Abstract
β-Catenin-independent Wnt signaling pathways have been implicated in the regulation of planar cell polarity (PCP) and convergent extension (CE) cell movements. Prickle, one of the core proteins of these pathways, is known to asymmetrically localize proximally at the adherens junction of Drosophila melanogaster wing cells and to locally accumulate within plasma membrane subdomains in cells undergoing CE movements during vertebrate development. Using mass spectrometry, we have identified the Ste20 kinase Mink1 as a Prickle-associated protein and found that they genetically interact during the establishment of PCP in the Drosophila eye and CE in Xenopus laevis embryos. We show that Mink1 phosphorylates Prickle on a conserved threonine residue and regulates its Rab5-dependent endosomal trafficking, a process required for the localized plasma membrane accumulation and function of Prickle. Mink1 also was found to be important for the clustering of Vangl within plasma membrane puncta. Our results provide a link between Mink and the Vangl-Prickle complex and highlight the importance of Prickle phosphorylation and endosomal trafficking for its function during Wnt-PCP signaling.
INTRODUCTION
Planar cell polarity (PCP), which is orthogonal to the cell apicobasal axis, is important to coordinate cellular orientation at the tissue level (15, 41, 56, 57, 63). The stereotypical orientation of the hairs at the distal tip of Drosophila melanogaster wing cells and the organization of the photoreceptor cells in the fly compound eye are two particularly striking manifestations of PCP that have been exploited as model systems for its study. Genetic screens have identified a core set of proteins that are important for the establishment and maintenance of PCP. These proteins include Frizzled (Fz) (1, 55), Flamingo (Fmi) (53), Van Gogh (Vangl) (50, 61), Dishevelled (Dsh) (39), Diego (16), and Prickle (Pk) (18). The underlying molecular mechanisms of how the core PCP proteins regulate cell polarity are poorly understood. At the cellular level, the asymmetric localization of the PCP core proteins on the apical cortex is thought to be key for PCP establishment (45). In the Drosophila wing, the seven-transmembrane protein Fz along with the cytosolic proteins Dsh and Diego are distally localized, whereas the four-transmembrane protein Vangl and the membrane-associated protein Pk are proximally localized. The asymmetric localization of these core proteins is thought to result from intracellular feedback interactions between proximal Vangl-Pk and distal Fz-Dsh protein complexes. One emerging mechanism that has been proposed to control the asymmetric distribution of PCP proteins involves the polarized control of membrane-trafficking events. The microtubule-dependent polarized transport of Fmi and Fz (42, 46), as well as the regulation of Fz and Fmi endocytosis (32, 46, 47), have been shown to play key roles in their localized accumulation on the cortex. Furthermore, it has been demonstrated recently that the cytoplasmic PCP core proteins Dsh, Pk, and Dgo are required to promote the clustering of asymmetric complexes into puncta producing local domains of asymmetry (47).
In vertebrates, the cochlear epithelium is a striking example in which PCP proteins are found to asymmetrically localize and where they were shown to be important for the correct patterning of the tissue (31, 33). Another vertebrate-specific function for the same core PCP proteins is the control of convergent extension (CE) cell movements during gastrulation (14, 20, 25, 36, 48, 49, 54) that result in the anterior-posterior elongation of the body axis. In that context, localized plasma membrane accumulation of Pk has been observed in the zebrafish neural keel (10), zebrafish dorsal mesoderm (62), and ascidian (26) and mouse neuroectoderm (33).
The core PCP protein Pk is a prenylated protein (30) composed of one Prickle, Espinas, and Testin (PET) domain and three Lin11, Isl-1, and Mec-3 (LIM) domains that have been characterized to be important for its function (49). Loss-of-function studies performed in Drosophila (19), Xenopus laevis (49), and zebrafish (9, 54) have established Pk as a core protein of β-catenin-independent Wnt pathways. In Drosophila, the pk gene encodes three transcripts (19) (pk, sple, and pkM) yielding protein isoforms of 870 (Pk), 936 (PkM), and 1,206 (Sple) amino acids that share the conserved PET and LIM domains. The pkpk and pksple mutants have severe planar polarity defects in distinct adult tissues, and gain of function for the Pk and Sple isoforms in the fly eye leads to typical PCP defects, such as symmetric ommatidia and chirality defects (19, 24)
The molecular mechanism underlying the asymmetrical localization of Pk remains poorly understood. In this study, we have used an unbiased immunoprecipitation and mass spectrometry approach to identify Misshapen-like kinase 1 (Mink1) as a novel protein associating with Pk. Mink1 belongs to the Ste20 group of kinases and is further classified as a member of the germinal center kinase (GCK) family (12). The Mink1 Drosophila homologue Misshapen (Msn) and vertebrate Mink1 have been linked to the PCP pathway in Drosophila (35), CE during Xenopus gastrulation (29), and the control of neural tube closure in zebrafish (28). In flies, Msn was found to act downstream of Fz and Dsh to regulate eye and wing polarity (35). In Xenopus, Mink1 was shown to be required for the plasma membrane recruitment of Dsh during β-catenin-independent Wnt signaling (29). However, precisely how Msn and Mink1 perform their functions is incompletely defined.
Our study uncovers an evolutionary conserved physical and genetic interaction between Mink1 and Pk during PCP in Drosophila and CE in Xenopus embryos. We establish that Mink1 phosphorylates Pk and regulates its localization within puncta, two processes that are intimately linked to control Pk function. Our results also suggest that Mink1 affects the localization of Vangl within Pk-containing puncta, indicating that the regulation of the cytoplasmic PCP components by Mink1 likely is part of the mechanism by which they modulate the clustering of the membrane components.
MATERIALS AND METHODS
Plasmid constructs and reagents.
cDNA for human PRICKLE1 and PRICKLE2 were obtained from J. L. Wrana. The cDNA for Mink1 was obtained from clone MGC:21111. cDNAs for Msn, Pkpk, and PkSple were obtained from D. Gubb and Y. Y. Lin. cDNAs for mouse Tnik and M4k4 were obtained from A. C. Gingras. The expression plasmids coding for the various enhanced green fluorescent protein (EGFP)-tagged Rab proteins were obtained from S. Ferguson. The various cDNAs were cloned into the pIRES-puro-GLUE, pIRES-puro-FLAG, pIRES-puro-CHERRY, or pIRES-puro-VENUS plasmid as needed (6). All PCR-amplified regions were verified by sequencing. Detailed descriptions of the different plasmids and sequences will be provided upon request and are posted on the laboratory website (http://phm.utoronto.ca/angers/). Antibodies purchased were mouse anti-FLAG M2 (Sigma), mouse anti-HA.11 (clone 16B12; Covance), and rat antihemagglutinin (anti-HA) (clone 3F10; Roche).
Tissue culture and transfection.
HEK293T cells were grown in Dulbecco's modified Eagle medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and penicillin-streptomycin (Bioshop) in a 37°C humidified incubator with 5% CO2. Stable cell lines were generated either by calcium phosphate transfection or polyethylenimine (PEI) transfection.
Immunopurification and mass spectrometry.
HEK293T cells (1 × 108 cells) expressing either FLAG-PRICKLE1 or FLAG-PRICKLE2 were used for the affinity purification procedure. Briefly, cells were lysed and solubilized in tandem affinity purification (TAP) lysis buffer (0.1% Igepal CA 630, 10% glycerol, 50 mM HEPES-NaOH, pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM dithiothreitol [DTT], 10 mM NaF, 0.25 mM NaOVO3, 50 mM β-glycerophosphate, and protease inhibitor cocktail [EMD]). After 30 min of centrifugation at 40,000 × g (18,000 rpm in a Beckman JA20 rotor), the soluble fraction was incubated overnight at 4°C with anti-FLAG M2 beads (Sigma). Beads were washed with TAP lysis buffer and then by 50 mM ammonium bicarbonate. Finally, proteins were eluted twice from the beads using 200 μl of 500 mM ammonium hydroxide at pH 11.0. The eluted fraction was evaporated by a speed vacuum, and the remaining pellet was washed with 100 μl of MilliQ water. The pellet was resuspended in 50 mM ammonium bicarbonate, reduced with 25 mM DTT, alkylated with 100 mM iodoacetamide (Sigma), and supplemented with 1 mM CaCl2 (Bioshop) prior to digestion with 1 μg of sequencing-grade trypsin (Promega). The resulting peptide mixture was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an LTQ-XL linear ion trap mass spectrometer (Thermo Scientific). The acquired tandem mass spectra were searched against a FASTA file containing the human NCBI sequences using a normalized implementation of SEQUEST running on the Sorcerer platform. The resulting peptide identifications returned by SEQUEST were filtered and assembled into protein identifications using the Peptide and Protein Prophet suite (ISB, Seattle, WA). To identify specific Pk-associated proteins, we subtracted a background list of contaminating proteins populated using control FLAG immunoprecipitation (IP) from wild-type (WT) lysates and data sets from six irrelevant FLAG-tagged proteins (Table 1).
Table 1.
List of proteins found in PRICKLE1 and PRICKLE2 complexes by LC-MS/MSa
| Gene no. | Gene name | Protein name | Results of LC-MS/MS for: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FLAG-PRICKLE1 |
FLAG-PRICKLE2 |
|||||||||
| No. of unique peptides | Total no. of peptides | % Coverage | No. of pulldowns | No. of unique peptides | Total no. of peptides | % Coverage | No. of pulldowns | |||
| 144165 | PRICKLE1 | 57 | 2,031 | 2 | ||||||
| 166336 | PRICKLE2 | 22 | 649 | 30.2 | 2 | |||||
| 81839 | VANGL1 | Vang-like 1 (van Gogh, Drosophila) | 3 | 3 | 7.6 | 1 | 2 | 2 | 5.3 | 1 |
| 57216 | VANGL2 | Vang-like 2 (van Gogh, Drosophila) | 1 | 1 | 2.3 | 1 | ||||
| 1460 | CSNK2B | Casein kinase 2, beta subunit | 7 | 24 | 32.1 | 1 | 4 | 7 | 40 | 2 |
| 1457 | CSNK2A1 | Casein kinase 2, alpha 1 subunit | 18 | 85 | 46.8 | 1 | 13 | 31 | 34 | 2 |
| 1459 | CSNK2A2 | Casein kinase 2, alpha prime subunit | 8 | 34 | 38.6 | 1 | 9 | 29 | 36.9 | 2 |
| 55914 | ERBB2IP | Erbb2 interacting protein | 15 | 25 | 16.7 | 2 | 17 | 31 | 18.3 | 2 |
| 613 | BCR | Breakpoint cluster region | 59 | 694 | 49.3 | 2 | 38 | 248 | 32.2 | 2 |
| 50488 | MINK1 | Misshapen-like kinase 1 | 17 | 25 | 18.2 | 2 | 6 | 12 | 7.1 | 2 |
| 85461 | TANC1 | Tetratricopeptide repeat, ankyrin repeat, and coiled-coil containing 1 | 27 | 46 | 22.7 | 2 | 3 | 3 | 2.5 | 2 |
| 26115 | TANC2 | Tetratricopeptide repeat, ankyrin repeat, and coiled-coil containing 2 | 18 | 38 | 13.8 | 2 | 12 | 18 | 9.3 | 2 |
| 400745 | SH2D5 | SH2 domain containing 5 | 13 | 44 | 52 | 2 | 6 | 18 | 26.8 | 2 |
| 253260 | RICTOR | RPTOR independent companion of MTOR, complex 2 | 8 | 18 | 9 | 2 | 7 | 14 | 6.3 | 2 |
Peptides from trypsin-digested PRICKLE1- and PRICKLE2-associated protein complexes were analyzed by LC-MS/MS. The peptides and resulting proteins were identified using an integrated version of SEQUEST and analyzed with the Peptide and Protein Prophet software suite (ISB, Seattle, WA). The unique peptides column represents the total number of unique peptides successfully identified. Total peptides corresponds to the total number of peptides matching a given protein identification. Percent coverage is the percentage of the protein sequence covered by the peptide sequences identified. Two independent experiments were performed for both PRICKLE1 and PRICKLE2, and the frequency with which a protein was identified is designated the number of pulldowns.
Affinity purification, immunoprecipitation, and Western blotting.
Forty-eight h posttransfection, cells were lysed with TAP lysis buffer and incubated at 4°C for 1 h to solubilize proteins. Affinity purification and immunoprecipitations were performed using either streptavidin resin (GE Healthcare) or anti-FLAG-M2 beads (Sigma) for 3 h at 4°C. After extensive washes with lysis buffer, proteins were eluted with 2× Laemmli sample buffer and heated at 95°C for 5 min in the presence of β-mercaptoethanol (Sigma). Whole-cell lysates or purified protein samples were resolved by SDS-PAGE and transferred onto Biotrace NT nitrocellulose transfer membranes (Pall Corporation). Western blotting was performed with antibodies as indicated in the figure legends, followed by chemiluminescent detection using appropriate horseradish peroxidase (HRP)-conjugated antibodies and the SuperSignal West Pico (Thermo Scientific) reagent.
Fly strains and genetics.
Fly strains used were yw, Sev-Gal4 (obtained from K. Basler via A. Jenny), UAS-pksple (obtained from A. Jenny), and UAS-msn RNA interference (RNAi) (no. 101517) (obtained from the Vienna Drosophila RNA; Center [VDRC]) (13). pk and msn interaction was tested using the Gal4/UAS system (8) by driving the appropriate combinations of pksple or msn RNAi with Sev-Gal4 at 29°C. Ommatidia were scored for polarity across four eyes of each genotype tested.
Histology.
Tangential sections of adult Drosophila eyes were prepared as previously described (51).
Xenopus injection and explants.
Xenopus embryos were injected at the 4-cell stage into two dorsal blastomeres. For morpholino oligonucleotide (MO)-xPrickle1, 40 ng/blastomere was used, and for MO-xMink1 either 8.7 ng/blastomere (convergent extension experiment) or 13 ng/blastomere (fluorescence microscopy) was used. For Venus-Pk1, 500 pg/blastomere was injected, and 100 pg/blastomere of membrane-tethered RFP was injected as a plasma membrane marker (23). At stage 10+, dorsal marginal zone (DMZ) explants were dissected and cultured in 1× modified Barth's solution until stage 17. Explants were mounted in glass-bottom dishes and then analyzed by confocal microscopy using a 40× oil immersion objective.
The MOs were obtained from Genetools, and the sequences were the following: Mink1, 5′-AGCTGGTGGGTCTGATGCCATGATC-3; Pk1, 5′-CTTCTGATCCATTTCCAAAGGCATG-3′.
Immunocytochemistry and confocal microscopy.
HEK293T cells were grown on poly-d-lysine (Sigma)-treated coverslips and transfected as described in the figure legends. Forty-eight h posttransfection, cells were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS), and the nonspecific sites were saturated with a 3% bovine serum albumin (BSA) (Bioshop) solution in PBS. Samples were immunostained with anti-FLAG M2 (Sigma) or anti-HA clone 3F10 (Roche) antibody in 3% BSA. Cells subsequently were labeled with secondary (Invitrogen) goat anti-mouse or goat anti-rat antibodies conjugated to Alexa Fluor 594 and Alexa Fluor 647, respectively. Coverslips were mounted and sealed onto slides using Vectashield mounting medium (Vector Laboratories). Cells then were visualized and images acquired with a Carl Zeiss LSM510 confocal microscope using a Plan-Apochromat 63× oil immersion objective. Lasers at 488, 543, and 633 nm were triggered independently using the multitrack function of LSM510. Uncompressed images were processed using Zeiss LSM Image browser version 4.2 and ImageJ 1.37a software.
In vitro kinase assay.
MINK1 and MINK1-KD were purified from HEK293T cells stably expressing FLAG-MINK1 or FLAG-MINK1-KD and were eluted from the beads using FLAG peptides. FLAG-MINK or FLAG-MINK-KD was incubated with recombinant glutathione S-transferase (GST)–PRICKLE1 proteins purified from Escherichia coli and immobilized on glutathione beads. Phosphorylation reactions were performed in kinase buffer (25 mM HEPES, pH 7.4, 25 mM β-glycerophosphate, 25 mM MgCL2, 0.1 mM Na3VO4, 0.5 mM DTT) supplemented with 20 μM ATP and 5 μCi of [γ-32P]ATP at 37°C for 1 h. Reactions were stopped by five successive washes with kinase buffer and the addition of 4× Laemmli sample buffer. Proteins were resolved by SDS-PAGE and analyzed by autoradiography.
Mass spectrometry identification of phosphorylation site.
In vitro phosphorylation reactions were performed as described above but with cold ATP. Reactions were stopped by five successive washes with kinase buffer and twice with 50 mM ammonium bicarbonate. Proteins then were digested with trypsin, and the resulting peptide mixture was analyzed by LC-MS/MS. Peptide and protein identifications were performed as described above using a peptide mass tolerance of 3 amu, but search parameters were modified to allow for the presence of one or more phosphate groups on serine, threonine, or tyrosine residues. MS/MS spectra attributed to a phosphorylated peptide in Pk were manually inspected and validated to locate the position of the phosphoamino acid.
Statistical analysis.
Data, reported as ± standard errors of the means (SEM), were analyzed by analysis of variance (ANOVA) followed by Tukey's correction using GraphPad Prism 5 software. Statistical significance was measured at P < 0.05 for all analyses.
RESULTS
Mink1 interacts with Prickle.
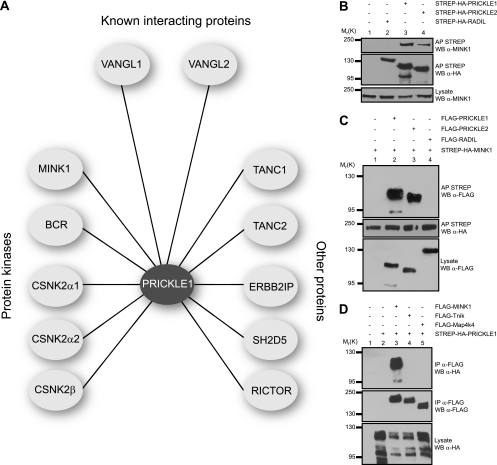
To identify potential regulators of vertebrate Pk proteins, we used a variation of a previously described methodology (2, 4–6) based on LC-MS/MS analysis of FLAG immunoprecipitates from HEK293T cells expressing FLAG-PRICKLE1 and FLAG-PRICKLE2. Several proteins were identified that interact with both PRICKLE1 and PRICKLE2 (Fig. 1A and Table 1), including VANGL1 and VANGL2, which are known to associate with Pk (24). Among the Pk-interacting proteins identified, we focused on the serine/threonine kinase MINK1, since the Drosophila homologue Msn was previously associated with PCP signaling (35) and another study implicated Mink1 in Wnt signaling during vertebrate CE (29). To confirm the interaction, we affinity purified PRICKLE1 or PRICKLE2 and probed for their association with endogenous MINK1 by Western blotting (Fig. 1B). We also confirmed this interaction in reverse by showing that Pk can selectively interact with affinity-purified MINK1 (Fig. 1C). To control for the interaction, we showed that the non-PCP protein RADIL (2) does not interact with MINK1. Mink1 is a member of the Ste20 protein kinase family that also includes Map4k4 (NIK, for Nck-interacting kinase) and Tnik (for Traf2- and Nck-interacting kinase). Since human MINK1 is 65.7% homologous to MAP4K4 and 66% to TNIK, we tested for the selectivity of binding to Pk. While MINK1 was coaffinity purified with PRICKLE1, no interaction could be detected with Map4K4 or Tnik (Fig. 1D). These results showed that the Ste20 kinase MINK1 specifically interacts with Pk.
Fig 1.
MINK1 interacts with PRICKLE. (A) Immunoprecipitation (IP) and MS experiments were performed on HEK293T cells stably expressing FLAG-PRICKLE1 or FLAG-PRICKLE2. The analysis of the FLAG immunoprecipitates for PRICKLE1 and PRICKLE2 using mass spectrometry revealed a protein-protein interaction network composed of known interacting proteins, protein kinases, and other proteins. (B and C) MINK1 interacts with PRICKLE1 and PRICKLE2. HEK293T cells were transfected with the indicated expression plasmids, and the association between PRICKLE and MINK was assessed by coaffinity purifications and Western blotting (WB). (D) PRICKLE selectively interacts with MINK1. HEK293T cells were transfected with expression vectors coding for STREP-HA-PRICKLE1 and FLAG-MINK1 (lane 3), FLAG-Tnik (lane 4), or FLAG-Map4k4 (lane 5). Lysates from these cells were subjected to immunoprecipitation with FLAG antibodies, and the association of PRICKLE1 was assessed by Western blotting using HA antibodies.
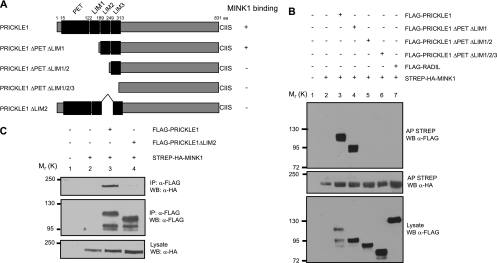
We then mapped the interaction domain of Pk underlying MINK1 binding. Using mutations of human PRICKLE1 that progressively remove the PET and individual LIM domains (Fig. 2A), we identified the second LIM domain as being essential for MINK1 binding (Fig. 2B, compare lanes 3 and 4 to lane 5). We confirmed that the deletion of the LIM2 domain in the context of full-length Pk is sufficient to inhibit Pk binding (Fig. 2C, compare lanes 3 and 4). Taken together, these results suggest that the second LIM domain of Pk is required for MINK1 binding.
Fig 2.
Identification of the PRICKLE1 domain involved in MINK1 binding. (A) Schematic representation of the different PRICKLE1 mutants. (B) FLAG-PRICKLE1 mutant proteins were expressed together with full-length STREP-HA-MINK1 in HEK293T cells. MINK1 was isolated using streptavidin affinity chromatography, and the binding to the PRICKLE mutants was determined using Western blotting with anti-FLAG antibodies. The full-length wild-type PRICKLE1 and the ΔPETΔLIM1 mutant interact with MINK1 (lanes 3 and 4), but the association is lost when the LIM2 domain is missing (lanes 5 and 6). FLAG-RADIL was used as a negative control (lane 7). (C) FLAG-PRICKLE1ΔLIM2 was expressed together with STREP-HA-MINK1 in HEK293T cells. PRICKLE1 was isolated using anti-FLAG immunoprecipitation, and the binding of MINK1 was determined using Western blotting with anti-HA antibodies.
Pk is a substrate for MINK1.
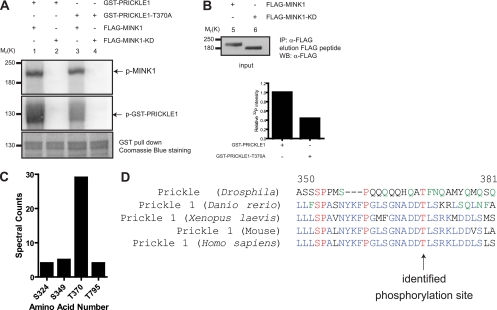
The identification of the serine/threonine kinase protein MINK1 as a Pk-interacting protein raises the possibility that Pk is phosphorylated by MINK1. We performed an in vitro phosphorylation assay using recombinant Pk proteins as the substrate and showed that MINK1, but not a kinase domain mutant of MINK1 (MINK1-KD), phosphorylates Pk (Fig. 3A, compare lanes 1 and 2). We confirmed that MINK-KD still is able to interact with PRICKLE1 by coimmunoprecipitation (data not shown). Using tandem mass spectrometry, we identified four candidate phosphopeptides that are all localized downstream of the Pk LIM domains. Spectral counts from the LC-MS/MS experiments (Fig. 3C) and manual validation of the spectra hinted that the phosphopeptide containing the phosphothreonine at position 370 is the preferred and a bona fide target site for MINK1. Interestingly, this residue is evolutionarily conserved in all species examined (Fig. 3D). We mutated threonine 370 to alanine and confirmed that the phosphorylation of this mutant by MINK1 was reduced by 60% (Fig. 3A, compare lanes 1 and 3, and B). These findings suggest that MINK1 directly phosphorylates Pk on T370.
Fig 3.
PRICKLE1 is a substrate for the serine/threonine kinase MINK1. (A) Recombinant GST-PRICKLE1 and GST-PRICKLE1 (T370A) were used as substrates for in vitro phosphorylation assays using purified MINK1 or MINK1-KD. The top panel (lanes 1 to 4) shows the autophosphorylation of MINK1 bound to recombinant PRICKLE1 and also indicates that equal amounts of kinase were used. In the middle panel, recombinant PRICKLE1 is phosphorylated by MINK1 (lane 1) but not by MINK1-KD (lane 2). The phosphorylation of PRICKLE1 (T370A) is dramatically reduced (lane 3). The bottom panel represents the loading of PRICKLE1 and PRICKLE (T370A) assessed using Coomassie blue staining. The input of purified MINK1 and MINK1-KD is shown in lanes 5 and 6 using Western blotting. (B) Quantification of PRICKLE1 phosphorylation by MINK1. The mutation of threonine 370 to alanine inhibits Pk phosphorylation by 57%. (C) Mass spectrometry analysis of the in vitro kinase reactions revealed four potential phosphorylation sites in PRICKLE1. Spectral counts represent the total number of independent spectra assigned to the indicated phosphopeptides. A total number of 1,819 peptides were surveyed for PRICKLE1 in three independent experiments. (D) Alignments of human PRICKLE1 with the mouse, Xenopus laevis, Danio rerio, and Drosophila homologues show that T370 is evolutionarily conserved.
Mink1 is required for Pk localization.
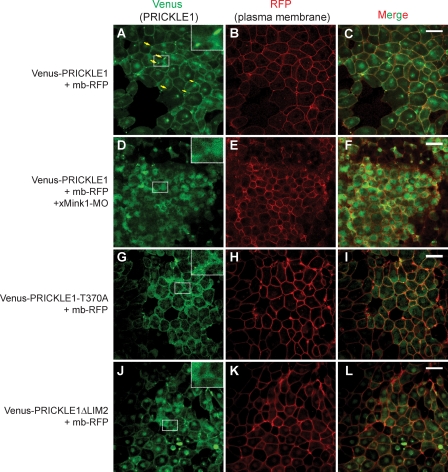
Pk has been shown to be asymmetrically localized in the zebrafish neural keel (10) during gastrulation in the zebrafish dorsal mesoderm (62) and the ascidian notochord (26). Therefore, we studied the cellular localization of Pk in dorsal marginal zone (DMZ) explants from Xenopus embryos injected with Venus-PRICKLE1 mRNA. Similarly to what is observed during zebrafish CE (10, 62), Venus-PRICKLE1 is largely cytoplasmic during early developmental stages and becomes distributed within plasma membrane puncta at the onset of CE (stage 10.5) (Fig. 4A, yellow arrows). Interestingly, the punctum localization of Pk was completely lost in Mink1-depleted embryos, with Pk redistributing to the cytoplasm (Fig. 4D). These results established that Mink1 and Pk functionally interact during Xenopus CE and that Mink1 is required for the punctum localization of Pk during this process. To determine if Pk regulation by Mink1 is important for its cellular localization and function, we asked if the Pk mutants deficient for Mink1 binding or phosphorylation had compromised localization in the DMZ explants undergoing CE. Unlike that of wild-type Pk, the punctum localization of both PRICKLE1-T370A and PRICKLE1ΔLIM2 was robustly inhibited (Fig. 4A to C and G to L). Arguing that the folding of the mutant proteins was not grossly affected, we confirmed that they could still interact with DSH2 and VANGL2 (data not shown). These findings suggest that Pk phosphorylation by Mink1 is required for its localization into plasma membrane puncta.
Fig 4.
Mink1 is required for the localized accumulation of Pk in the plasma membrane. Venus-PRICKLE1 mRNA (500 pg) and membrane-bound RFP (mb-RFP) mRNA (100 pg) were injected in dorsal blastomeres of 4-cell Xenopus embryos. The cellular localization of Pk was monitored in dorsal explants at stage 11 using a confocal microscope under a 40× oil immersion objective. The plasma membrane is highlighted using membrane-bound RFP (mb-RFP). (A to C) During convergence and extension, PRICKLE1 localizes at the membrane and accumulates in plasma membrane subdomains (yellow arrows). (D to F) Downregulation of xMink1 using 13 ng of MO-xMink1 leads to an absence of PRICKLE1 puncta. PRICKLE1-T370A (G to I) or PRICKLE1ΔLIM2 (J to L) does not localize efficiently to puncta. Images shown are representative of two (PRICKLEΔLIM2) or four (PRICKLE1, xMink1-MO, and PRICKLE1-T370A) independent experiments. Scale bars represent 50 μm.
Mink1 is required for Prickle function during CE.
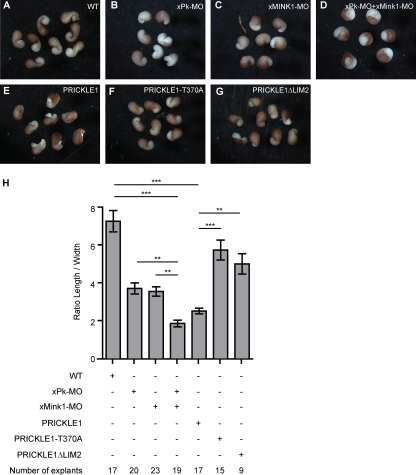
Pk is required for CE in zebrafish and Xenopus (9, 49, 54). Similarly, interfering with Mink1 function also affects CE during Xenopus gastrulation (29). To test if Mink1 is important for the previously characterized function of Pk during CE, we carried out a genetic interaction experiment using DMZ explants of Xenopus embryos that undergo CE movements. We first identified subthreshold doses of individual Mink1 and Pk morpholino oligonucleotides (MOs) that yielded moderate CE defects in this assay when injected separately into two dorsal blastomeres of 4-cell embryos (Fig. 5B and C). When coinjected, the same doses of MOs strongly inhibited the narrowing and elongation of the explants (Fig. 5D). Explants derived from embryos injected with the same total amount of control MO were indistinguishable from noninjected animals (data not shown). Since CE is characterized by the mediolateral narrowing and anterior-posterior elongation of the body axes, we quantified the morphant phenotypes as a ratio of the width to the length of the explants (Fig. 5H).
Fig 5.
Mink1 and Prickle functionally interact during Xenopus CE. (B to D) xPrickle1-MO (40 ng) and xMink1-MO (8.7 ng) were injected separately or together in dorsal blastomeres of 4-cell embryos. (E to G) Five hundred ng of mRNA expressing PRICKLE1, PRICKLE1-T370A, or PRICKLE1ΔLIM2 was independently injected in dorsal blastomeres of 4-cell Xenopus embryos. (H) DMZ explants were dissected at stage 10 and cultured until stage 17. The quantification was performed by determining the ratio of the length to the width of the explants. The downregulation of Pk or Mink1 leads to CE defects. The coinjection of both MOs exacerbated the effect obtained with either MO injected alone. The injection of PRICKLE1 mRNA leads to strong CE defects, whereas equivalent doses of PRICKLE1ΔLIM2 or PRICKLE1-T370A had milder defects. Error bars indicate ±SEM. Statistical analysis was performed using one-way ANOVA followed by Tukey postanalysis (**, P < 0.01; ***, P < 0.001).
To directly test if the punctum localization of Pk at the plasma membrane is required for the function of Pk and is important for CE movements, we used the overexpression phenotype of Pk to assess its activity. Indeed, the loss and gain of function for most PCP proteins, including Pk (24, 49), leads to similar defects. Unlike that of WT Pk (Fig. 5E), the expression of the PRICKLE1-T370A (Fig. 5F) and PRICKLE1ΔLIM2 (Fig. 5G) mutants minimally affected the elongation of the explants (Fig. 5H). Taken together, these results led us to conclude that the phosphorylation of Pk on T370 by Mink1 is a key event regulating Pk localization within plasma membrane subdomains, and that this is critical for its activity during β-catenin-independent Wnt signaling.
Mink is sufficient to localize Pk into plasma membrane puncta.
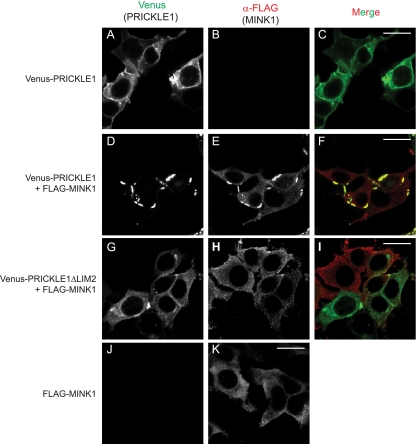
The requirement of Mink1 for the localization of Pk in Xenopus DMZ explants led us to examine more closely the effect of Mink1 gain of function on Pk localization in cultured cells. When transfected, Venus-PRICKLE1 is uniformly localized in the cytoplasm and the plasma membrane (Fig. 6A). Interestingly, the overexpression of MINK1 led to a robust localized accumulation of Pk within plasma membrane patches (Fig. 6D to F), which is reminiscent of its punctum localization observed during convergent extension in Xenopus embryos (Fig. 4A). When expressed alone, MINK1 is uniformly distributed in the cytoplasm (Fig. 6K) but colocalizes with Pk in plasma membrane patches when Pk is coexpressed (Fig. 6D to F). We next confirmed that the Pk mutant PRICKLE1ΔLIM2, which lacks the interaction domain for Mink1, is unable to be redistributed into localized plasma membrane puncta upon MINK1 overexpression (Fig. 6H and I).
Fig 6.
Subcellular localization of PRICKLE1 is controlled by MINK1. Shown are HEK293T cells expressing Venus-PRICKLE1 alone (A to C) or coexpressed together with FLAG-MINK1 (D to F). (A to C) PRICKLE1 is expressed throughout the cytosol and at the plasma membrane. (D to F) In the presence of overexpressed MINK1, PRICKLE1 accumulates in localized plasma membrane patches. (G to I) The coexpression of PRICKLE1ΔLIM2 with MINK1 leads to no change in PRICKLE distribution. (K) When expressed alone, wild-type MINK1 localizes in the cytoplasm. Images shown are representative of cells analyzed in three independent experiments. Scale bars represent 20 μm.
Mink regulates the punctum localization of the Vangl-Pk complex.
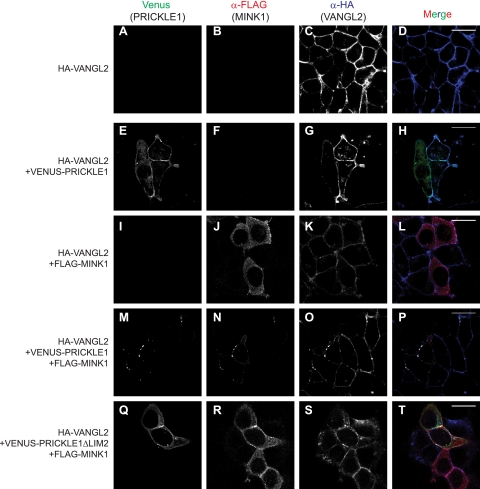
Vangl and Pk are known to interact physically and to be mutually dependent for their colocalization in plasma membrane puncta, a process requiring the PET and LIM domains of Pk (24), which we show is the Mink1 interacting region. Furthermore, the cytoplasmic components of PCP (Dsh and Pk) were recently demonstrated to be required for the clustering of PCP complexes in membrane subdomains (47). Based on these data, we hypothesized that Mink1, through Pk phosphorylation, is an important factor for the subcellular localization of the Vangl-Pk complex during PCP. To test this, we expressed Pk and Mink1 separately or together in a cell line stably expressing HA-VANGL2 and monitored the subcellular localization of each protein. In this context, the expression of Mink1 alone had minimal influence on the subcellular localization of VANGL2 (Fig. 7I to L), but the coexpression of both Pk and Mink1 led to the robust relocalization of all three proteins in plasma membrane puncta (Fig. 7M to P). As a control for this experiment, we coexpressed MINK1 and PRICKLE1ΔLIM2 and showed that the localization of Pk and Vangl remained unchanged (Fig. 7Q to T). We conclude that Mink1 promotes the distribution of the Vangl-Pk complex into plasma membrane puncta.
Fig 7.
MINK1 regulates the localization of the Pk-VANGL2 complex. HEK293T cells stably expressing HA-VANGL2 (A to D) were transfected with plasmids coding for Venus-PRICKLE1 (E to H), FLAG-MINK1 (I to L), or with both together (M to P). (A to D) VANGL2 is expressed at the plasma membrane. Coexpression with either Pk (E to H) or MINK1 (I to L) alone does not influence its cellular localization. (M to P) The coexpression of Pk and MINK1 clusters VANGL2 in plasma membrane puncta. (Q to T) As a control, the coexpression of MINK1 with the PRICKLE1ΔLIM2 mutant (which does not associate with MINK1) does not influence VANGL2 subcellular localization. Images shown are representative of cells analyzed in three independent experiments. Scale bars represent 20 μm.
Endosomal trafficking is required for the punctum localization of Pk.
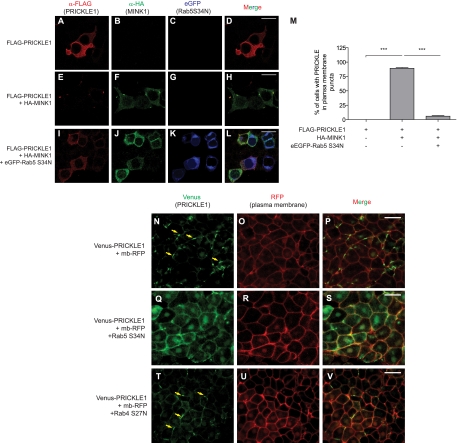
Rab5 specifically and endosomal trafficking in general have previously been linked to the establishment of PCP in Drosophila (11, 21, 32, 42, 46) and to the regulation of Wnt11- and E-cadherin-dependent cell cohesion during zebrafish gastrulation (52). Rab23 also has been found to interact with Pk and to control, albeit modestly, the cortical localization of PCP proteins (38). We hypothesized that endocytic trafficking was important for the accumulation of Pk within plasma membrane puncta, and that MINK1 could regulate that process. To test this, we asked if interfering with endocytic trafficking using a dominant-negative Rab5 mutant protein could inhibit the MINK1-induced redistribution of Pk. MINK1 overexpression led to the accumulation of Pk into membrane puncta in 89% ± 5% of the cells that were examined (Fig. 8E to H and M). In contrast, the coexpression of the Rab5-S34N dominant-negative mutant efficiently rescued this localization to levels nearing the expression of Pk alone (6% ± 4%) (Fig. 8I to M).
Fig 8.
Rab5 is involved in PRICKLE1 trafficking. FLAG-PRICKLE1 was expressed alone (A to D), in the presence of HA-MINK1 (E to H), or in the presence of HA-MINK1 and a dominant-negative version of Rab5, eGFP-Rab5 (S34N) (I to L). Expressed alone, PRICKLE1 is localized in the cytoplasm and evenly at the plasma membrane (A); when MINK1 is expressed, PRICKLE1 is redistributed within plasma membrane patches (E), whereas this membrane accumulation is inhibited by the overexpression of a dominant-negative mutant of Rab5 (I). Scale bars represent 20 μm for panels A to L. (M) The quantification of PRICKLE1-containing plasma membrane patches in cells expressing PRICKLE1 alone, PRICKLE1, and MINK1 or within cells coexpressing PRICKLE1, MINK1, and Rab5 (S34N). Error bars indicate ±SEM. Statistical analysis was performed using one-way ANOVA followed by Tukey postanalysis (***, P < 0.001). In the absence of MINK1, PRICKLE1 was never present in plasma membrane patches (n = 83). The overexpression of MINK1 led to 89% ± 5% (n = 244) of the cells displaying punctum accumulation of PRICKLE1. This patchy localization of PRICKLE1 induced by MINK1 was inhibited by the overexpression of the dominant-negative mutant of Rab5, with 6% ± 4% (n = 196) harboring these structures. The images and the quantification shown are representative and tallied from three independent experiments. The punctum localization of Pk during Xenopus CE requires Rab5. Venus-PRICKLE1 mRNA (500 pg) and membrane-bound RFP (mb-RFP) mRNA (100 pg) were injected alone (N to P) or together with 1 ng of dominant-negative Rab5 (S34N) (Q to S) or Rab4 (S27N) (T to V) in dorsal blastomeres of 4-cell Xenopus embryos. Pk cellular localization was monitored in DMZ explants at stage 11. (T to V) In the presence of dominant-negative Rab4 (S27N), PRICKLE1 localization is not affected. Yellow arrows indicate plasma membrane puncta containing Venus-Pk. (Q to S) The coexpression of Rab5 (S34N) inhibits the localized accumulation of PRICKLE1 at the plasma membrane. Scale bars represent 50 μm in panels N to V.
Since Mink1 is required for the localized plasma membrane accumulation of Pk at the onset of gastrulation in Xenopus embryos (Fig. 4A), we tested if Rab5-dependent membrane traffic also was implicated for the localization of Pk in this physiological context. We expressed dominant-negative Rab5-S34N in Xenopus embryos and found that it largely prevented the plasma membrane punctum localization of Pk (Fig. 8N and Q). As a control, we expressed dominant-negative Rab4-S27N and showed that Pk localization was unaffected (Fig. 8N and T), suggesting that recycling through either the fast Rab4 or slower Rab11 recycling pathway is sufficient, which is similar to what has been reported for Fz and Fmi (46, 47). We conclude that Pk employs a Rab5-dependent trafficking pathway for its localized accumulation within plasma membrane puncta, and that MINK1 positively regulates this process.
Msn and Pk physically and genetically interact during PCP establishment in Drosophila.
Much of our understanding of PCP comes from genetic studies of Drosophila. In the compound eye, PCP is reflected in the mirror-symmetric arrangement of ommatidia across the dorsoventral midline. PCP defects are easily detected by the loss of mirror-image symmetry, with ommatidia failing to rotate and the photoreceptors adopting the wrong chirality or remaining symmetric (15, 43, 56). Given our vertebrate interaction data, we tested if the Pk-Msn association was evolutionarily conserved and began to assess its functional relevance during Drosophila PCP. Using coimmunoprecipitation, we first established that Msn could physically interact with two Pk Drosophila gene products, Prickle (Pk) and Spiny-legs (Sple) (19) (Fig. 9A). We next tested for a functional interaction between msn and pksple. As previously described (24), the overexpression of the pksple isoform in Drosophila ommatidia using the sev enhancer (sev>pksple) led to typical but mild PCP defects, i.e., symmetric ommatidia as well as chirality and rotation defects (Fig. 9B, F, and J). To test for genetic interaction, we crossed sev>pksple to flies expressing msn RNAi under the expression of the sev enhancer. To enable the detection of genetic interaction, we used an msn RNAi that had little or no polarity defect (Fig. 9C, G, and J) rather than msn mutant animals that show strong polarity defects (35). While the knockdown of msn alone did not cause any appreciable PCP phenotypes, driving msn RNAi in pksple-overexpressing ommatidia strongly enhanced the severity of the sev>pksple phenotype, with 14.3% (49/342) of ommatidia showing a symmetric arrangement compared to 3.8% (19/497) when pksple was expressed alone (Fig. 9D, H, and J). We conclude that the Pk-Mink1 protein interaction is evolutionarily conserved, and that these proteins act in concert during PCP establishment in Drosophila.
Fig 9.
Drosophila Pk and Msn physically associate and genetically interact during PCP establishment in Drosophila ommatidia. (A) Association between the Mink1 Drosophila homologue Msn and the fly Prickle variants Pk and Sple. Epitope-tagged fly proteins were expressed in HEK293T cells, and their interactions were determined by coimmunoprecipitation. FLAG-Msn coprecipitates with HA-Pk (lane 3) and HA-Sple (lane 4) but not the control protein HA-RADIL (lane 5). (B to I) Tangential sections of adult eyes expressing sev-Gal4 and UAS-pksple (B), UAS-msn RNAi (C), both pksple and msn RNAi (D), or pksple and a control RNAi (E). Ommatidia are arranged in a mirror-symmetric pattern with respect to the midline. The dorsal and ventral polarity of ommatidia are diagrammed in the bottom panels (F to I) with green and red arrows, respectively. Yellow arrows represent achiral/symmetrical ommatidum. (J) Quantification of ommatidia exhibiting polarity defects (n = 1,332 from three independent experiments). Error bars indicate ±SEM. Statistical analysis was performed using one-way ANOVA followed by Tukey postanalysis (*, P < 0.05; **, P < 0.01). Overexpressing pksple while knocking down msn leads to a higher incidence of symmetric ommatidia.
DISCUSSION
The tissue-wide asymmetric localization of the core PCP proteins is well established in Drosophila and vertebrates. Vertebrate Pk is known to localize asymmetrically in cells undergoing convergence and extension in the mouse (33), ascidian (26), and zebrafish (10, 62). In this study, we similarly noted that Venus-Pk accumulates within discrete plasma membrane regions during Xenopus gastrulation and discovered that this localization depends on its phosphorylation by Mink1. In the context of Xenopus DMZ explants, however, we could not reliably determine if Pk localization was anteriorly biased, as has been observed in zebrafish and mouse tissues.
A previous study described that Pk and Vangl both are required for their distribution into patches at the cell membrane in Xenopus animal caps (24). In this study, the C terminus of Pk was showed to be sufficient for its recruitment at the plasma membrane by Vangl2, but the PET and LIM domains of Pk, which are located in the N-terminal portion of the protein, were found to be required for their redistribution and clustering in puncta. Our results provide a potential mechanism for this observation, since Mink1 interacts with the LIM2 domain of Pk, and we found that this interaction and the phosphorylation of Pk by Mink1 are required for the punctum localization. Evidence supporting a causal link between the fine regulation of localized membrane accumulation, phosphorylation, and activity exist for other core PCP proteins, such as Dsh during Drosophila PCP (7) and CE in Xenopus (37, 40) and for Vangl2 in mouse limb morphogenesis (17). In addition, the tyrosine phosphorylation of Dsh by Abl kinase was reported to be required for its recruitment at the plasma membrane and activity during PCP establishment in flies (44). Furthermore, whether the asymmetric localization of PCP proteins is instructive or simply a consequence of polarity establishment still remains elusive. In the case of Pk, we now identify two mutants that perturb either the interaction with Mink or its phosphorylation, and we showed that their localization and activity are impaired. This suggests a direct relationship between the phosphorylation, the punctum localization, and the function of Pk. Determining the roles of protein phosphorylation for the control of localized membrane accumulation of proteins implicated in β-catenin-independent Wnt signaling and the identification and regulation of the various kinases implicated will provide additional insights into these processes.
Our results also uncover a role for endocytic trafficking for the punctum accumulation of Pk. Although well established for the coordination of apicobasal polarity (58), intracellular trafficking is emerging as an important process during PCP. For instance, Frizzled and Fmi have been observed in rapidly moving intracellular vesicles undergoing polarized transport on microtubules, and the functional inhibition of endosomal trafficking was shown to alter their asymmetrical localization in flies (42, 46). Interestingly, the regulation of Fmi traffic into clathrin-coated vesicles by the membrane protein Echinoid was shown to be essential for ommatidial PCP (22). Adding to this mechanism, Rab5 was implicated for the endocytic trafficking and apical localization of Fmi and the establishment of PCP in the Drosophila wing (32), as well as for the localization of E-cadherin during zebrafish gastrulation (52). Another Rab protein, Rab23, was identified in a screen for genes affecting PCP in the fly wing (38). Recent findings obtained in Drosophila have further established that endocytic trafficking contributes to stably localizing Fmi to junctions, and that the clustering of PCP complexes into puncta depends on the recruitment of the cytoplasmic components Dsh and Pk (47). Whether endocytic trafficking is solely implicated for the accumulation of Fmi at junctions or also is required for the dynamic regulation of PCP complex localization within puncta has not been addressed. Indeed, changes in the localization of PCP proteins that are accompanied by reduced punctum size and the loss of cellular asymmetry (3, 11) occur during junctional remodeling and tissue morphogenesis. Our data demonstrate that Rab5 is required for the Mink1-promoted localization of Pk within puncta. Since Pk is anchored at the plasma membrane through prenylation, it may be undergoing endocytic trafficking on its own. However, we think that this is unlikely, since a prenylation mutant of Pk is able to rescue the pknull eye phenotype in Drosophila (24). Given that Vangl and Pk are mutually required for their localization in puncta (24), another possibility is that Pk is cotrafficked with the transmembrane Vangl protein. One model that is consistent with our findings is that the cotrafficking and clustering of the Vangl-Pk complex into plasma membrane puncta is modulated by the Mink1-dependent phosphorylation of Pk. In support of this model, the coexpression of PRICKLE1 with Mink1 was found to be sufficient to cluster Vangl and Pk into puncta, whereas the expression of the PRICKLE1ΔLIM2 mutant, which does not interact with Mink1, had no effect on the localization of Pk or Vangl (Fig. 7).
Some questions remain regarding the precise mechanisms directing Pk localization. How local Wnt signaling and other extrinsic factors, such as cell-cell and cell-matrix interactions, are integrated and translated into the regulation of Mink1 activity and therefore into Pk punctum localization remains to be understood. Indeed, β-catenin-independent signaling can affect morphogenesis through the modulation not only of cell polarity but also of cell adhesion. In zebrafish, the interaction of Wnt11, Fz7, Dsh, and Fmi increases the persistence of cell-cell contacts in the embryo (60), and E-cadherin-dependent cell adhesion is increased by Pk (34). Interfering with such a function could affect CE (27) in the absence of polarized protein localization.
Finally, whether membrane trafficking plays a ubiquitous role for the asymmetric localization of proteins during all contexts of β-catenin-independent Wnt signaling remains to be examined. Recently, a study reported that cancer cells respond cell autonomously to a noncanonical Wnt5A signal to asymmetrically polarize an intracellular macrostructure named W-RAMP, which contains adhesion receptors and cytoskeletal proteins (59). Interestingly, the polarization of W-RAMP in response to the Wnt cues was shown to involve endosomal trafficking. In summary, our study provides biochemical and functional evidence linking the phosphorylation of Pk by Mink1 and its endocytic trafficking in plasma membrane subdomains, a process that appears to be integral to its function. A further biochemical understanding of the function and regulation of the proteins implicated in β-catenin-independent Wnt signaling will continue to shed light on the wealth of genetic data that functionally implicated these proteins in these pathways.
ACKNOWLEDGMENTS
We are grateful to Anne-Claude Gingras and Brian Raught for help with the phosphoproteomic experiments and to members of the Angers laboratory for discussions and feedback on the manuscript.
S.A. holds the Canada Research Chair in Functional Architecture of Signal Transduction. This work was supported by grants from the Canadian Institute for Health Research (CIHR 171619) to S.A. and from the Canadian Cancer Society Ontario Division (019355) to R.W.
Footnotes
Published ahead of print 28 October 2011
REFERENCES
- 1. Adler PN, Vinson C, Park WJ, Conover S, Klein L. 1990. Molecular structure of frizzled, a Drosophila tissue polarity gene. Genetics 126:401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed SM, Daulat AM, Meunier A, Angers S. 2010. G protein betagamma subunits regulate cell adhesion through Rap1a and its effector Radil. J. Biol. Chem. 285:6538–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aigouy B, et al. 2010. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142:773–786 [DOI] [PubMed] [Google Scholar]
- 4. Angers S. 2008. Proteomic analyses of protein complexes in the Wnt pathway. Methods Mol. Biol. 468:223–230 [DOI] [PubMed] [Google Scholar]
- 5. Angers S, et al. 2006. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443:590–593 [DOI] [PubMed] [Google Scholar]
- 6. Angers S, et al. 2006. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 8:348–357 [DOI] [PubMed] [Google Scholar]
- 7. Axelrod JD. 2001. Unipolar membrane association of Dishevelled mediates Frizzled planar cell polarity signaling. Genes Dev. 15:1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415 [DOI] [PubMed] [Google Scholar]
- 9. Carreira-Barbosa F, et al. 2003. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development 130:4037–4046 [DOI] [PubMed] [Google Scholar]
- 10. Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. 2006. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Classen AK, Anderson KI, Marois E, Eaton S. 2005. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev. Cell 9:805–817 [DOI] [PubMed] [Google Scholar]
- 12. Dan I, Watanabe NM, Kusumi A. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220–230 [DOI] [PubMed] [Google Scholar]
- 13. Dietzl G, et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156 [DOI] [PubMed] [Google Scholar]
- 14. Djiane A, Riou J, Umbhauer M, Boucaut J, Shi D. 2000. Role of frizzled 7 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development 127:3091–3100 [DOI] [PubMed] [Google Scholar]
- 15. Fanto M, McNeill H. 2004. Planar polarity from flies to vertebrates. J. Cell Sci. 117:527–533 [DOI] [PubMed] [Google Scholar]
- 16. Feiguin F, Hannus M, Mlodzik M, Eaton S. 2001. The ankyrin repeat protein Diego mediates Frizzled-dependent planar polarization. Dev. Cell 1:93–101 [DOI] [PubMed] [Google Scholar]
- 17. Gao B, et al. 2011. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev. Cell 20:163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gubb D, Garcia-Bellido A. 1982. A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. J. Embryol. Exp. Morphol. 68:37–57 [PubMed] [Google Scholar]
- 19. Gubb D, et al. 1999. The balance between isoforms of the prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes Dev. 13:2315–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heisenberg CP, et al. 2000. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 405:76–81 [DOI] [PubMed] [Google Scholar]
- 21. Hermle T, Saltukoglu D, Grunewald J, Walz G, Simons M. 2010. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr. Biol. 20:1269–1276 [DOI] [PubMed] [Google Scholar]
- 22. Ho YH, et al. 2010. Echinoid regulates Flamingo endocytosis to control ommatidial rotation in the Drosophila eye. Development 137:745–754 [DOI] [PubMed] [Google Scholar]
- 23. Iioka H, Ueno N, Kinoshita N. 2004. Essential role of MARCKS in cortical actin dynamics during gastrulation movements. J. Cell Biol. 164:169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jenny A, Darken RS, Wilson PA, Mlodzik M. 2003. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 22:4409–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jessen JR, et al. 2002. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 4:610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang D, Munro EM, Smith WC. 2005. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr. Biol. 15:79–85 [DOI] [PubMed] [Google Scholar]
- 27. Kim SK, et al. 2010. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science 329:1337–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Köppen M, Fernandez BG, Carvalho L, Jacinto A, Heisenberg CP. 2006. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development 133:2671–2681 [DOI] [PubMed] [Google Scholar]
- 29. Lee RH, et al. 2007. XRab40 and XCullin5 form a ubiquitin ligase complex essential for the noncanonical Wnt pathway. EMBO J. 26:3592–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maurer-Stroh S, et al. 2007. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 3:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montcouquiol M, et al. 2006. Asymmetric localization of Vangl2 and Fz3 indicate novel mechanisms for planar cell polarity in mammals. J. Neurosci. 26:5265–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mottola G, Classen AK, Gonzalez-Gaitan M, Eaton S, Zerial M. 2010. A novel function for the Rab5 effector Rabenosyn-5 in planar cell polarity. Development 137:2353–2364 [DOI] [PubMed] [Google Scholar]
- 33. Narimatsu M, et al. 2009. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 137:295–307 [DOI] [PubMed] [Google Scholar]
- 34. Oteiza P, et al. 2010. Planar cell polarity signalling regulates cell adhesion properties in progenitors of the zebrafish laterality organ. Development 137:3459–3468 [DOI] [PubMed] [Google Scholar]
- 35. Paricio N, Feiguin F, Boutros M, Eaton S, Mlodzik M. 1999. The Drosophila STE20-like kinase misshapen is required downstream of the Frizzled receptor in planar polarity signaling. EMBO J. 18:4669–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park M, Moon RT. 2002. The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat. Cell Biol. 4:20–25 [DOI] [PubMed] [Google Scholar]
- 37. Park TJ, Gray RS, Sato A, Habas R, Wallingford JB. 2005. Subcellular localization and signaling properties of dishevelled in developing vertebrate embryos. Curr. Biol. 15:1039–1044 [DOI] [PubMed] [Google Scholar]
- 38. Pataki C, et al. 2010. Drosophila Rab23 is involved in the regulation of the number and planar polarization of the adult cuticular hairs. Genetics 184:1051–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perrimon N, Mahowald AP. 1987. Multiple functions of segment polarity genes in Drosophila. Dev. Biol. 119:587–600 [DOI] [PubMed] [Google Scholar]
- 40. Rothbächer U, et al. 2000. Dishevelled phosphorylation, subcellular localization and multimerization regulate its role in early embryogenesis. EMBO J. 19:1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seifert JR, Mlodzik M. 2007. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat. Rev. Genet. 8:126–138 [DOI] [PubMed] [Google Scholar]
- 42. Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T. 2006. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev. Cell 10:209–222 [DOI] [PubMed] [Google Scholar]
- 43. Simons M, Mlodzik M. 2008. Planar cell polarity signaling: from fly development to human disease. Annu. Rev. Genet. 42:517–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh J, Yanfeng WA, Grumolato L, Aaronson SA, Mlodzik M. 2010. Abelson family kinases regulate Frizzled planar cell polarity signaling via Dsh phosphorylation. Genes Dev. 24:2157–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strutt DI. 2002. The asymmetric subcellular localisation of components of the planar polarity pathway. Semin. Cell Dev. Biol. 13:225–231 [DOI] [PubMed] [Google Scholar]
- 46. Strutt H, Strutt D. 2008. Differential stability of flamingo protein complexes underlies the establishment of planar polarity. Curr. Biol. 18:1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strutt H, Warrington SJ, Strutt D. 2011. Dynamics of core planar polarity protein turnover and stable assembly into discrete membrane subdomains. Dev. Cell 20:511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tada M, Concha ML, Heisenberg CP. 2002. Non-canonical Wnt signalling and regulation of gastrulation movements. Semin. Cell Dev. Biol. 13:251–260 [DOI] [PubMed] [Google Scholar]
- 49. Takeuchi M, et al. 2003. The prickle-related gene in vertebrates is essential for gastrulation cell movements. Curr. Biol. 13:674–679 [DOI] [PubMed] [Google Scholar]
- 50. Taylor J, Abramova N, Charlton J, Adler PN. 1998. Van Gogh: a new Drosophila tissue polarity gene. Genetics 150:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tomlinson A, Ready DF. 1987. Cell fate in the Drosophila ommatidium. Dev. Biol. 123:264–275 [DOI] [PubMed] [Google Scholar]
- 52. Ulrich F, et al. 2005. Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev. Cell 9:555–564 [DOI] [PubMed] [Google Scholar]
- 53. Usui T, et al. 1999. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98:585–595 [DOI] [PubMed] [Google Scholar]
- 54. Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13:680–685 [DOI] [PubMed] [Google Scholar]
- 55. Vinson CR, Conover S, Adler PN. 1989. A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature 338:263–264 [DOI] [PubMed] [Google Scholar]
- 56. Vladar EK, Antic D, Axelrod JD. 2009. Planar cell polarity signaling: the developing cell's compass. Cold Spring Harb. Perspect. Biol. 1:a002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Y, Nathans J. 2007. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development 134:647–658 [DOI] [PubMed] [Google Scholar]
- 58. Weisz OA, Rodriguez-Boulan E. 2009. Apical trafficking in epithelial cells: signals, clusters and motors. J. Cell Sci. 122:4253–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Witze ES, Litman ES, Argast GM, Moon RT, Ahn NG. 2008. Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science 320:365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Witzel S, Zimyanin V, Carreira-Barbosa F, Tada M, Heisenberg CP. 2006. Wnt11 controls cell contact persistence by local accumulation of Frizzled 7 at the plasma membrane. J. Cell Biol. 175:791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wolff T, Rubin GM. 1998. Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development 125:1149–1159 [DOI] [PubMed] [Google Scholar]
- 62. Yin C, Kiskowski M, Pouille PA, Farge E, Solnica-Krezel L. 2008. Cooperation of polarized cell intercalations drives convergence and extension of presomitic mesoderm during zebrafish gastrulation. J. Cell Biol. 180:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zallen JA. 2007. Planar polarity and tissue morphogenesis. Cell 129:1051–1063 [DOI] [PubMed] [Google Scholar]