D195R), or empty vector (pMSCV). (C) MEFs expressing Ku70 with S155A/D156A substitutions show increased survival following IR exposure. Clonogenic assay results are presented as described for panel A. Error bars represent the standard deviations. (D) Representative Western blot analysis of Ku70−/− MEFs or Ku70−/− MEFs expressing wild-type Ku70 (WT) and Ku70 S155A/D156A. The blot was analyzed with antibodies to Ku80, Ku70, and PCNA as indicated. The asterisk indicates the position of a nonspecific (NS) band migrating above Ku80.
Fig 3.
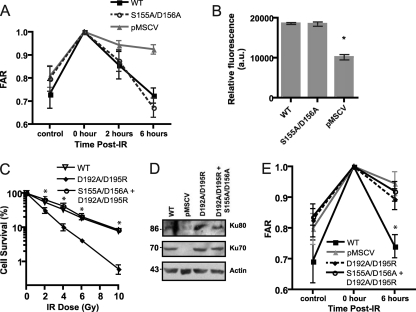
Ku70 S155A/D156A mutation does not affect DNA repair. (A) The S155A/D156A mutation does not interfere with the repair of IR-induced genomic DNA damage. PFGE analysis was performed on genomic DNA from Ku70−/− MEFs expressing wild-type Ku70 (WT), Ku70 S155A/D156A, or empty pMSCV either untreated (control) or immediately after IR treatment (0 h) or 2 or 6 h following IR treatment. For all samples, FAR (fraction of activity released) was averaged from three independent experiments, with error bars representing standard errors of the means. (B) The Ku S155A/D156A mutation does not interfere with the repair of extrachromosomal DNA breaks. Ku70−/− MEFs expressing wild-type Ku70 (WT), mutant S155A/D156A, or empty vector (pMSCV) were transfected with a linearized pGL3-Promoter plasmid and control pRL-SV40 plasmid and assayed for luciferase activity 48 h later. Data represent the average firefly luciferase values normalized to the renilla luciferase values for three separate experiments, with error bars representing SD (*, P < 0.01). (C) Ku70 S155A/D156A mutation rescues the IR sensitivity conferred by the D192A/D195R substitution. Clonogenic survival assay of Ku70−/− MEFs expressing wild-type Ku70 (WT), Ku70 bearing the substitutions D192A/D195R, or the double mutant D192A/D195R, S155A/D156A. Results are averaged from three experiments, and the error bars represent the SD. (D) Western blot analysis of Ku70−/− MEFs with empty vector (pMSCV) or Ku70−/− MEFs expressing wild-type Ku70 (WT) and Ku70 mutants as indicated. The blot was analyzed with antibodies to Ku80, Ku70, and actin. (E) Ku70 S155A/D156A substitutions do not rescue the DNA repair defect conferred by the Ku70 D192A/D195R mutation. PFGE analysis was done as described for panel A with genomic DNA from Ku70−/− MEFs expressing wild-type Ku70 (WT), D192A/D195R, the double mutant D192A/D195R, S155A/D156A, or empty pMSCV either untreated (control) or immediately after (0 h) and 6 h following IR treatment. FAR was averaged from three independent experiments, with error bars representing SEM (∗, P < 0.05).
MEFs expressing α-helix 3 and α-helix 4 Ku70 mutant constructs produced survival curves not significantly different from that of the wild type, suggesting that these residues are not essential for Ku function in response to IR (see Fig. S1 in the supplemental material). An α-helix 5 mutation (D192A/D195R) was designed based on the previously identified DNA repair defect associated with the mutation of the corresponding residues in yeast Ku70 (56). The Ku70 D192A/D195R-expressing cells showed a dramatic decrease in survival following IR treatment, displaying a survival curve that more closely matched that of the Ku70−/− cells than that of the wild type (Fig. 2B). For example, at 4 Gy, 34% of Ku70 wild-type-expressing MEFs formed colonies, whereas only 4.5% of cells expressing empty vector and 10% of cells expressing Ku70 D192A/D195 did so. At 6 Gy, 20% of wild-type cells survived versus 4% of Ku70 D192A/D195R and 1.5% of cells expressing empty vector. This suggests that these residues are functionally conserved between yeast and human. Intriguingly, a Ku70 mutant bearing alanine substitutions of residues S155 and D156 located in a loop region between α-helices 4 and 5 consistently conferred a 40 to 50% increase in survival compared to that of the wild type (at 4 Gy, 60 versus 33%; at 6 Gy, 30 versus 19%) (Fig. 2C). This was unexpected, and as the expression of this mutant was similar to that of the Ku70 wild type (Fig. 2D), it suggested that the mutation enhanced viability in response to IR treatment.
S155A/D156A mutation does not affect DNA repair efficiency.
Since Ku's prominent documented function is to recruit DNA repair factors to DSBs and promote C-NHEJ, we first considered the possibility that the increase in survival conferred by the Ku70 S155A/D156A mutant was due to an improved capacity for DNA repair. To test this possibility, we employed pulsed-field gel electrophoresis (PFGE) to analyze DNA repair. The analysis was performed with samples processed immediately following IR to measure the total amount of genomic DNA breakage, 2 h later, when DNA repair is ongoing, and then 6 h after IR treatment, at which time most DNA breaks are already repaired (33, 61). A comparison of Ku70 wild-type and Ku70 S155A/D156A mutant cells revealed no significant differences in their abilities to repair genomic DNA damaged by IR (Fig. 3A). To confirm this result, an in vivo plasmid repair assay was employed, which measures the cell's ability to recircularize a transfected linearized luciferase expression plasmid by measuring luciferase activity. Ku wild-type and Ku70 S155A/D156A cells showed no significant difference in their abilities to repair restriction enzyme-cut extrachromosomal plasmid DNA (Fig. 3B), suggesting that the mutated residues did not enhance the DNA repair function of Ku. To substantiate the potential for the Ku S155A/D156A mutation to increase survival independently of DNA repair, we tested whether this mutation could rescue the survival defect conferred by a Ku repair mutation. To this end, we introduced the D192A/D195R substitution in α-helix 5 in the Ku70 S155A/D156A construct. The survival of cells expressing the double mutant Ku70 S155A/D156A, D192A/D195R was completely rescued (Fig. 3C), suggesting that the mutation of the S155/D156 residues can compensate for the defects imparted by a DNA repair deficiency. To confirm this result, we analyzed DNA repair in cells expressing Ku70 D192A/D195R and that of the double mutant Ku70 S155A/D156A, D192A/D195R using PFGE (Fig. 3E). As expected, Ku70 D192A/D195R-expressing cells displayed a marked repair defect not statistically different from that of the Ku70-deficient cells. A similar repair defect was observed in cells expressing the Ku70 S155A/D156A, D192A/D195R substitutions, providing evidence that S155A/D156A substitutions do not affect Ku's function in DNA end joining and confer a survival advantage that is independent of DNA repair.
Ku70 S155A/D156A mutant cells display decreased activation of apoptosis.
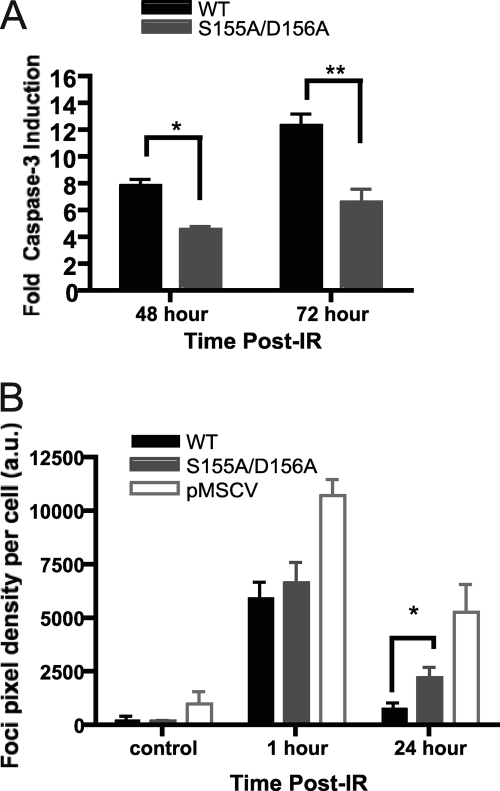
The S155A/D156A mutation did not affect DNA repair, suggesting that it could interfere with DNA damage response pathways and prevent the execution of apoptosis. To determine whether apoptosis was affected in Ku70 S155A/D156A-expressing cells, we tested the endpoint of apoptosis using a caspase-3 assay. While wild-type Ku70 MEFs showed a strong caspase-3 activity at 48 and 72 h after IR treatment, the mutant cells displayed significantly lower caspase-3 activity and, therefore, decreased apoptotic activation (Fig. 4A). We then compared the ability of wild-type Ku70- and Ku70 S155A/D156A-expressing cells to form γ-H2AX foci, an early marker of the DNA damage response (33). Ku70 S155A/D156A cells showed no significant difference in basal focus levels in the absence of IR treatment, whereas Ku70−/− cells had increased γ-H2AX focus levels indicative of unrepaired endogenous DSBs. Early focus formation 1 h following IR treatment again was similar in wild-type Ku70- and S155A/D156A-expressing cells, suggesting that the mutation did not interfere with the initial phosphorylation events. However, Ku70 S155A/D156A cells displayed persistent foci 24 h following IR treatment, whereas Ku70 wild-type cells did not (Fig. 4B), suggesting an abnormally prolonged DNA damage response. Taken together, these results suggested that the mutation interfered with the activation of apoptosis and resulted in a persistent DNA damage response.
Fig 4.
Ku70 S155A/D156A-expressing cells exhibit DNA damage signaling defects. (A) Analysis of IR-induced apoptosis in Ku70 S155A/D156A-expressing cells. Irradiated or mock-treated Ku70−/− MEFs expressing wild-type Ku70 (WT) or Ku70 S155A/D156A were assayed for caspase-3 activity at the times indicated. The fold activation of caspase-3 activity is shown relative to that of the unirradiated control and averaged for four experiments, with error bars representing the SEM (∗∗, P < 0.01; ∗, P < 0.05). (B) S155A/D156A Ku70 mutant cells display prolonged H2AX serine 139 phosphorylation (γ-H2AX) 24 h after IR. As described for panel A, cells were irradiated with 4 Gy of IR or mock treated, fixed at the time points indicated, and subjected to analysis with a γ-H2AX antibody and DAPI. Foci were quantified based on pixel intensity and averaged for the number of cells present (a.u., arbitrary units). Data represent averages from four separate experiments, each assessing approximately 500 cells, and error bars represent SEM (∗, P < 0.05).
Ku70 S155A/D156A mutant cells display altered transcriptional regulation in response to DNA damage response.
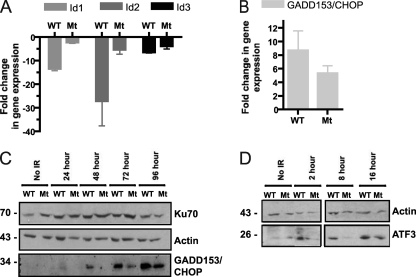
DNA damage-induced apoptosis is regulated largely at the transcriptional level (51, 60), so we thought to investigate whether Ku S155A/D156A could interfere with the transcriptional regulation of genes involved in the IR-induced apoptotic response. To examine global gene expression differences between S155A/D156A and wild-type Ku70-expressing MEFs following IR treatment, Affymetrix GeneChip analysis was performed using RNA prepared from unirradiated control cells and from cells at 8 and 24 h after IR treatment (unpublished data). We noticed that the change of expression of several genes induced or repressed by IR was reduced in Ku70 S155A/D156A-expressing cells. The genes included the inhibitor of differentiation (Id) genes (Id1, Id2, and Id3), activating transcription factor 3 (ATF3), and growth arrest and DNA damage-inducible gene 153 (GADD153), also known as DNA damage-inducible transcript 3 (Ddit3) and C/EBP-homologous protein (CHOP), which previously have been characterized as participating in the regulation of apoptosis (see Table S2 in the supplemental material) (31, 48, 62, 69). Id1 was shown to be downregulated by ATF3 during stress (29, 31), whereas GADD153/CHOP was found to be upregulated by ATF3 to induce cell death programs in response to stress (28, 73). A change of the expression of these genes was confirmed using quantitative RT-PCR analysis. The downregulation of Id1 and Id2 expression in response to IR was found to be severely inhibited in Ku70 S155A/D156A cells, while Id3 also showed a decreased inhibition, albeit less pronounced (Fig. 5A). The induction of GADD153/CHOP also was affected 24 h after IR in cells expressing Ku70 S155A/D156A (Fig. 5B). Western blot analysis further confirmed the reduction in GADD153/CHOP protein expression in Ku70 S155A/D156A mutant cells compared to that of the wild type (Fig. 5C). Since ATF3 activation would be expected to precede that of its target genes Id1 and GADD153/CHOP, we tested ATF3 protein levels in Ku70 wild-type and S155A/D156A mutant cells by Western blotting at earlier time points following IR treatment. A marked decrease in ATF3 induction in cells expressing Ku70 S155A/D156A was detected 2 and 8 h after IR treatment and was still noticeable at 16 h after IR, confirming that Ku70 S155A/D156A interferes with and/or delays ATF3 activation (Fig. 5D). Taken together, these results suggest that the Ku70 S155A/D156A mutation impairs a signaling pathway that affects ATF3 and its target genes in response to IR. ATF3 itself is regulated by ATF2, a transcription factor of the same family (34, 45). Interestingly, we identified additional genes known to be regulated by ATF2 and whose expression is altered by stress or DNA damage that were differentially expressed in Ku wild-type and S155A/D156A cells in response to IR (see Table S2 in the supplemental material). This suggested that the Ku70 S155/D156 residues function to modulate the activation of an ATF2-dependent pathway in response to IR.
Fig 5.
S155A/D156A Ku70-expressing cells show the altered expression of ATF3, GADD153/CHOP, and Id1, Id2, and Id3 in response to IR treatment. (A) RT-PCR analysis of Id family genes Id1, Id2, and Id3. RNA samples from Ku70 S155A/D156A (Mt)-expressing and wild-type Ku70 (WT)-expressing MEFs 24 h after treatment with 6 Gy of irradiation or unirradiated control cells were analyzed by RT-quantitative PCR with primers for the indicated Id genes (Id1, Id2, and Id3). The fold change of gene expression relative to that of unirradiated control samples is shown with error bars indicating SEM (P < 0.05 between WT and Mt for all samples). (B) RT-PCR analysis of proapoptotic GADD153/CHOP expression. RNA samples were processed as described for panel A, and RT-qPCR was performed using primers specific for GADD153/CHOP (P < 0.05). (C) Western blot analysis of GADD153/CHOP protein levels in Ku70 S155A/D156A (Mt) and Ku70 wild-type (WT) MEFs following IR. Shown is a representative Western blot analysis of cells that were irradiated at 6 Gy or mock treated (control), and the extracts were taken at the time points indicated. The membrane was hybridized with the indicated antibodies. (D) Western blot analysis of ATF3 expression. Cells were treated and harvested as described for panel C at the times indicated, and the blot was hybridized with ATF3 and actin antibodies. Both panels are from the same blot, but a longer exposure is shown for ATF3 in the left panel (no IR, 2 h) due to weaker ATF3 signal intensity.
Ku70 S155A/D156A inhibits ATF2 phosphorylation and focus formation.
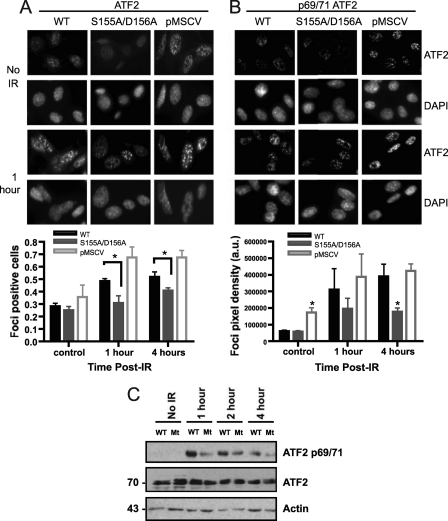
ATF2 expression is not altered in response to IR, but ATF2 is rapidly recruited to IR-induced foci that colocalize with γ-H2AX (8). Focus formation by ATF2 is dependent on the phosphorylation of C-terminal residues (490 and 498) (8). In addition, the activation of ATF2 transcriptional activity in response to DNA damage and other forms of stress is dependent on the phosphorylation of two residues in the N-terminal region (Thr69/71) (45). Thus, to determine whether the Ku70 S155A/D156A mutation interfered with ATF2 activation in response to IR, we first analyzed ATF2 focus formation in Ku70−/− pMSCV MEFs and cells reexpressing wild-type Ku70 and the Ku70 S155A/D156A mutant. While all three cell lines displayed equivalent background levels of ATF2 foci in untreated cells, IR-induced ATF2 focus formation was much stronger in Ku-deficient cells than in cells reexpressing wild-type Ku70 (Fig. 6A). Focus formation was reduced in cells expressing Ku70 S155A/D156A compared to that of the wild type at both 1 and 4 h after IR treatment. To determine whether Ku also modulated ATF2 phosphorylation at the N-terminal sites (Thr69/71) that activate ATF2 transcriptional function, we then analyzed phospho-ATF2 at the 69/71 residues using immunocytochemistry. IR-induced phospho-69/71 ATF2 staining appeared mostly diffuse but exhibited small foci (Fig. 6B). Similarly to ATF2 focus formation, phospho-69/71 was enhanced in Ku−/− cells compared to that in Ku70 wild-type MEFs and was reduced in Ku70 S155A/D156A at both time points tested. To confirm the difference in phosphorylation at the ATF2 69/71 site between Ku70 wild-type and S155A/D156A mutant MEFs, we analyzed phospho-ATF2 by Western blotting. Consistently with the immunofluorescence result, a noticeable reduction in ATF2 phosphorylation was observed at all times points between 1 and 4 h after IR in Ku70 S155A/D156A MEFs compared to levels for the wild type, suggesting that substitutions at S155/D156 impaired ATF2 69/71 phosphorylation (Fig. 6C). Taken together, these results suggest that Ku functions to repress ATF2 activation in response to DNA damage and that the S155A/D156A mutation further enhances this repression.
Fig 6.
Deficient ATF2 activation in response to DNA damage in Ku70 S155/D156-expressing cells. (A) Representative images of ATF2 focus formation in response to IR in Ku70−/− MEFs expressing wild-type Ku70 (WT), Ku70 S155A/D156A, and empty vector (pMSCV). Fixed cells either untreated (no IR) or 1 h after IR (6 Gy) treatment were stained with an ATF2 antibody and DAPI. The graphs show the quantification of focus formation done for unirradiated cells (control) or cells processed 1 or 4 h after IR treatment as described in Materials and Methods, and the results were averaged from four experiments (about 250 cells/experiments), with error bars representing SEM (∗, P < 0.05). (B) Cells were analyzed as described for panel A with a phospho-ATF2 (69/71) antibody. Representative images are shown at the top, with the quantification of phospho-ATF2 staining intensity from three separate experiments being shown below (a.u., arbitrary units of signal intensity). (C) Western blot analysis of phospho-ATF2 (69/71) in Ku70−/− MEFs expressing wild-type Ku70 (WT) and Ku70 S155A/D156A (Mt). Cells were left untreated (no IR) or were subjected to 10 Gy and collected at the time points indicated to prepare nuclear extracts. Western blot analysis was done with a phospho-ATF2 69/71 antibody (p69/71) or with an ATF2 antibody to determine total ATF2 protein and actin expression as indicated.
Increased survival in response to IR is dependent on the mutation of Ku70 S155.
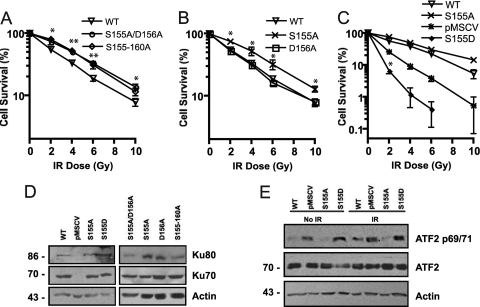
The Ku70 S155/D156 residues are present in a loop region between α-helix 4 and α-helix 5 of the Ku70 vWA domain (72). The effect of the mutation could be due to the disruption of a key phosphorylation event on Ser155 or could simply be disrupting a protein-protein interaction surface. To address the latter possibility, we produced a Ku70 mutant containing alanine substitutions across the entire loop (amino acids [aa] 155 to 160), reasoning that extending the mutated surface could amplify the effect observed with the S155A/D156A mutation. The analysis of IR survival curves revealed no significant difference in survival between MEFs expressing the Ku70 155-160A mutant and those expressing the S155A/D156A mutant, suggesting that S155A/D156A alone conferred maximal increased resistance to IR (Fig. 7A). We next tested the effect of the single S155A and D156A substitutions on cell survival in response to IR. Ku70 D156A expression resulted in a survival profile not significantly different from that of wild-type Ku70. The Ku70 S155A mutant, however, conferred enhanced survival that was similar to that of the 155-160A mutant and the double S155A/D156A mutation (Fig. 7B). This suggests that the DNA damage signaling events that modulate cell survival in response to IR are solely dependent on Ku70 S155.
Fig 7.
Ku70 S155A substitution is required and sufficient to confer increased survival following IR. (A) Clonogenic assay of Ku70−/− MEFs expressing Ku70 with alanine substitutions at positions 155 to 160 (155-160A) and S155A/D156A and Ku70−/− expressing wild-type Ku70 (WT). Survival is expressed as the number of colonies present at each IR dose relative to the unirradiated control, averaged for three experiments, with error bars representing the SD (∗∗, P < 0.01; ∗, P < 0.05). (B) Clonogenic assay comparing the survival of MEFs expressing wild-type Ku70 (WT) and Ku70 D156A and S155A substitutions. Survival is expressed as described for panel A (∗, P < 0.01). (C) Clonogenic assay of MEFs expressing wild-type Ku70 (WT), Ku70 S155A, and empty pMSCV vector (KO) compared to Ku70 bearing the phosphomimetic S155D substitution. Survival is expressed as described for panel A. WT, S155A, and pMSCV are significantly different from each other at all time points (P < 0.05), but asterisks to indicate that significance were omitted for clarity. Significance is indicated for S155D compared to pMSCV (∗, P < 0.001). (D) Western blot analysis of Ku subunit expression in Ku70−/− MEFs expressing empty vector (pMSCV), wild-type Ku70 (WT), or Ku70 mutants as indicated. (E) Western blot analysis of phospho-ATF2 (69/71) in Ku70−/− MEFs expressing wild-type Ku70 (WT), Ku70 S155A, and Ku70 S155D or empty vector (pMSCV). Cells were either left untreated (no IR) or subjected to 10 Gy and collected 2 h later. Western blot analysis was done with a phospho-ATF2 69/71 antibody (p69/71), an ATF2 antibody to determine total ATF2 protein expression, and actin as indicated.
As this residue is a serine, it suggested the possibility that S155 is targeted for phosphorylation to modulate DNA damage signaling, and that the S-to-A substitution prevented this crucial modification. To test this possibility, we generated an aspartic acid mutant, S155D, to assess the effect of a phosphomimetic substitution on cell survival in response to IR. Cells expressing Ku70 S155D appeared fragile and susceptible to cell death. Also, while we confirmed the expression of Ku70 S155D (Fig. 7D), the protein levels appeared to decline quickly in the first few passages following drug selection, and only about 65% of MEFs were found to express this mutant, whereas more than 90% of Ku70 wild-type and other mutant MEFs were expressed (data not shown; also see Table S1 in the supplemental material). Clonogenic assays revealed that the Ku70 S155D mutation conferred a pronounced hypersensitivity to IR, as cells expressing Ku70 S155D displayed even greater radiosensitivity than the Ku-deficient cells (Fig. 7C). To determine the effect of the S155D substitution on ATF2 phosphorylation, we compared ATF2 69/71 phosphorylation in cells expressing Ku70 S155D, Ku70 S155A, wild-type Ku70, and Ku70−/− pMSCV MEFs. In control unirradiated cells, both Ku70-deficient MEFs and S155D mutant MEFs showed strong background levels of phospho-ATF2 compared to those of the Ku70 wild type and S155A mutant (Fig. 7E). In response to IR, as expected, phospho-ATF2 was enhanced in Ku-deficient cells and reduced in Ku70 S155A compared to that of the Ku70 wild type. Also, Ku70 S155D-expressing cells displayed increased levels of phospho-ATF2 compared to that of the Ku70 wild type. Thus, these results are consistent with a phosphomimetic effect of the Asp substitution and support the notion that Ku70 S155 phosphorylation in response to IR is an important event that activates apoptotic pathways in response to DNA damage.
DISCUSSION
This study identifies a novel function for Ku in regulating signaling pathways leading to apoptosis in response to DNA damage. This regulation occurs through a previously uncharacterized region near α-helix 4 in the Ku70 vWA domain. Amino acid substitutions in this region, while not affecting DNA repair, compromise the activation of apoptosis and alter the transcriptional profile of genes regulated by an ATF2/ATF3 pathway.
Previous studies have shown the involvement of the Ku70 vWA domain in C-NHEJ. A recent study demonstrated that Ku has a 5′ lyase activity that is conferred by specific residues in the Ku70 vWA, supporting a direct role for Ku in end processing (59). This activity was found to be dependent on an N-terminal active site (aa 4 to 34) and on three lysine residues within the Ku70 N-terminal domain. In yeast, Ku70 α-helix 5 was found to convey crucial C-NHEJ functions (56). We show here that substitutions in the corresponding human Ku70 α-helix 5 residues (D192A/D195R) caused a survival defect in MEFs that is consistent with a C-NHEJ defect. α-Helix 5 is an exposed α-helix facing toward the DNA terminus that is well conserved from yeast to humans (Fig. 1) (56). While the underlying cause of this DNA repair defect still is unknown, this implies that the function of this Ku region is evolutionarily conserved. Mutations in α-helices 3 and 4 did not cause any obvious defects. Our results are consistent with previous findings in yeast showing that α-helix 4 mutations do not affect C-NHEJ (56). Additionally, it should be pointed out that α-helix 3 is positioned away from the DNA and therefore may be less likely to function in the DNA repair process (56, 72).
In contrast to the aforementioned involvement of vWA regions in C-NHEJ, the Ku70 S155A/D156A mutation is fully functional for DNA repair, suggesting that these residues are not involved in the interaction of Ku with C-NHEJ factors. In particular, it also indicates that these substitutions do not interfere with the overall DNA-PK kinase activity, which is known to be required for C-NHEJ and defects in which result in IR sensitivity (14, 47). Since the DNA-PKcs region of interaction with Ku lies in the Ku80 C-terminal domain, it is unlikely to be affected by a Ku70 N-terminal substitution. However, the possibility remains that S155A/D156A interferes with the phosphorylation of specific targets by DNA-PK.
The increased survival of the MEFs expressing Ku70 S155A/D156A correlated with a marked decrease in apoptosis, as measured by caspase-3 activation. In contrast to wild-type cells, persistent γ-H2AX foci were present 24 h following IR, which is suggestive of the presence of residual unrepaired DNA breaks. Since Ku70 S155A/D156A does not confer any repair defects, the persistence of γ-H2AX foci suggests defects in DNA damage signaling. γ-H2AX foci present at 24 h may indicate DSBs that were unable to be repaired and normally would trigger apoptotic pathway activation to eliminate the damaged cells. We postulate that in Ku wild-type cells, the activation of apoptotic pathways allows the return of γ-H2AX foci to background levels, whereas defects in signaling to apoptosis in Ku70 S155A/D156A delays or impedes focus disappearance in these cells.
Since Ku70 S155A/D156A resulted in apoptotic defects, we first investigated whether it could affect p53, since this factor is a major regulator of apoptotic pathways in response to DNA damage (60). The analysis of p53 response to IR revealed that p53 expression is not induced in either Ku wild-type or Ku70 S155A/D156A cells (data not shown), suggesting that the immortalization of the Ku70−/− MEFs disrupted p53 regulation, an event that frequently occurs in the process of MEF immortalization (22). However, p53 was efficiently phosphorylated at Ser 15 (Ser 18 in mouse) in response to IR, suggesting that the ATM-dependent signaling which is responsible for p53 phosphorylation at this site is intact in these cells (35). Importantly, no difference in the efficiency of phosphorylation was observed between wild-type and Ku70 S155A/D156A cells (data not shown), suggesting that the effect of Ku on signaling to apoptosis does not affect p53 response, and it does not appear to be p53 dependent.
The altered expression of several genes involved in an ATF2/ATF3 signaling pathway in Ku70 S155A/D156A cells led us to speculate that Ku functions to regulate this signaling pathway in response to IR. ATF3 is a basic-region leucine zipper (bZIP) transcription factor member of the ATF/CREB superfamily that is rapidly upregulated by a variety of stress signals, including DNA damage (23, 34, 69, 70). ATF3 can function both to activate and repress transcription, depending on its dimerization partner and the promoter context. Several studies have demonstrated a crucial role for ATF3 activity in inducing apoptosis and the suppression of tumorigenesis (38, 46, 69). Furthermore, ATF3 is directly involved in downregulating Id1 expression while also contributing to GADD153/CHOP transcriptional activation (28, 31). While Id1 has been shown to be directly regulated by ATF3, Id2 and Id3 expression is not well characterized; however, there is evidence that they are regulated coordinately (54, 62). Id proteins function as dominant-negative antagonists of the basic helix-loop-helix (bHLH) family of transcription factors and play roles in development, tumorigenesis, and cell cycle by promoting cell survival and proliferation (54, 62). The overexpression of Id proteins correlates with tumorigenesis, and the downregulation of Id1 expression sensitizes cells to apoptotic agents (50, 54, 62, 76). Finally, previous studies have shown that Id1 is downregulated in response to stress and DNA damage in an ATF3-dependent manner (29, 31). Consistent with these studies, our RT-PCR analyses showed a strong downregulation of Id1 following IR treatment that was concurrent with that of Id2 and Id3. In Ku70 S155A/D156A cells, this repression was substantially lessened, correlating with the reduced activation of ATF3. Thus, the combined IR-induced regulation of these transcription factors converge toward the regulation of apoptosis, and their dysregulation in Ku70 S155A/D156A cells is consistent with the reduced activation of apoptosis observed in response to DNA damage.
ATF3 activation is mediated by several factors and pathways depending on the activating stimulus (69, 73). DNA damage activation of ATF3 has been suggested to depend on an ATM-NBS1 pathway and on ATF2 (34). Recent studies have implicated ATF2 in the DDR (8, 40). In response to DNA damage, ATF2 transcriptional activity is activated by phosphorylation at N-terminal residues T69/71 by p38 and Jun-N-terminal kinase (JNK) in an ATM-dependent manner (7, 34, 75). In addition, ATF2 is phosphorylated at the C-terminal S490/498 residues by ATM, resulting in its accumulation at IR-induced foci that colocalize with γ-H2AX and the MRN complex (8). The mutation of these residues results in the loss of ATF2 focus formation and defective DNA damage response, and it confers increased sensitivity to IR and tumor susceptibility in mice (8, 40).
The analysis of ATF2 IR-induced focus formation and T69/71 phosphorylation showed that both were affected by Ku expression and Ku70 S155A/D156A mutation. Previously, it was determined that while the mutation of ATF2 S490/498 prevented focus formation, T69/71 phosphorylation was dispensable for ATF2 localization into foci, and phosphorylations at both sites were suggested to be independent events (7, 8). The relationship between these two phosphorylation events still is unclear, as ATM is required for ATF2 T69/71 phosphorylation (34), but whether T69/71 phosphorylation is an independent event or is contingent on S490/498 modification has not been determined. We found that phospho-T69/71-ATF2 is localized into IR-induced foci. This infers the existence of an ATF2 population that is phosphorylated at both N-terminal and C-terminal motifs, suggesting that phosphorylation at both sites is not exclusive and that transcriptionally active ATF2 is present at DNA breaks. Thus, our results suggest that Ku functions to modulate both events whether or not they are independent of one another.
The dysregulation of several genes directly or indirectly dependent on ATF2 transcriptional activity in Ku70 S155A/D156A cells suggests that this mutation can interfere with the transcriptional activity of ATF2 mediated by T69/71 phosphorylation. The phosphorylation of T69/71 is induced in Ku70−/− cells but not in wild-type cells, suggesting that Ku plays an inhibitory role in ATF2 transcriptional activation in response to IR. Since ATF2 T69/71 phosphorylation initiates signaling cascades, leading to the activation of apoptosis, one explanation is that the Ku-mediated inhibition of ATF2 activation is linked to Ku's ability to activate DNA repair. The Ku-mediated assembly of a functional C-NHEJ complex and/or completion of DNA repair could prevent ATF2 activation. In the case of overwhelming DSBs, Ku may be present at the break; however, it may not be able to assemble a functional repair complex because of the limiting availability of other C-NHEJ factors, thus allowing ATF2 phosphorylation and the establishment of a signaling pathway leading to the activation of apoptosis. The increased repression of ATF2 phosphorylation by Ku70 S155A suggests that this mutation disrupts an event that normally allows ATF2 activation when DNA repair is not completed. As ATF2 phosphorylation and activation in response to DNA damage is ATM dependent (8, 34), the Ku70 vWA region may function to link Ku to ATM signaling and modulate an ATM-dependent pathway.
We demonstrated that S155 is the essential residue implicated in this regulation of cell survival. As serine/threonine kinases are an integral part of the DNA damage response signaling pathway, the S155A mutation could be preventing an important posttranslational signaling event in the regulation of apoptosis. However, S155 has not been previously identified as a DNA-PK phosphorylation site on Ku70 (10), and it is not located in any canonical kinase phosphorylation motif (as determined using NetworKIN) (44). Interestingly, a recent proteomic study analyzing site-specific phosphorylation after IR treatment identified a new DNA damage-related phosphorylation motif, SXXQ, which was overrepresented among phosphorylation sites regulated within 1 h following IR treatment (5). Peptides with this motif were found to follow a profile of phosphorylation similar to that of SQ motifs, suggesting that this site could be targeted by ATM or DNA-PK. S155 is located in an SXXQ motif (SDVQ in Fig. 1), thus making these two kinases prime candidates for a phosphorylation event at this site. Future experiments will have to determine whether ATM or DNA-PK can phosphorylate this site and whether the phosphorylation of S155 is indeed responsible for the modulation of cell survival through the regulation of an ATF2-dependent pathway.
The idea that S155 is a phosphorylation site is supported by our experimental results showing that the survival advantage conferred by the S155A substitution is completely overturned by the S155D mutation. This suggests that this phosphomimetic substitution provides for a constitutive activation of apoptosis irrespective of DNA repair. Somewhat surprisingly, cells expressing Ku70 S155D displayed an even more pronounced hypersensitivity to IR than cells lacking Ku. ATF2 phosphorylation appeared similar in Ku-deficient and S155D mutant cells within the limit of sensitivity of the Western blot analysis. Thus, it is possible that Ku70 phosphorylation at S155 not only potentiates ATF2 phosphorylation but also has additional effects that contribute to the further activation of downstream apoptotic pathways.
The apoptotic pathway regulated by Ku identified in this study seems independent from the control of Bax by Ku70 described in previous studies (55). First, the Bax-Ku70 interaction that has been described relies on the acetylation of several residues in the Ku70 C-terminal domain, thus in a region that is quite distinct from S155 in the N-terminal domain (11). Second, Bax regulation by Ku70 hinges on the modulation of an interaction between Bax and Ku that has been suggested to occur in the cytoplasm (19), while the regulation that we have uncovered here involves the interplay between proteins that form DNA-induced foci at DNA breaks. However, further investigations will be needed to determine whether Ku70 S155 modulates the activation of apoptosis through other pathways, and in particular whether Ku-Bax interaction is affected by this regulation.
Finally, it is interesting to consider the positioning of the α-helix 5 residues D192 and D195, the mutation of which confers a DNA repair defect in yeast and that we have confirmed here to severely impair viability, and the loop region adjacent to α-helix 4 where S155 is located. Both are facing outwards toward the DNA break (Fig. 1B) and are in proximity to one another, suggesting the potential for interactions or cross-talk between the two regions. Because of its presence at the DNA break and its primary function in C-NHEJ, Ku is well positioned to act as a sensor of DNA repair. Therefore, the proximity of α-helix 5, which appears essential for DNA repair, to the S155 residues suggests that Ku functions to relay signals from the repair machinery to nearby regulators of signaling pathways that control apoptosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Matsuyama for the Ku70 knockout cells. We also thank C. Shen, M. Widlicki, and S. Hershenfeld for technical help; C. Brandl, M. Davey, and G. Gloor for valuable comments and suggestions; and C. Brandl for critical comments on the manuscript.
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (355799-2008) to C.S.P.
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abraham RT. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amsterdam) 3:883–887 [DOI] [PubMed] [Google Scholar]
- 2. Andrews NC, Faller DV. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aravind L, Koonin EV. 2000. SAP–a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112–114 [DOI] [PubMed] [Google Scholar]
- 4. Aravind L, Koonin EV. 2001. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11:1365–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bennetzen MV, et al. 2010. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell Proteomics 9:1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bertinato J, Schild-Poulter C, Hache RJ. 2001. Nuclear localization of Ku antigen is promoted independently by basic motifs in the Ku70 and Ku80 subunits. J. Cell Sci. 114:89–99 [DOI] [PubMed] [Google Scholar]
- 7. Bhoumik A, Lopez-Bergami P, Ronai Z. 2007. ATF2 on the double–activating transcription factor and DNA damage response protein. Pigment Cell Res. 20:498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhoumik A, et al. 2005. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol. Cell 18:577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cann KL, Hicks GG. 2007. Regulation of the cellular DNA double-strand break response. Biochem. Cell Biol. 85:663–674 [DOI] [PubMed] [Google Scholar]
- 10. Chan DW, Ye R, Veillette CJ, Lees-Miller SP. 1999. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry 38:1819–1828 [DOI] [PubMed] [Google Scholar]
- 11. Cohen HY, et al. 2004. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell 13:627–638 [DOI] [PubMed] [Google Scholar]
- 12. Collis SJ, Deweese TL, Jeggo PA, Parker AR. 2005. The life and death of DNA-PK. Oncogene 24:949–961 [DOI] [PubMed] [Google Scholar]
- 13. Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. 2007. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amsterdam) 6:712–722 [DOI] [PubMed] [Google Scholar]
- 14. Dobbs TA, Tainer JA, Lees-Miller SP. 2010. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair 9:1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Downs JA, Jackson SP. 2004. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5:367–378 [DOI] [PubMed] [Google Scholar]
- 16. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amsterdam) 3:959–967 [DOI] [PubMed] [Google Scholar]
- 18. Fulop GM, Phillips RA. 1990. The scid mutation in mice causes a general defect in DNA repair. Nature 347:479–482 [DOI] [PubMed] [Google Scholar]
- 19. Gama V, et al. 2009. Hdm2 is a ubiquitin ligase of Ku70-Akt promotes cell survival by inhibiting Hdm2-dependent Ku70 destabilization. Cell Death Differ. 16:758–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. U. S. A. 94:8076–8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gu Y, et al. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7:653–665 [DOI] [PubMed] [Google Scholar]
- 22. Hahn WC, Weinberg RA. 2002. Modelling the molecular circuitry of cancer. Nat. Rev. Cancer 2:331–341 [DOI] [PubMed] [Google Scholar]
- 23. Hai T, Hartman MG. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273:1–11 [DOI] [PubMed] [Google Scholar]
- 24. Harris R, et al. 2004. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR). J. Mol. Biol. 335:573–582 [DOI] [PubMed] [Google Scholar]
- 25. Hartlerode AJ, Scully R. 2009. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 423:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helleday T, Lo J, van Gent DC, Engelward BP. 2007. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amsterdam) 6:923–935 [DOI] [PubMed] [Google Scholar]
- 27. Hill R, Lee PW. 2010. The DNA-dependent protein kinase (DNA-PK): more than just a case of making ends meet? Cell Cycle 9:3460–3469 [DOI] [PubMed] [Google Scholar]
- 28. Jiang H-Y, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24:1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang Y, Chen C-R, Massagué J. 2003. A self-enabling TGF[beta] response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11:915–926 [DOI] [PubMed] [Google Scholar]
- 30. Karagiannis TC, El-Osta A. 2004. Double-strand breaks: signaling pathways and repair mechanisms. Cell. Mol. Life Sci. 61:2137–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kashiwakura Y, et al. 2008. Down-regulation of inhibition of differentiation-1 via activation of activating transcription factor 3 and Smad regulates REIC/Dickkopf-3-induced apoptosis. Cancer Res. 68:8333–8341 [DOI] [PubMed] [Google Scholar]
- 32. Kim J-S, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AMR, Yokomori K. 2005. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 170:341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kinner A, Wu W, Staudt C, Iliakis G. 2008. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36:5678–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kool J, et al. 2003. Induction of ATF3 by ionizing radiation is mediated via a signaling pathway that includes ATM, Nibrin1, stress-induced MAP kinases and ATF-2. Oncogene 22:4235–4242 [DOI] [PubMed] [Google Scholar]
- 35. Kurz EU, Lees-Miller SP. 2004. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair 3:889–900 [DOI] [PubMed] [Google Scholar]
- 36. Lavin MF, Kozlov S. 2007. ATM activation and DNA damage response. Cell Cycle 6:931–942 [DOI] [PubMed] [Google Scholar]
- 37. Lee JH, Paull TT. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26:7741–7748 [DOI] [PubMed] [Google Scholar]
- 38. Lee SH, Bahn JH, Whitlock NC, Baek SJ. 2010. Activating transcription factor 2 (ATF2) controls tolfenamic acid-induced ATF3 expression via MAP kinase pathways. Oncogene 29:5182–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li GC, et al. 1998. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 2:1–8 [DOI] [PubMed] [Google Scholar]
- 40. Li S, et al. 2010. Radiation sensitivity and tumor susceptibility in ATM phospho-mutant ATF2 mice. Genes Cancer 1:316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79:181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lieber MR, Ma Y, Pannicke U, Schwarz K. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4:712–720 [DOI] [PubMed] [Google Scholar]
- 43. Lieber MR, Yu K, Raghavan SC. 2006. Roles of nonhomologous DNA end joining, V(D)J. recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amsterdam) 5:1234–1245 [DOI] [PubMed] [Google Scholar]
- 44. Linding R, et al. 2008. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 36:D695–D699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopez-Bergami P, Lau E, Ronai Z. 2010. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer 10:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu D, Wolfgang CD, Hai T. 2006. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J. Biol. Chem. 281:10473–10481 [DOI] [PubMed] [Google Scholar]
- 47. Mahaney BL, Meek K, Lees-Miller SP. 2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417:639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maytin EV, Ubeda M, Lin JC, Habener JF. 2001. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp. Cell Res. 267:193–204 [DOI] [PubMed] [Google Scholar]
- 49. Meek K, Dang V, Lees-Miller SP. 2008. DNA-PK: the means to justify the ends, p. 33–58 In Frederick WA. (ed.), Advances in immunology. Academic Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 50. Mern DS, Hasskarl J, Burwinkel B. 2010. Inhibition of Id proteins by a peptide aptamer induces cell-cycle arrest and apoptosis in ovarian cancer cells. Br. J. Cancer 103:1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Norbury CJ, Zhivotovsky B. 2004. DNA damage-induced apoptosis. Oncogene 23:2797–2808 [DOI] [PubMed] [Google Scholar]
- 52. Nussenzweig A, Nussenzweig MC. 2007. A backup DNA repair pathway moves to the forefront. Cell 131:223–225 [DOI] [PubMed] [Google Scholar]
- 53. Nussenzweig A, Sokol K, Burgman P, Li L, Li GC. 1997. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. U. S. A. 94:13588–13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Perk J, Iavarone A, Benezra R. 2005. Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 5:603–614 [DOI] [PubMed] [Google Scholar]
- 55. Rathaus M, Lerrer B, Cohen HY. 2009. DeubiKuitylation: a novel DUB enzymatic activity for the DNA repair protein, Ku70. Cell Cycle 8:1843–1852 [DOI] [PubMed] [Google Scholar]
- 56. Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. 2007. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat. Struct. Mol. Biol. 14:301–307 [DOI] [PubMed] [Google Scholar]
- 57. Riches LC, Lynch AM, Gooderham NJ. 2008. Early events in the mammalian response to DNA double-strand breaks. Mutagenesis 23:331–339 [DOI] [PubMed] [Google Scholar]
- 58. Riha K, Heacock ML, Shippen DE. 2006. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 40:237–277 [DOI] [PubMed] [Google Scholar]
- 59. Roberts SA, et al. 2010. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 464:1214–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roos WP, Kaina B. 2006. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12:440–450 [DOI] [PubMed] [Google Scholar]
- 61. Rothkamm K, Kruger I, Thompson LH, Lobrich M. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23:5706–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ruzinova MB, Benezra R. 2003. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13:410–418 [DOI] [PubMed] [Google Scholar]
- 63. Schild-Poulter C, Haché RJG, Soubeyrand S. 2004. Ku antigen: a versatile DNA binding protein with multiple cellular functions, p. 257–284 In Parisi V, De Fonzo V, Aluffi-Pentini F. (ed.), Recent research developments in dynamical genetics, Research Signpost, Transworld Research Network [Google Scholar]
- 64. Schild-Poulter C, et al. 2007. DNA-PK phosphorylation sites on Oct-1 promote cell survival following DNA damage. Oncogene 26:3980–3988 [DOI] [PubMed] [Google Scholar]
- 65. Sekiguchi JM, Ferguson DO. 2006. DNA double-strand break repair: a relentless hunt uncovers new prey. Cell 124:260–262 [DOI] [PubMed] [Google Scholar]
- 66. Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25:3934–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smith GC, Jackson SP. 1999. The DNA-dependent protein kinase. Genes Dev. 13:916–934 [DOI] [PubMed] [Google Scholar]
- 68. Su TT. 2006. Cellular responses to DNA damage: one signal, multiple choices. Annu. Rev. Genet. 40:187–208 [DOI] [PubMed] [Google Scholar]
- 69. Thompson M, Xu D, Williams B. 2009. ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. 87:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Turchi L, et al. 2008. Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death Differ. 15:1472–1480 [DOI] [PubMed] [Google Scholar]
- 71. Valerie K, Povirk LF. 2003. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22:5792–5812 [DOI] [PubMed] [Google Scholar]
- 72. Walker JR, Corpina RA, Goldberg J. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607–614 [DOI] [PubMed] [Google Scholar]
- 73. Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34:7–11 [DOI] [PubMed] [Google Scholar]
- 74. Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin a domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xia Y, Ongusaha P, Lee SW, Liou Y-C. 2009. Loss of Wip1 sensitizes cells to stress- and DNA damage-induced apoptosis. J. Biol. Chem. 284:17428–17437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang X, Ling MT, Wong YC, Wang X. 2007. Evidence of a novel antiapoptotic factor: role of inhibitor of differentiation or DNA binding (Id-1) in anticancer drug-induced apoptosis. Cancer Sci. 98:308–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang Z, et al. 2004. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure (Cambridge) 12:495–502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.