Abstract
Living cells are adaptive self-sustaining systems. They strictly depend on the sufficient supply of oxygen, energy, and nutrients from the outside in order to sustain their internal organization. However, as autonomous entities they are able to monitor and appropriately adapt to any critical fluctuation in their environment. In the case of insufficient external nutrient supply or augmented energy demands, cells start to extensively digest their own interior. This process, known as macroautophagy, comprises the transport of cytosolic portions and entire organelles to the lysosomal compartment via specific double-membrane vesicles, called autophagosomes. Although extensively upregulated under nutrient restriction, a low level of basal autophagy is likewise crucial in order to sustain the cellular homeostasis. On the other hand, cells have to avoid excessive and enduring self-digestion. The delicate balance between external energy and nutrient supply and internal production and consumption is a demanding task. The complex protein network that senses and precisely reacts to environmental changes is thus mainly regulated by rapid and reversible posttranslational modifications such as phosphorylation. This review focuses on the serine/threonine protein kinases AMP-activated protein kinase, mammalian target of rapamycin (mTOR), and unc-51-like kinase 1/2 (Ulk1/2), three interconnected major junctions within the autophagy regulating signaling network.
AMPK: THE ENERGY-SENSING KINASE
AMP-activated protein kinase (AMPK) was initially identified as a serine/threonine kinase that negatively regulates several key enzymes of the lipid anabolism (30). Meanwhile, AMPK is regarded as the major energy-sensing kinase that activates a whole variety of catabolic processes in multicellular organisms such as glucose uptake and metabolism, while simultaneously inhibiting several anabolic pathways, such as lipid, protein, and carbohydrate biosynthesis (reviewed in reference 30).
AMPK is a heterotrimeric protein complex that is precisely regulated in different ways. First, the phosphorylation of a conserved threonine residue (T172) in the activation loop of the catalytic α-subunit by upstream kinases is a prerequisite for the activity of AMPK. Several AMPK-phosphorylating kinases have been identified thus far. In addition to the ubiquitously expressed and constitutively active kinase LKB1 (31, 109), Ca2+-activated Ca2+/calmodulin-dependent kinase kinase β (CaMKKβ) (32, 43, 108) and transforming growth factor β-activated kinase-1 (TAK1) (80) are both known as activators of AMPK. Second, AMPK activity can further be modulated by allosteric binding to the regulatory β- and γ-subunit. Since the ratio of AMP to ATP represents the most accurate way to precisely measure the intracellular energy level, both AMP and ATP are able to oppositely regulate the activity of AMPK. While AMP binding to the γ-subunit allosterically enhances AMPK kinase activity and prevents the dephosphorylation of T172, ATP is known to counteract the activating properties of AMP (30). Although ADP does not allosterically activate AMPK, it could be shown very recently that it also binds to AMPK and enhances phosphorylation at T172 (83, 111).
AMPK is an evolutionarily conserved energy-sensing kinase that is activated by metabolic stress or ATP consumption and that globally promotes catabolic processes. In accordance with that, AMPK could also be linked to the regulation of autophagy. Initially, the yeast ortholog of AMPK (SNF1) was identified as a positive regulator of autophagy (42, 107). The essential role of AMPK for the regulation of autophagic proteolysis in mammalian cells was confirmed subsequently, mainly by addressing long-lived protein degradation in HT-29 human colon cancer and HeLa cells (77). In addition to AMPK's activation by low cellular energy levels, presumably via LKB1 and high AMP concentrations, it has been suggested that a variety of other non-starvation-related autophagy-inducing stimuli primarily act through the activation of AMPK even under normal energy levels (39, 40). Autophagy induction, observed after the rise in intracellular Ca2+ concentrations in human breast cancer and cervix carcinoma cells, has been linked to CaMKKβ-mediated enhancement of AMPK activity (39). Similarly, TRAIL-induced cytoprotective autophagy in nontransformed epithelial cells has been reported to depend on TAK1-mediated AMPK activation, and it has been argued that this may contribute to the differential cell death response of nontransformed versus tumor cells after TRAIL treatment (36). However, although the expression of a dominant-negative form of AMPK completely inhibited autophagic proteolysis in HT-29 and HeLa cells under harsh starvation conditions (Hanks balanced salt solution [HBSS]), transfection with a constitutively active form of AMPK did not affect the rate of autophagy (77). Thus, it is still not clear whether AMPK activation alone is sufficient to induce autophagy in mammalian cells or whether the basal activity of AMPK, although essential for autophagy induction, must be accompanied by additional cell stress pathways such as those triggered by TRAIL, an increase in intracellular [Ca2+], or nutrient and growth factor withdrawal (reviewed in reference 58).
As long as unicellular organisms live in the land of plenty, they will grow and divide. In multicellular organisms, cell growth and proliferation must also be tightly regulated by growth factor signaling to avoid neoplasm. However, cell growth and division only make sense when the cell is sufficiently supplied with energy and nutrients that serve as building blocks for biosynthesis. A key enzyme that balances diverse anabolic processes such as cell growth, proliferation, and protein synthesis in response to those growth-inducing stimuli is the target of rapamycin (TOR). As it turns out, the growth factor-regulated and nutrient-sensing kinase TOR and the energy-sensing kinase AMPK act in concert to control autophagy induction.
mTOR: THE NUTRIENT-SENSING KINASE
TOR was initially identified as a target for the antifungal properties of rapamycin, which leads to growth inhibition in Saccharomyces cerevisiae (35). Later, it was shown that TOR is also involved in the regulation of autophagy and that rapamycin is able to induce autophagy in yeast even under nutrient-rich conditions (82). TOR is an evolutionary highly conserved serine/threonine protein kinase and hence not only found in yeast but also in metazoans such as flies (dTOR) and mammals (mammalian target of rapamycin [mTOR]). Drosophila mutants deficient in dTOR exhibit reduced cell and body size (85, 120), as well as a massive accumulation of autophagic vesicles in the fat body even under fed conditions (92).
TOR proteins interact with several binding partners to form at least two functionally distinct complexes, called TOR complex 1 (TORC1) and TORC2 in yeast. Notably, rapamycin solely binds to TORC1, and only TORC1 deficiency resembles the effects of rapamycin treatment (110). The corresponding mammalian rapamycin-sensitive mTOR complex 1 (mTORC1) comprises mTOR, mLST8 (or GβL), PRAS40, and the regulatory-associated protein of TOR (raptor) (53). mTORC2, in contrast, is unaffected by rapamycin treatment since it comprises the rapamycin-insensitive companion of TOR (rictor) instead of raptor (91) (reviewed in references 94 and 110). Notably, while rapamycin is a strong autophagic stimulus in yeast, it poorly induces autophagy in mammalian cells. It could be demonstrated that this compound, although fully inhibiting the mTORC1-dependent phosphorylation of S6K, only partially inhibits the phosphorylation of other known mTORC1 substrates. Some mTORC1-dependent functions such as autophagy inhibition might thus be partially unaffected by rapamycin (101, 102). Using the ATP-competitive mTOR inhibitor Torin1, it could be shown that the above mentioned discrepancy to yeast is mainly due to those rapamycin-resistant functions of mTORC1 (101, 102). It has been argued that this might be the main reason for the limited success of rapamycin treatment in anticancer therapy (101).
While AMPK is activated under energy-low conditions, leading to autophagy induction, mTORC1 activity depends on diverse positive signals such as high energy levels, normoxia, amino acids, or growth factors that all result in the inhibition of autophagy.
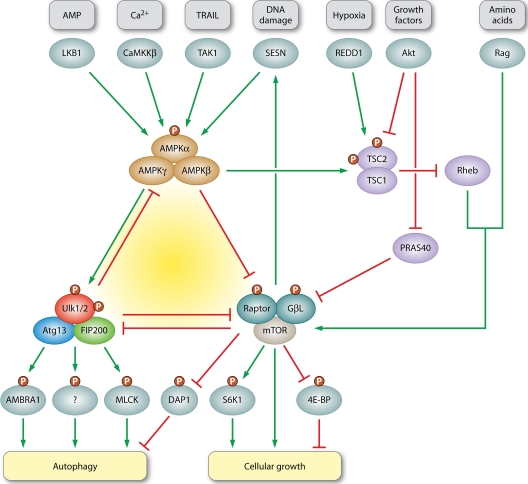
Growth factors activate the PI3K/Akt pathway through receptor tyrosine kinases (RTK). Upon growth factor binding, Akt is recruited to the plasma membrane, where it is activated through phosphorylation by PDK1. Activated Akt in turn phosphorylates TSC2 (tuberous sclerosis complex 2), which prevents formation of the inhibitory TSC1/TSC2 heterodimer. Since TSC2 serves as a GTPase-activating protein (GAP) and inactivates the small GTPase Rheb, its inhibition subsequently allows Rheb to directly activate mTORC1 (41, 71, 72, 119). In addition, the mTORC1 component PRAS40 has been identified as a direct Akt substrate, and the data suggest that this phosphorylation is responsible for mTORC1 activation via insulin signaling (105). Hence, growth factor withdrawal would finally lead to mTORC1 inactivation and in turn to the induction of autophagy.
In addition to growth factor signaling, nutrient availability contributes to the positive regulation of mTORC1 activity. It has been suggested that amino acids activate mTORC1 via the Rag family of small GTPases (54, 90), whose activity is regulated by amino acids. However, unlike Rheb, Rag GTPases are not able to activate mTORC1 activity in vitro (90). This discrepancy might have been solved recently, since Sancak et al. could show that mTORC1 is recruited to lysosomes in a Rag-dependent manner upon amino acid stimulation, where it is finally activated by Rheb GTPases (89). This would explain how growth factor and amino acid signaling are integrated to fully activate mTORC1. In addition, mTORC1 activity has been shown to be regulated by oxygen concentrations (2). Hypoxia-induced inhibition of mTOR is even dominant over mTOR-activating signals such as growth factors and nutrients (2). It likewise seems to act via the TSC1/TSC2 complex and depends on transcriptional upregulation of REDD1 (7).
Energy levels are mainly sensed via the above described AMPK pathway. Activated AMPK was thought to inhibit mTORC1 activity primarily in the opposite way as growth factors stimulate it, mainly by phosphorylation and activation of the negative regulator TSC2 (44). However, the fact that TSC2-deficient cells still respond to decreasing energy levels led to the investigation of additional mechanisms. Raptor has meanwhile been identified as a direct substrate of AMPK. It could be demonstrated that this phosphorylation generates a docking site for inhibitory 14-3-3 proteins and is required for AMPK-mediated inhibition of mTORC1 (27). It appears that AMPK possesses at least two different ways to release the mTORC1-mediated repression on autophagy induction in case of alarming energy states.
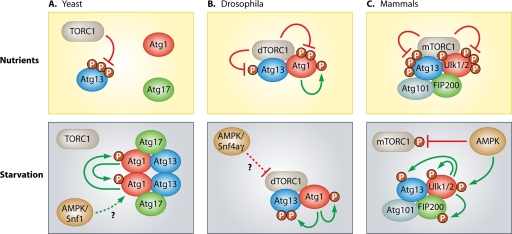
The mechanism by which TORC1 negatively regulates the autophagic machinery has first been described in yeast. Genetic screenings for autophagy defective mutants led to the identification of more than 30 essential autophagy-related genes (Atg) (34, 81). It turned out that most of these Atg proteins are recruited to a single preautophagosomal structure (PAS) in a hierarchical manner upon starvation (97, 98). These proteins can be classified into several groups depending on their function and interdependency (81). Most upstream is a protein complex that comprises the serine/threonine kinase Atg1, as well as two accessory proteins Atg13 and Atg17. This complex is directly regulated by TORC1 in a nutrient-dependent manner (14, 49, 50). Although counterparts of this complex could be identified in mammals and flies over the last years, their regulation seems to differ in several ways from the situation in yeast (Fig. 1).
Fig 1.
Regulation of autophagy induction by the TOR and AMPK complex in yeast, flies, and mammals. (A) Under nutrient-rich conditions, activated TORC1 inhibits autophagy induction in yeast through direct phosphorylation of Atg13. This hyperphosphorylation of Atg13 prevents binding to Atg1. Inactivation of TORC1, as induced by starvation or rapamycin treatment, results in rapid dephosphorylation of Atg13, leading to the formation of the active Atg1-Atg13-Atg17 kinase complex (50, 51). The Atg13-mediated dimerization of Atg1 has recently been described to be essential for the autophosphorylation and subsequent activation of Atg1 (114). Furthermore, Snf1, a yeast ortholog of mammalian AMPK, has been found to be a positive effector of autophagy, presumably through regulation of Atg1 and Atg13 (107). (B) In contrast to yeast, the Atg1-Atg13 complex is stable under nutrient-rich conditions in Drosophila and dTORC1 phosphorylates both Atg13 and Atg1. In starved animals or when dTORC1 is specifically inhibited, these sites are dephosphorylated, Atg1 kinase activity is elevated, thus leading to autophosphorylation and phosphorylation of Atg13 (10, 11). Snf4aγ, a Drosophila ortholog of the mammalian AMPK gamma subunit, was found to be an inducer of autophagy; nevertheless, the exact mechanism remains elusive (68). (C) In mammals, two orthologs of yeast Atg1, termed Ulk1 and Ulk2, have been linked to starvation induced autophagy. Both are found in a stable complex with Atg13, FIP200 (5, 26, 28, 29, 37, 47), and Atg101, an additional binding partner of Atg13 that has no ortholog in yeast (38, 78). In contrast to yeast, the composition of this complex does not change with the nutrient status. Under fed conditions mTORC1 phosphorylates Ulk1/2 and Atg13, thereby inhibiting the kinase complex (26, 37, 47). In response to starvation, the mTORC1-dependent phosphorylation sites in Ulk1/2 are rapidly dephosphorylated, and Ulk1/2 autophosphorylates and phosphorylates Atg13 and FIP200. Alternatively, Ulk1/2 is phosphorylated by AMPK and thereby activated (20, 54). In addition, AMPK indirectly leads to the induction of autophagy by inhibiting mTORC1 through phosphorylation of raptor (27, 64). Note, however, that this is a schematic overview, and we do not provide any determination regarding the association between AMPK and the Ulk1/2-Atg13-FIP200 complex due to the conflicting results published in the recent past. For further details, see the legend to Fig. 3. (This figure was adopted and modified with permission from reference 11.)
ULK1/2: THE AUTOPHAGY INITIATORS
Under optimal growth conditions, activated TORC1 inhibits autophagy induction in yeast by direct phosphorylation of Atg13 at several serine residues (Fig. 1). This hyperphosphorylation prevents binding to Atg1 and, hence, Atg1-Atg13-Atg17 complex formation (51). TORC1 inactivation, as resulting from nutrient deprivation or rapamycin treatment, in turn leads to immediate dephosphorylation of Atg13, to complex assembly and finally to the enhancement of Atg1 kinase activity (50, 51). The resulting redistribution of Atg1-Atg13-Atg17 to the PAS is followed by the recruitment of other Atg proteins (97, 98). Although the Atg13-mediated dimerization of Atg1 is essential and presumably even sufficient for the autophosphorylation and subsequent activation of Atg1, it does not require Atg17 (114). Atg17, however, seems to be most fundamental in PAS organization and serves as a scaffold that determines the site to which the other Atg proteins—including Atg13 and Atg1—are recruited (12, 98). Although the subsequent localization of most of the other Atg proteins does not depend on the catalytic activity of Atg1, the data suggest that it is nonetheless essential for the formation of proper autophagosomes (12, 52). Interestingly, the expression of an Atg13 mutant lacking the TORC1-dependent phosphorylation sites is sufficient to induce autophagy in yeast, even under optimal growth conditions, presumably by uncontrolled complex formation and enhancement of Atg1 kinase activity (51). However, although several proteins have been suggested as potential Atg1 in vitro substrates after a global phosphorylation analysis in yeast (86), the exact kinase-dependent function of Atg1 and its direct downstream in vivo targets are still unknown.
The essential role of Atg1 orthologs for autophagy induction has been confirmed in several species such as Caenorhabditis elegans (84) and Drosophila melanogaster (92), all of which possess only one Atg1 gene. Vertebrates in contrast have at least five different kinases closely related to Atg1 in their N-terminal kinase domain. The most highly related unc-51-like kinase 1 (Ulk1) and Ulk2 were first identified as close homologs of C. elegans uncoordinated-51 (unc-51), showing 78% amino acid identity in their catalytic domains (61, 103, 112, 113). Initially, it was suggested that primarily Ulk1 is responsible for autophagy induction since the single knockdown of Ulk1 is able to inhibit this process in several cell lines (8, 26). However, in contrast to the knockout of other essential autophagy-related genes (56, 59), ulk1−/− and ulk2−/− mice are born viable and are able to survive the short starvation period after birth (13, 60, 62). Nevertheless, Ulk1-deficient mice exhibit a compromised clearance of mitochondria during reticulocyte maturation (60) arguing for a specific role of Ulk1 in the selective engulfment of mitochondria (mitophagy) during erythropoiesis. In addition, ulk2−/− murine embryonic fibroblasts (MEFs) have been reported to display normal autophagy induction after starvation in HBSS, and only the additional knockdown of Ulk1 was able to inhibit autophagy induction (62). Furthermore, while ulk1−/− MEFs displayed normal LC3 lipidation in response to glucose starvation (60), these cells did not respond with autophagy induction to rapamycin treatment (47). Collectively, these results indicate that Ulk1 and Ulk2, although they seem to possess unique and cell type-specific roles, have partially redundant functions in starvation-induced autophagy. However, the discordance concerning the role of Ulk1 and Ulk2 in mammalian autophagy induction might simply reflect the heterogeneity of the experimental settings, the autophagic stimuli, and the respective autophagic readout used.
Notably, in contrast to the respective single knockouts, the recently generated ulk1−/− ulk2−/− mice do display early neonatal lethality (13). MEFs from these mice show a complete blockage of autophagy induction upon amino acid withdrawal but did respond to increasing ammonia concentrations, as a result from enhanced amino acid metabolism after long-term glucose withdrawal (13). Thus, it is well conceivable that autophagy in vertebrates can be induced either selectively or simultaneously by several partially overlapping signaling pathways, depending on the exact autophagic stimulus. Some of these pathways might even be independent of Ulk1 and Ulk2, as shown for ulk1−/− ulk2−/− MEFs (13) and ulk1−/− ulk2−/− DT40 cells by our group (1).
Although there is no clear Atg17 homolog in higher eukaryotes, the focal adhesion kinase (FAK) family-interacting protein of 200 kDa (FIP200), also called retinoblastoma 1-inducible coiled-coil 1 (RB1CC1), has been proposed as the functional counterpart of Atg17 (28, 29). FIP200 is a multifunctional protein that possesses several interaction partners and regulates diverse cellular processes such as cell growth, proliferation, cell spreading, and migration (reviewed in reference 25). FIP200 additionally indirectly interacts with Ulk1 and Ulk2, promotes Ulk1 kinase activity, translocates to the pre-autophagosomal membrane after starvation, and is essential for autophagy induction (29). A weakly conserved mammalian homolog of yeast Atg13 was first predicted by a sequence homology search (75), and its relevance for autophagy induction was subsequently confirmed by Chan et al. (10).
However, in contrast to the yeast Atg1-Atg13-Atg17 complex, the binding affinity between unc-51-like kinase 1/2 (Ulk1/2), Atg13, and FIP200 is largely unaffected by the nutrient status (Fig. 1). Instead, these proteins form a large and stable complex of ∼3 MDa whose composition is not altered after starvation (10, 26, 29, 37, 38, 47). The product of the mammalian C12orf44 gene has been identified as an additional component of the Ulk1/2-Atg13-FIP200 complex and as direct binding partner of Atg13 (38, 78). It has no obvious homolog in yeast and was accordingly named Atg101. Although its exact function is unknown thus far, the data suggest that it is crucial for basal phosphorylation of Atg13 and Ulk1 and that it prevents the proteasomal degradation of Atg13 (38, 78).
There is a significant body of evidence that the phosphorylation status within the Ulk1/2-Atg13-FIP200 complex dramatically changes with the nutrient status. Under normal growth conditions, mTORC1 associates with the Ulk1/2-Atg13-FIP200 complex, via direct interaction between raptor and Ulk1/2 (37). The active mTOR phosphorylates Atg13 and Ulk1/2 (26, 37, 47), thereby suppressing Ulk1/2 kinase activity. Under starvation conditions or when mTORC1 activity is pharmacologically inhibited, these sites are rapidly dephosphorylated by yet unknown phosphatases. Using stable isotope labeling with amino acids in cell culture (SILAC), Shang et al. could recently identify several serine and threonine residues in human Ulk1 whose phosphorylation was decreased after starvation (see Fig. 3). The most prominent decrease was detected at S638 and S758 and could be verified after rapamycin treatment and mTOR knock-down (95). Notably, S757 in mouse Ulk1 (corresponding to S758 in human Ulk1) was additionally identified as mTOR site by Kim et al. (55). The activated Ulk1/2 autophosphorylates and phosphorylates both FIP200 and Atg13, which in turn leads to translocation of the entire complex to the pre-autophagosomal membrane and to autophagy induction (8, 10, 26, 29, 37, 47) (Fig. 1). However, the functional relevance of Ulk1/2-mediated phosphorylation of Atg13 and FIP200 for this recruitment and the relevant phosphorylation sites have not been verified yet. Notably, although our group was able to identify five Ulk1-dependent in vitro phosphorylation sites in human Atg13, the mutation of these sites did not affect Atg13 function in DT40 cells (1). Furthermore, the mTORC1-dependent phosphorylation sites in mammalian Atg13 are still unknown and a recent mass spectrometric approach failed to clearly identify nutrient-regulated in vivo phosphorylation sites in human Atg13 (96). Thus, it is still an exciting task to identify the exact in vivo phosphorylation sites within this complex and to explore their functional relevance. Interestingly, another Ulk1-dependent phosphorylation site in human Atg13 (S318) has been identified recently (46). The authors of that study could show that the Hsp90-Cdc37 chaperone complex selectively stabilizes and activates Ulk1. The activation of Ulk1 leads to phosphorylation of Atg13 and in turn to the independent translocation of Atg13 to depolarized mitochondria. However, although the expression of a nonphosphorylatable S318A mutant inhibited the selective clearance of damaged mitochondria in a dominant-negative way, it did not affect basal or starvation-induced autophagy (46). This mechanism might further explain the specific role of Ulk1 for mitochondrial clearance during reticulocyte maturation, as described above.
Fig 3.
Mutual phosphorylation of Ulk1, mTORC1, and AMPK. (A) Ulk1 is a hyperphosphorylated protein that is massively dephosphorylated upon starvation. Under normal growth conditions, mTORC1 has been shown to directly bind to and negatively regulate Ulk1/2 kinase activity by direct phosphorylation (26, 37, 47). A total of 16 phosphorylation sites in mouse Ulk1, purified from HEK293T cells under fed conditions, were first mapped by Dorsey et al. (18). These authors additionally identified two sites, differentially phosphorylated in wild-type versus kinase-dead Ulk1 (S1043 and S1047). While the one was suggested as direct target for autophosphorylation, the other might represent a putative PKA site (18). Shang et al. (95) quantitatively analyzed the differential phosphorylation status in human Ulk1 purified from HEK293T cells between fed versus starved conditions (HBSS containing 1% rich medium) using stable isotope labeling with amino acids in cell culture (SILAC). A total of 13 sites were identified, with the strongest dephosphorylation at S638 and S758 (corresponding to S637 and S757 in mouse Ulk1) showing a >10-fold decrease after starvation, although with different kinetics. The same decrease was seen after rapamycin treatment and mTOR knockdown (95). Interestingly, phosphorylation at S638 was affected by the knockdown of AMPKα and AMPKβ (95). Shang et al. found that AMPK was associated with Ulk1 under fed conditions and proposed that the dephosphorylation of S758 seen after starvation is critical for the dissociation of AMPK/Ulk1. In contrast, Lee et al. (64) proposed that the interaction between AMPK and Ulk1 is essential for the induction of autophagy and that AMPK activity both recruits 14-3-3 proteins to the complex and leads to inactivation of mTORC1 activity by AMPK-mediated phosphorylation of raptor at S792 (27, 64). Egan et al. (21) additionally identified Ulk1 both as an AMPK substrate and as a 14-3-3 binding protein. This group found S467, S555, T574, and S637 of Ulk1 to be phosphorylated after phenformin treatment and confirmed these sites in an AMPK in vitro kinase assay (21). Notably, Bach et al. (4) could meanwhile confirm the AMPK-dependent phosphorylation at S555 and that this induces the binding to 14-3-3 proteins. This group additionally identified a critical phosphorylation site in the Ulk1 activation loop (T180), as well as a potential Akt phosphorylation site (S774) whose phosphorylation is increased after insulin treatment. The phosphorylation and differential regulation of Ulk1 by mTORC1 and AMPK has also been reported by Kim et al. (55). This group identified S757 in mouse Ulk1 as a direct mTOR site, the same identified by Shang et al. in human Ulk1 (95). However, Kim et al., in contrast, suggest that phosphorylation at S757 prevents the interaction between Ulk1 and AMPK. mTORC1 inhibition would thus lead to an association of AMPK and Ulk1. In line with that, the data suggest an activating effect of AMPK on Ulk1 kinase activity by direct phosphorylation at S317 and S777. These authors found that both sites are required for Ulk1 activation after glucose starvation (55). Notably, neither of these two sites has been identified by one of the other groups. All phosphorylation sites identified by the five groups (4, 18, 21, 55, 95) are shown in single-letter code and refer to mouse Ulk1. Proposed kinases are either indicated by name or otherwise labeled with question marks (?). (B) Dunlop et al. (19) and Jung et al. (48) identified raptor as a direct substrate of Ulk1 and Ulk1 thereby as a negative regulator of either mTORC1 activity (48) or substrate binding (19). After overexpression of Ulk1, Dunlop et al. observed an increase in the in vivo phosphorylation of raptor at S696, T706, S792, S855, S859, and S863 using phospho-specific antibodies and could subsequently confirm the latter four sites as directly phosphorylated by Ulk1 (19). The strongest phosphorylation was seen on S859. Interestingly, S792 is the AMPK and 14-3-3 binding site identified by Gwinn et al. (27), by which AMPK negatively regulates mTORC1 activity, while S863 is known to be phosphorylated by mTOR and to promote mTORC1 activity (23, 106). (C) Löffler et al. (70) identified all three subunits of AMPK as a direct substrate of Ulk1 and Ulk2 and mapped several Ulk1-dependent in vitro phosphorylation sites in AMPKα1, -β2, and -γ1 (residues refer to rat AMPK, for some peptides the phospho-acceptor sites could not be distinguished: S360/T368, S486/T488, and S260/T262). The Ulk1-dependent phosphorylation of AMPK has been proposed to negatively regulate AMPK kinase activity, thus constituting a negative regulatory feedback loop (70). (This figure was adopted and modified with permission from reference 88.)
Furthermore, the existence of a mechanism for the selective elimination of damaged mitochondria (reviewed in reference 116) that can occur even under nutrient-rich conditions generally increases the complexity of autophagy regulation, especially since mitochondria are the major site of ATP and ROS production and the loss of mitochondrial membrane potential is an important trigger of apoptotic cell death pathways (99). Thus, it is conceivable that some in vitro conditions, widely considered as starvation, do not resemble physiological starvation conditions in vivo and instead massively interfere with mitochondrial homeostasis.
In yeast, autophagosomes originate from a single pre-autophagosomal structure. Although an equivalent structure seems to be absent from mammalian cells, a special subdomain in the endoplasmic reticulum (ER) termed the “omegasome” has been suggested as a putative origin of autophagosomes (3, 33, 115). This structure is enriched in PI(3)P, a product of the phosphatidylinositol 3-kinase (PI3K) class III complex. A hierarchical analysis of the mammalian Atg proteins could recently confirm the recruitment of Ulk1 proximal to these omegasomes (45). The translocation of Ulk1, presumably in a complex with Atg13 and FIP200, is the initial step of autophagosome biogenesis and is completely abrogated in FIP200−/− MEFs (45). The subsequent recruitment of the PI3K class III complex depends on Ulk1 and its kinase activity (45, 74).
Interestingly, various subunits of the AMPK complex have been identified as additional components of the Ulk1 kinase network (5), and several studies could meanwhile confirm the direct interaction between AMPK and Ulk1 as well as a positive regulation of Ulk1 activity through AMPK-dependent phosphorylation (21, 55, 64, 95). The AMPK-mediated phosphorylation of Ulk1 and the partially conflicting results have recently been discussed in a detailed commentary by Roach (88) and are summarized in Fig. 3. This further enlarges the range of possibilities for AMPK to induce autophagy, in addition to the more indirect AMPK-mediated regulation of mTORC1 activity via phosphorylation of Raptor and TSC2 (27, 44). Bach et al. could recently integrate some of the above mentioned findings (21, 95) by confirming S555 in Ulk1 as a major AMPK-dependent phosphorylation and 14-3-3 binding site (4). Interestingly, these authors could further identify S774 in Ulk1 as a potential Akt phosphorylation site, whose phosphorylation was enhanced by insulin treatment (4). Thus, Ulk1 (and presumably Ulk2) are direct targets of multiple kinases that might affect Ulk1/2 function in multiple, not mutually exclusive ways. It might induce conformational changes, thereby regulating Ulk1/2 kinase activity, the interaction with other regulatory components, such as 14-3-3 proteins, AMPK, and mTORC1, and/or affect the subcellular localization of the Ulk1/2-Atg13-FIP200 complex.
Although both mTORC1 and AMPK are able to oppositely regulate Ulk1 (and Ulk2) kinase activity by direct phosphorylation, the intriguing question remains how exactly Ulk1 activity is linked to autophagy induction. Two alternative mechanisms have been proposed recently. One report suggests that Ulk1 directly phosphorylates AMBRA1, a Beclin1-interacting protein and regulatory component of the PI3K class III complex (17, 22). Under normal growth conditions, the PI3K complex associates with the dynein motor complex via direct interaction between AMBRA1 and dynein light chain 1 (DLC1). Upon starvation, activated Ulk1 phosphorylates AMBRA1; thereupon, the PI3K complex is released and translocates to the ER, where it initiates autophagosome formation (17). Another group reported that dAtg1 and Ulk1 are able to regulate the actin motor protein myosin II (100). In Drosophila, dAtg1 directly phosphorylates and activates the myosin light-chain kinase (MLCK) Spaghetti-squash activator (Sqa). The depletion of Sqa and its mammalian homolog zipper-interacting protein kinase (ZIPK, also known as DAPK3), which is likewise phosphorylated by Ulk1, attenuated myosin II activation and starvation-induced autophagy (100). Notably, ZIPK knockdown and myosin II inhibition in addition significantly inhibited the redistribution of mAtg9 from the trans-Golgi network (TGN) to a peripheral pool upon starvation (100). The Ulk1- and Atg13-dependent cycling of the transmembrane protein mAtg9 from a juxtanuclear to a dispersed cytosolic pool in response to nutrient deprivation has been reported before (9, 117). This is similar to the situation in yeast, where Atg9 cycles from peripheral structures to the PAS in an Atg1- and Atg17-dependent manner upon autophagy induction (87, 93). Although Atg9 is essential for autophagy induction, its exact function is still unknown. However, it has been suggested that shuttling of mAtg9 from the Golgi apparatus helps to provide membranes for the newly formed autophagosomes (117). These two mechanisms are not mutually exclusive, since Ulk1 might act at several stages of autophagy initiation and regulate both mAtg9 trafficking and recruitment of the PI3K class III complex to the ER in order to initiate autophagosome generation.
mTOR AND ULK1/2: SPLIT PERSONALITIES?
Like any stable system, living cells strictly depend on negative feedback loops to retain the internal control. Although autophagy constitutes a rescue mechanism for starving cells and helps to get rid of superfluous material, prolonged and immoderate consumption of intracellular components necessarily causes severe side effects. Indeed, while mTOR is inactivated at early stages of autophagy induction, long-term starvation leads to reactivation of mTOR activity by enhanced autophagolysosomal generation of nutrients. This in turn results in a slow-down of the autophagic machinery (118). Prolonged inhibition of autophagy on the other hand results in accumulation of protein aggregates and damaged organelles, causing pathological disorders such as neuro- and myodegenerative diseases (67). In Drosophila, chronic activation of dTOR hence increases FoxO transcription factor-mediated sestrin (dSesn) expression. The resulting AMPK-dependent inhibition of TOR in turn helps to prevent pathological long-term TOR hyperactivation (63).
Resembling the ambivalent nature of autophagy, mTORC1 not only inactivates the Ulk1/2-Atg13-FIP200 complex as described above, thereby inhibiting autophagy, but also simultaneously phosphorylates and inactivates the protein DAP1. Astonishingly, DAP1 turned out to be a repressor of autophagy, since its knockdown enhances the autophagic flux (57). During starvation, the mTORC1-dependent sites are rapidly dephosphorylated and DAP1 antagonizes autophagy induction in an as-yet-unknown manner (57). Thus, the current data suggest that mTORC1 inhibition, although activating the autophagic machinery, simultaneously contributes to the prevention of unrestrained autophagic degradation.
In line with that, our group could show that activated Ulk1 directly phosphorylates AMPK and inhibits its activation, thus providing another potential negative-feedback loop on autophagy induction (70).
Several studies could demonstrate that Ulk1 in addition directly interferes with mTORC1 signaling and negatively regulates S6K1 activity, both in Drosophila and mammalian cells (65, 92). While phosphorylation of the dTOR-downstream target S6K was almost completely inhibited after Atg1 overexpression (92), it was strongly enhanced in Atg1-deficient fly mutants (65). Recently, two groups found evidence for the mechanism by which Ulk1 and Ulk2 negatively regulate mTORC1 signaling. It was already known that mTORC1 associates with the Ulk1/2-Atg13-FIP200 complex, via direct interaction between raptor and Ulk1/2, and negatively regulates Ulk1/2 kinase activity by direct phosphorylation (10, 26, 37, 47, 55). Vice versa, the phosphorylation of raptor is strongly enhanced after overexpression of Ulk1 (19, 48), presumably by direct phosphorylation of raptor at numerous sites (19). Interestingly, one of these residues (T792) is the above-mentioned effector site through which AMPK negatively regulates mTORC1 activity (27). The multiple Ulk1-dependent phosphorylation of raptor, which either results in direct inhibition of mTORC1 kinase activity (48) or interferes with raptor-substrate interaction (19), thus finally leads to reduced phosphorylation of mTORC1 downstream targets.
THERAPEUTIC IMPLICATIONS
Recently, autophagy has evolved as one of the central topics in cancer research. However, the role of autophagy during tumorigenesis and tumor progression is far from being completely understood. Interestingly, both the induction and the inhibition of autophagy have been reported to be beneficial for the patient. Whereas the inhibition of autophagy appears to increase the responsiveness of tumor cells toward conventional anticancer drugs, the induction of autophagy may induce cell death in tumor cells with high intrinsic apoptosis resistance. The ambivalent role of autophagy in cancer biology and the emergence of autophagy pathways as novel targets for drug development in anticancer therapy have been extensively reviewed in recent publications (1a, 6, 16, 66, 69, 73, 104).
Regarding the interplay between AMPK, mTOR, and Ulk1/2 described above, these kinases represent an attractive target for therapeutic treatment. To date, especially mTOR has been in the focus of anticancer therapeutic strategies (reviewed in references 20 and 79). However, although mTOR signaling is frequently dysregulated in cancer, rapamycin analogs do not show any considerable activity as single agents in many tumor types and thus are often used in combination therapy (79). This might be partially due to the rapamycin-resistant functions of mTORC1, as mentioned previously (20, 101, 102), and could thus be overcome by inhibitors that directly target the catalytic activity of mTOR (20). However, keeping the complex network of regulation involving cross talks and feedbacks, as well as the ambivalent nature of autophagy in mind, both the activation and the inhibition of these three kinases (kinase complexes) might be desirable, and a better understanding of their interplay during the regulation of autophagy will hopefully allow us to design tumor-specific and tumor stage-specific therapies.
CONCLUSION
The orchestration of the cellular metabolism is not conducted by a single leader; the complex protein network itself precisely senses demand and supply and appropriately responds even to conflicting requirements by self-organization. Effective teamwork, however, depends on frequent and extensive communication within the team. Collectively, the above-described results point out an astonishingly complex interplay between AMPK, Ulk1/2-Atg13-FIP200, and mTORC1, comprising both positive- and negative-feedback regulation. mTORC1 and AMPK are able to oppositely regulate Ulk1/2 kinase activity by direct phosphorylation. On the other hand, Ulk1 can both negatively regulate mTORC1 activity by direct phosphorylation of raptor and negatively regulate AMPK activity by multiple phosphorylation of all three AMPK subunits (Fig. 2 and 3). Although we are just beginning to understand the details of this mutual regulation, the extensive cross talk might help to accurately coordinate cell growth and the increasing demand of energy and nutrients with regard to fluctuations in their external supply and the need of internal production by autophagic degradation.
Fig 2.
Fine adjustment of autophagy by the AMPK-mTORC1-Ulk1/2 kinase network. The two protein complexes AMPK and mTORC1 are known to oppositely regulate the autophagy inducing complex Ulk1/2-Atg13-FIP200. Under sufficient supply of growth factors and nutrients, the active mTORC1 stimulates growth related processes such as protein translation, e.g., by phosphorylation of S6K1 and 4E-BP, while simultaneously inhibiting self-consuming processes such as autophagy (94, 110). mTORC1 activity is positively regulated by growth factor signaling via the PI3K-Akt pathway. Akt activates mTORC1 by inhibition of TSC1/2 (41, 71, 72, 119) or PRAS40 (105), two negative regulators of mTORC1 activity that both antagonize the Rheb-mediated activation. Hypoxia counteracts mTORC1 activation via the TSC1/2-Rheb pathway, e.g., by upregulation of REDD1 (7). Amino acids, in contrast, stimulate the Rag-GTPase-dependent recruitment of mTORC1 to lysosomes and its subsequent activation by Rheb-GTPases (54, 89, 90). The catalytic activity of AMPK crucially depends on phosphorylation by upstream kinases, such as the constitutively active LKB1. AMPK activity is further enhanced by decreasing ATP/AMP ratios (30). In addition, the other two known upstream kinases, CaMKKβ and TAK1, have been implicated in AMPK-mediated autophagy induction by intracellular [Ca2+] and TRAIL treatment, respectively (36, 39). Under low-energy conditions, AMPK positively regulates autophagy induction (77) through inhibition of mTORC1. This releases the negative regulation of mTORC1 on the Ulk1/2-Atg13-FIP200 complex, especially on Ulk1/2 kinase activity (26, 37, 47). AMPK inhibits mTORC1 either via the TSC1/2-Rheb pathway (44) or by direct phosphorylation of raptor (27). However, AMPK is also able to bind, phosphorylate, and directly activate Ulk1/2 (5, 21, 55, 64, 88, 95). Again, this interaction is counteracted by mTORC1 (55). For a more detailed discussion see Roach (88) and Fig. 3. Prolonged TORC1 activation, on the other hand, leads to the accumulation of Sestrin (SESN) in Drosophila, a DNA damage-inducible protein that suppresses TOR activity by AMPK activation (63). Furthermore, mTORC1 not only inhibits autophagy by suppressing Ulk1/2 kinase activity, it also simultaneously inhibits DAP1, a negative regulator of autophagy (57). mTORC1 inhibition thus leads to both autophagy induction via Ulk1/2-Atg13-FIP200 and to its restriction via DAP1. Ulk1 kinase activity might be linked to autophagy induction in several ways. Two downstream targets of Ulk1 have been proposed thus far. First, Ulk1 directly phosphorylates AMBRA1, a Beclin1-interacting protein and regulatory component of the PI3K class III complex (17) and, second, it phosphorylates and activates a distinct myosin light chain kinase (MLCK) in mammals (ZIPK) and Drosophila (Sqa) (100). Two Ulk1-dependent feedback loops additionally help to fine-tune the autophagic response. Ulk1 has been shown to phosphorylate and inhibit both of its upstream regulators AMPK and mTORC1. While phosphorylation of raptor might help to maintain mTORC1 inhibition when nutrients are limited (19, 48), the inhibition of AMPK activity by Ulk1 antagonizes this action and restricts the autophagic response (70). This perplexingly complex network of mutual activation and inhibition will ultimately establish an appropriate response to conflicting demands.
The growth of unicellular organisms such as yeast largely depends on the external availability of oxygen and suitable carbon and nitrogen sources. Autophagy therefore represents an accurate way to rapidly adapt to decreasing external concentrations. The growth and proliferation of cells from multicellular organisms, however, that benefit from a relatively constant supply with nutrients is additionally regulated by hormones and growth factors (Fig. 2). These factors regulate the differential uptake of nutrients by specific cell types, the apoptotic cell death of individual cells, and the cell type-specific fate during development. This impressively reflects the subordination of the individual cell fate to the need of the whole organism. Furthermore, the autophagic machinery, once invented by unicellular eukaryotes, has been adopted for additional purposes during evolution, such as development (76), as well as adaptive and innate immunity (15, 24). Autophagy regulation in metazoans is thus by far more complex and inextricably interwoven with diverse signaling pathways that regulate cell death, cell survival, and tumor suppression. The obvious differences in regulation of autophagy induction between yeast, flies, and vertebrates (Fig. 1) and the existence of several partially redundant isoforms in vertebrates can thus be explained by the need to integrate and consider further variables. Although negative-feedback circuits prevent the system from overriding, redundancy in protein function and signaling pathways might provide a safety mechanism that prevents the system from failure. However, since multicellular organisms comprise hundreds of cell types with specialized function, this redundancy might simply reflect the different routes preferentially taken by different cell types.
ACKNOWLEDGMENTS
We thank Stephan Mumm for carefully reading the manuscript.
This study was supported by grants from the Deutsche Forschungsgemeinschaft SFB 773 and GRK 1302 and from the Interdisciplinary Center of Clinical Research, Faculty of Medicine, Tübingen, Germany (Nachwuchsgruppe 1866-0-0).
Footnotes
Published ahead of print 24 October 2011
REFERENCES
- 1. Alers S, et al. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a. Apel A, Zentgraf H, Buchler MW, Herr I. 2009. Autophagy: a double-edged sword in oncology. Int. J. Cancer 125: 991–995 [DOI] [PubMed] [Google Scholar]
- 2. Arsham AM, Howell JJ, Simon MC. 2003. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 278: 29655–29660 [DOI] [PubMed] [Google Scholar]
- 3. Axe EL, et al. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem. J., in press [DOI] [PubMed] [Google Scholar]
- 5. Behrends C, Sowa ME, Gygi SP, Harper JW. 2010. Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brech A, Ahlquist T, Lothe RA, Stenmark H. 2009. Autophagy in tumour suppression and promotion. Mol. Oncol. 3: 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brugarolas J, et al. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18: 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan EY, Kir S, Tooze SA. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282: 25464–25474 [DOI] [PubMed] [Google Scholar]
- 9. Chan EY, Longatti A, McKnight NC, Tooze SA. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang YY, Neufeld TP. 2009. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang YY, Neufeld TP. 2010. Autophagy takes flight in Drosophila. FEBS Lett. 584: 1342–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheong H, Klionsky DJ. 2008. Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae. Autophagy 4: 724–726 [DOI] [PubMed] [Google Scholar]
- 13. Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. 2011. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. U. S. A. 108: 11121–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheong H, et al. 2005. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16: 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crotzer VL, Blum JS. 2010. Autophagy and adaptive immunity. Immunology 131: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. 2010. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy 6: 322–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Di Bartolomeo S, et al. 2010. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dorsey FC, et al. 2009. Mapping the phosphorylation sites of Ulk1. J. Proteome Res. 8: 5253–5263 [DOI] [PubMed] [Google Scholar]
- 19. Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. 2011. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 7: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Efeyan A, Sabatini DM. 2010. mTOR and cancer: many loops in one pathway. Curr. Opin. Cell Biol. 22: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Egan DF, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fimia GM, et al. 2007. Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- 23. Foster KG, et al. 2010. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J. Biol. Chem. 285: 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galluzzi L, Kepp O, Kroemer G. 2011. Autophagy and innate immunity ally against bacterial invasion. EMBO J. 30: 3213–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gan B, Guan JL. 2008. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 20: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ganley IG, et al. 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284: 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gwinn DM, et al. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hara T, Mizushima N. 2009. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy 5: 85–87 [DOI] [PubMed] [Google Scholar]
- 29. Hara T, et al. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell. Biol. 8: 774–785 [DOI] [PubMed] [Google Scholar]
- 31. Hawley SA, et al. 2003. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hawley SA, et al. 2005. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2: 9–19 [DOI] [PubMed] [Google Scholar]
- 33. Hayashi-Nishino M, et al. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- 34. He C, Klionsky DJ. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heitman J, Movva NR, Hall MN. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- 36. Herrero-Martín G, et al. 2009. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 28: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hosokawa N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hosokawa N, et al. 2009. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979 [DOI] [PubMed] [Google Scholar]
- 39. Høyer-Hansen M, et al. 2007. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell 25: 193–205 [DOI] [PubMed] [Google Scholar]
- 40. Høyer-Hansen M, Jäättelä M. 2007. AMP-activated protein kinase: a universal regulator of autophagy? Autophagy 3: 381–383 [DOI] [PubMed] [Google Scholar]
- 41. Huang J, Manning BD. 2009. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 37: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang KM, Snider MD. 1995. Isolation of protein glycosylation mutants in the fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 6: 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hurley RL, et al. 2005. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280: 29060–29066 [DOI] [PubMed] [Google Scholar]
- 44. Inoki K, Zhu T, Guan KL. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- 45. Itakura E, Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6: 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joo JH, et al. 2011. Hsp90-cdc37 chaperone complex regulates ulk1- and atg13-mediated mitophagy. Mol. Cell 43: 572–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jung CH, et al. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jung CH, Seo M, Otto NM, Kim DH. 2011. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 7: 1212–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kabeya Y, et al. 2005. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16: 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kamada Y, et al. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kamada Y, et al. 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 19: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim DH, et al. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- 54. Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Komatsu M, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koren I, Reem E, Kimchi A. 2010. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20: 1093–1098 [DOI] [PubMed] [Google Scholar]
- 58. Kroemer G, Marino G, Levine B. 2010. Autophagy and the integrated stress response. Mol. Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuma A, et al. 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- 60. Kundu M, et al. 2008. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuroyanagi H, et al. 1998. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics 51: 76–85 [DOI] [PubMed] [Google Scholar]
- 62. Lee EJ, Tournier C. 2011. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 7: 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee JH, et al. 2010. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327: 1223–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee JW, Park S, Takahashi Y, Wang HG. 2010. The association of AMPK with ULK1 regulates autophagy. PLoS One 5: e15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee SB, et al. 2007. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 8: 360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Levine B. 2007. Cell biology: autophagy and cancer. Nature 446: 745–747 [DOI] [PubMed] [Google Scholar]
- 67. Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lippai M, et al. 2008. SNF4Agamma, the Drosophila AMPK gamma subunit is required for regulation of developmental and stress-induced autophagy. Autophagy 4: 476–486 [DOI] [PubMed] [Google Scholar]
- 69. Liu B, Cheng Y, Liu Q, Bao JK, Yang JM. 2010. Autophagic pathways as new targets for cancer drug development. Acta Pharmacol. Sin. 31: 1154–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Löffler AS, et al. 2011. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 7: 696–706 [DOI] [PubMed] [Google Scholar]
- 71. Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15: 702–713 [DOI] [PubMed] [Google Scholar]
- 72. Long X, Ortiz-Vega S, Lin Y, Avruch J. 2005. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J. Biol. Chem. 280: 23433–23436 [DOI] [PubMed] [Google Scholar]
- 73. Mathew R, Karantza-Wadsworth V, White E. 2007. Role of autophagy in cancer. Nat. Rev. Cancer 7: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matsunaga K, et al. 2010. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190: 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. 2007. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3: 106–116 [DOI] [PubMed] [Google Scholar]
- 76. Melendez A, Neufeld TP. 2008. The cell biology of autophagy in metazoans: a developing story. Development 135: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Meley D, et al. 2006. AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 281: 34870–34879 [DOI] [PubMed] [Google Scholar]
- 78. Mercer CA, Kaliappan A, Dennis PB. 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5: 649–662 [DOI] [PubMed] [Google Scholar]
- 79. Meric-Bernstam F, Gonzalez-Angulo AM. 2009. Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 27: 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Momcilovic M, Hong SP, Carlson M. 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 281: 25336–25343 [DOI] [PubMed] [Google Scholar]
- 81. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell. Biol. 10: 458–467 [DOI] [PubMed] [Google Scholar]
- 82. Noda T, Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273: 3963–3966 [DOI] [PubMed] [Google Scholar]
- 83. Oakhill JS, et al. 2011. AMPK is a direct adenylate charge-regulated protein kinase. Science 332: 1433–1435 [DOI] [PubMed] [Google Scholar]
- 84. Ogura K, et al. 1994. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 8: 2389–2400 [DOI] [PubMed] [Google Scholar]
- 85. Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14: 2689–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ptacek J, et al. 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- 87. Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. 2004. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6: 79–90 [DOI] [PubMed] [Google Scholar]
- 88. Roach PJ. 2011. AMPK → ULK1 → autophagy. Mol. Cell. Biol. 31: 3082–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sancak Y, et al. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sancak Y, et al. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sarbassov DD, et al. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- 92. Scott RC, Schuldiner O, Neufeld TP. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7: 167–178 [DOI] [PubMed] [Google Scholar]
- 93. Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. 2009. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells 14: 525–538 [DOI] [PubMed] [Google Scholar]
- 94. Sengupta S, Peterson TR, Sabatini DM. 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shang L, et al. 2011. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl. Acad. Sci. U. S. A. 108: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shang L, Wang X. 2011. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy 7: 924–926 [DOI] [PubMed] [Google Scholar]
- 97. Suzuki K, et al. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Suzuki K, Kubota Y, Sekito T, Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- 99. Tait SW, Green DR. 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell. Biol. 11: 621–632 [DOI] [PubMed] [Google Scholar]
- 100. Tang HW, et al. 2011. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 30: 636–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Thoreen CC, et al. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Thoreen CC, Sabatini DM. 2009. Rapamycin inhibits mTORC1, but not completely. Autophagy 5: 725–726 [DOI] [PubMed] [Google Scholar]
- 103. Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. 1999. A mouse serine/threonine kinase homologous to Caenorhabditis elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24: 833–846 [DOI] [PubMed] [Google Scholar]
- 104. Turcotte S, Giaccia AJ. 2010. Targeting cancer cells through autophagy for anticancer therapy. Curr. Opin. Cell Biol. 22: 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. 2007. Insulin signaling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9: 316–323 [DOI] [PubMed] [Google Scholar]
- 106. Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. 2009. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J. Biol. Chem. 284: 14693–14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang Z, Wilson WA, Fujino MA, Roach PJ. 2001. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 21: 5742–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Woods A, et al. 2005. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2: 21–33 [DOI] [PubMed] [Google Scholar]
- 109. Woods A, et al. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13: 2004–2008 [DOI] [PubMed] [Google Scholar]
- 110. Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- 111. Xiao B, et al. 2011. Structure of mammalian AMPK and its regulation by ADP. Nature 472: 230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yan J, et al. 1998. Identification of mouse ULK1, a novel protein kinase structurally related to Caenorhabditis elegans UNC-51. Biochem. Biophys. Res. Commun. 246: 222–227 [DOI] [PubMed] [Google Scholar]
- 113. Yan J, et al. 1999. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene 18: 5850–5859 [DOI] [PubMed] [Google Scholar]
- 114. Yeh YY, Shah KH, Herman PK. 2011. An Atg13-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- 116. Youle RJ, Narendra DP. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell. Biol. 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Young AR, et al. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119: 3888–3900 [DOI] [PubMed] [Google Scholar]
- 118. Yu L, et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang H, et al. 2003. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Invest. 112: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14: 2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]