Abstract
Mammalian spermatogenesis is a highly regulated system dedicated to the continuous production of spermatozoa from spermatogonial stem cells, and the process largely depends on microenvironments created by Sertoli cells, unique somatic cells that reside within a seminiferous tubule. Spermatogenesis progresses with a cyclical program known as the “seminiferous epithelial cycle,” which is accompanied with cyclical gene expression changes in Sertoli cells. However, it is unclear how the cyclicity in Sertoli cells is regulated. Here, we report that Notch signaling, which is known to play an important role for germ cell development in Drosophila and Caenorhabditis elegans, is cyclically activated in Sertoli cells and regulates stage-dependent gene expression of Hes1. To elucidate the regulatory mechanism of stage-dependent Hes1 expression and the role of Notch signaling in mouse spermatogenesis, we inactivated Notch signaling in Sertoli cells by deleting protein O-fucosyltransferase 1 (Pofut1), using the cre-loxP system, and found that stage-dependent Hes1 expression was dependent on the activation of Notch signaling. Unexpectedly, however, spermatogenesis proceeded normally. Our results thus indicate that Notch signaling regulates cyclical gene expression in Sertoli cells but is dispensable for mouse spermatogenesis. This highlights the evolutionary divergences in regulation of germ cell development.
INTRODUCTION
Mammalian spermatogenesis is based on a highly specialized stem cell system that largely depends on microenvironments within a seminiferous tubule. Spermatogonia, immature male germ cells containing the stem cell population in mammalian testes, proliferate several times and then differentiate into meiotic spermatocytes. After meiosis, these cells sequentially transform to round spermatids and elongated spermatids and are finally released into the lumen as spermatozoa (25). Spermatogenesis progresses with a cyclical program known as the “seminiferous epithelial cycle” (33). During spermatogenesis, germ cells at different developmental stages form groups and synchronously differentiate. In the mouse, 12 germ cell groups, known as seminiferous epithelial stages I to XII, are identifiable and are arranged in order along the length of tubules. Sertoli cells, which are unique somatic cells in the seminiferous tubules, directly interact with a germ cell group and support the constant differentiation of these cells by creating microenvironments essential for self-renewal and differentiation (3, 22, 32). It has also been shown that Sertoli cells change their gene expression patterns in response to alterations in the interacting germ cell groups (40, 50), and such gene expression changes suggest that Sertoli cells create stage-specific microenvironments. It has been known that retinoic acid (RA) signaling is involved in the seminiferous epithelial cycle (43, 44). However, it remains unclear whether other signaling pathways are implicated in the cyclical gene expression change and how the cyclicity in Sertoli cells is regulated.
Notch signaling is an evolutionarily conserved system that is implicated in cell fate decisions and various developmental processes in invertebrates and vertebrates (2, 15, 20, 21). The activation of Notch signaling is induced by the interaction between the extracellular domain of Notch receptor and the ligands Delta and Jagged expressed on neighboring cells. This leads to the cleavage of the Notch intracellular domain (NICD) by gamma-secretase and its translocation to the nucleus, where it forms a complex with a transcriptional regulator RBP-jk and other coregulators. This, in turn, results in the transcriptional activation of specific target genes, such as Hes family genes (11). All four identified Notch receptors (Notch1 to -4) are modified posttranslationally by protein O-fucosyltransferase 1 (Pofut1), an enzyme that transfers fucose to epidermal growth factor (EGF) repeats of the extracellular domain and may act as a chaperone protein for Notch receptors (28, 29, 36). These regulatory mechanisms have been shown to be important for the cellular localization of Notch receptors and their binding to ligands, and the inactivation of Pofut1 thus causes severe Notch signaling defects (30, 31).
It has been reported previously that Notch signaling is important for germ cell development in Caenorhabditis elegans (13) and Drosophila (1, 14, 37, 46). In mammalian testes, other group of investigators have also reported the immunohistological detection of Notch receptors and ligands (5, 8, 23, 45), but these findings are not consistent. Furthermore, there have been no reports that have examined the significance of Notch signaling in mouse spermatogenesis using genetic approaches. Thus, it remains unclear whether Notch signaling is implicated in mouse spermatogenesis and whether there is any functional difference of Notch signaling in mammalian germ cell development compared to that in invertebrates. In our present study, we have found that Notch signaling is cyclically activated in Sertoli cells during stages VII to VIII and regulates stage-dependent expression of Hes1. However, inactivation of Notch signaling in Sertoli cells and germ cells by deletion of Pofut1 does not cause abnormal spermatogenesis during the periods we examined. These findings suggest that cyclical activation of Notch signaling in Sertoli cells regulates stage-dependent gene expression of Hes1 but that both are dispensable for mouse spermatogenesis.
MATERIALS AND METHODS
Animals.
The N1IP::CreHI Rosa-YFP, Pofut1 conditional knockout (cKO), Cag-Cat-GFP Nanos3-cre mice have been described previously (19, 30, 34, 38). For the generation of an Amh-cre transgenic mouse, the 720-bp promoter region and entire first intron of the Amh gene were amplified from the tail DNA of a C57BL6/j mouse by PCR, and this product was used to drive Cre recombinase as previously described (9). Transgenic mice were generated by pronuclear injection of the purified transgene into F2 hybrid oocytes from B6/C3H parents (CLEA Japan), and six founders were obtained. The wild-type mice used in this study were the MCH strain purchased from CLEA Japan. All mice were maintained in accordance with National Institute of Genetics (NIG) guidelines, and all procedures were carried out with approval from the Committee for Animal Care and Use in NIG.
Histology.
For histological analysis, mouse testes without tunica albuginea and epididymides were fixed in Bouin's solution at room temperature for more than 24 h and embedded in paraffin. Sections (4 μm thick) were stained using hematoxylin and the periodic acid-Schiff (PAS) method. Seminiferous epithelial stages were determined as described previously (33).
Immunostaining.
After removal of the tunica albuginea, the mouse testes were fixed with 4% paraformaldehyde (PFA) at 4°C overnight and embedded in paraffin or 22-oxacalcitriol (OCT) compound (Sakura Finetek). After a blocking step, sections were incubated with goat anti-Gata4 antibody (Santa Cruz), rabbit anti-Plzf antibody (Santa Cruz), or chick anti-green fluorescent protein (anti-GFP) antibody (Aves) overnight at 4°C. The resulting signals were detected by incubation with Alexa 488- or Alexa 594-conjugated IgG antibodies (Molecular Probes). For the detection of rabbit WT1 antibody (Abcam), rabbit anti-Notch1 ICD (N1ICD) antibody (Cell Signaling Technology), rabbit anti-β-galactosidase antibody (Serotec) or rabbit anti-Hes1 antibody (30), EnVision anti-rabbit antibody (Dako), and Tyramid Signal Detection Reagent (Perkin Elmer) were used. For the double immunostaining of N1ICD and WT1 and Hes1 staining, the appropriate antibodies were separately incubated after antigen retrieval (26). All sections were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and images were processed with Adobe Photoshop, version 10.
In situ hybridization.
In situ hybridizations were performed as previously described with some modifications (49). The probes for Notch1, Notch4, Jagged1, Jagged2, Dll3, and Dll4 have been previously described (18, 30), and those for Dll1 and for Notch2 and Notch3 were kindly provided by A. Gossler (Institute for Molecular Biology, Germany) and T. Gridly (The Jackson Laboratory), respectively. Lgals1 (0.5 kbp), Stra6 (2.3 kbp), and Stra8 (1.1 kbp) were subcloned from testis cDNA by PCR. Digoxigenin (DIG)-labeled cRNA probes were synthesized with RNA labeling mix (Roche). Mice were perfusion fixed with 4% PFA under anesthesia with Avertin, and their testes were dissected. After removal of the tunica albuginea, the testes were further fixed in 4% PFA at 4°C overnight and embedded in paraffin. Sections were hybridized with DIG-labeled probe and incubated with alkaline phosphatase (AP)-conjugated anti-DIG Fab fragments (Roche). Signals were then detected using nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) solution (Roche) mixed with 5% polyvinyl alcohol (Wako) to enhance the AP reaction (4).
Quantitative RT-PCR.
Seminiferous tubules during stages I to III, IV to VI, VII to VIII, and IX to XII were isolated under dissection microscopy as previously described (16). Total RNA was purified with an RNeasy kit (Qiagen), and cDNA was synthesized using oligo(dT) primer and SuperScript III (Invitrogen) according to manufacturer's instructions. Real-time reverse transcription-PCR (RT-PCR) was performed using a KAPA SYBR Fast qPCR kit (KAPA Biosystems), Thermal Cycler Dice (Takara), and the following PCR primers: Hes1, 5′-CCAGCCAGTGTCAACACGA-3′ and 5′-ATGCCGGGAGCTATCTTTCT-3′; Gapdh, 5′-CATGTTCCAGTATGACTCCACTC-3′ and 5′-GGCCTCACCCCATTTGATGT-3′. Signals were normalized by the Gapdh expression.
Preparation and injection of lentivirus.
The packaging plasmids pCAG-HIVgp and pCMV-VSV-G-RSV-Rev (where CMV is cytomegalovirus, VSV-G is vesicular stomatitis virus G protein, and RSV is Rous sarcoma virus) and the expression plasmids CSII-EF-MCS-IRES2-Venus (where EF is elongations factor, MCS is multiple cloning site, and IRES2 is internal ribosome entry site 2) and CSII-EF-Venus were kindly provided by H. Miyoshi (Riken BRC, Japan). Lentiviral vectors were prepared as previously described with some modifications (17, 41). A total of 5 × 106 HEK293T cells were seeded on a 10-cm dish 24 h prior to transfection. A total of 10 μg of plasmid DNA (2.7 μg of pCAG-HIVgp, 2.7 μg of pCMV-VSV-G-RSV-Rev, and 4.6 μg of transfer vector plasmid) mixed with 15 mM (based on monomer units) polyethylenimine (PEI) solution (Polysicence) was added to the culture medium. After 36 h of cultivation, the medium containing lentivirus vector was collected, and lentiviruses were purified with sequential centrifugation at 20,000 × g for 5 h and resuspended in Hanks' balanced salt solution (Invitrogen). Lentiviruses were injected into testes from postnatal day 7 (P7) to P10 via efferent ductules (10).
Isolation and culture of primary Sertoli cells.
Primary Sertoli cells were isolated as previously described with some modifications (12). Two- to three-week testes without tunica albuginea were sequentially treated with 0.5 mg/ml collagenase (WAKO), 1 mg/ml hyaluronidase (Sigma) plus 1 mg/ml collagenase, and 1 mg/ml hyaluronidase in Dulbecco's modified Eagle's medium (DMEM) containing DNase I. Small pieces of seminiferous tubules were removed via filtering through a 100-μm-pore-size filter, and isolated Sertoli cells were cultured with F12-DMEM mixed with 10 μg/ml insulin (Invitrogen), 5 μg/ml transferrin (Sigma), and 5 ng/ml epidermal growth factor (Becton Dickinson) at 34°C. Medium was changed at days 2 and 4, and genomic DNA was isolated at day 5.
Sperm count.
Cauda epididymides were dissected into small pieces and incubated in potassium simplex optimized medium (KSOM) at 37°C for 1 h under 5% CO2 to allow the sperm to exude. The collected sperm were then fixed with 4% PFA and counted with a hemacytometer.
RESULTS
Notch signaling is cyclically activated in stage VII to VIII Sertoli cells.
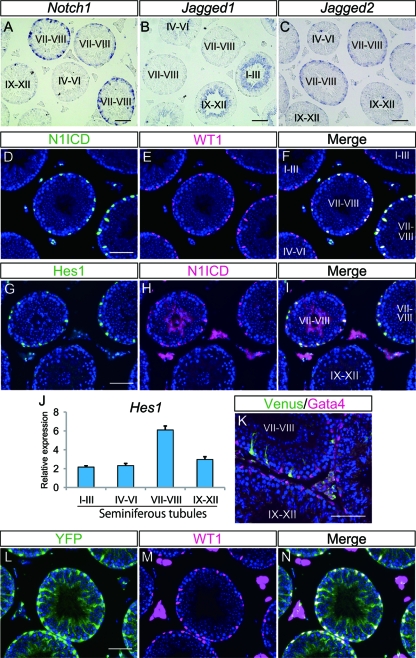
We first examined the expression pattern of each of the Notch receptors (Notch1 to -4) and Notch ligands (Jagged1 and -2 and Delta-like1, -3, and -4) in adult mouse testes by in situ hybridization and observed stage-dependent expression for some of the genes (Fig. 1A to C and data not shown). These expression profiles included Notch1 in stage VII to VIII Sertoli cells (Fig. 1A), Jagged1 in stage I to VI and IX to XII elongated spermatids (Fig. 1B), and Jagged2 in stage VII and VIII spermatogonia and preleptotene spermatocytes (Fig. 1C). We did not detect expression of genes encoding other Notch receptors and ligands. Since the genes encoding Notch1 receptor and ligands are expressed in adult mouse testes, we next examined the activation of Notch signaling using a specific antibody for the cleaved Notch1 intracellular domain (N1ICD) (26). We detected signals for cleaved N1ICD in the nuclei of stage VII to VIII tubules, and these nuclei also stained positively for WT1, a Sertoli cell-specific transcription factor in the adult testis, thus indicating Notch1 activation in Sertoli cells (Fig. 1D to F). Furthermore we found that Hes1, a well-known target of Notch signaling, was also expressed in stage VII to VIII Sertoli cells together with the N1ICD (Fig. 1G to I). This stage-dependent expression of Hes1 was also confirmed by real-time RT-PCR using stage-specific seminiferous tubules (Fig. 1J). These results suggest that simultaneous expression of Notch1 and Jagged2 in stage VII to VIII tubules leads to activation of the Notch signaling in stage-specific Sertoli cells.
Fig 1.
Expression pattern of Notch signaling-related genes and activation of Notch signaling in the adult mouse testis. (A to C) Expression of Notch1, Jagged1, and Jagged2 in adult mouse testes, examined by in situ hybridization. (D to F) Immunostaining of cleaved-N1ICD and WT1 and their merged signal in the adult mouse testis. (G to I) Immunostaining of Hes1 and cleaved-N1ICD and their merged signal. (J) Expression of Hes1 mRNA in stage-specific tubules. Gapdh was used as the internal control. Error bars, standard deviations. (K) Immunostaining of Venus (green) and Gata4 (magenta) in adult testis from a TP1-Venus transgenic mouse. (L to N) Immunostaining of YFP and WT1 and their merged signals in an N1IP::Cre; Rosa-YFP testis. Nuclei were stained with DAPI (blue). The seminiferous epithelial stages were determined by staining of serial sections with PAS and hematoxylin. Scale bar, 80 μm.
We also confirmed Notch activation using a Notch reporter system, the TP1-Venus transgenic mouse, in which Venus expression is induced upon activation of Notch signaling via RBP-jk-binding sequence (35). Consistent with the expression pattern of N1ICD, reporter expression in this animal model was detected exclusively in stage VII to VIII Gata4-positive Sertoli cells (Fig. 1K). We next examined N1IP::Cre knock-in mice, in which Cre-dependent recombination is induced in cells that experienced activation of Notch1 signaling (19). To visualize the cells in adult testes, N1IP::Cre mice were crossed with Rosa-YFP reporter mice in which yellow fluorescent protein (YFP) is expressed after Cre-mediated recombination, and we found YFP expression in all Sertoli cells but not in any germ cells (Fig. 1L to N). These findings also support the idea that Notch signaling is cyclically activated in stage VII to VIII Sertoli cells.
Activation of Notch signaling is spatially and temporally regulated during the first round spermatogenesis.
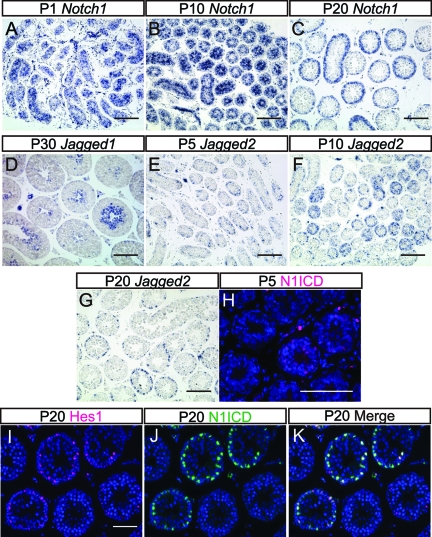
We next examined the time point at which Notch activation is established during mouse spermatogenesis. For this purpose, sections of mouse testes were prepared and subjected to in situ hybridization for Notch1, Jagged1, and Jagged2 at 5-day intervals from postnatal day 1 (P1) to P30 (Fig. 2A to G). Notch1 was detected in all Sertoli cells at P1 (Fig. 2A), and this expression was gradually restricted to some tubules through P10 to P20 and onward (Fig. 2B and C and data not shown). Jagged1 transcription appeared to start at P30 (Fig. 2D and data not shown) when elongated spermatids differentiate. Although Jagged2 transcripts were not detected in P1 testis (data not shown), they became weakly detectable in all seminiferous tubules at P5 (Fig. 2E), when the differentiation of gonocytes into spermatogonia takes place. At P10, just after initiation of meiosis, strong expression of Jagged2 became apparent in some tubules (Fig. 2F), but this number decreased at P20 and onward (Fig. 2G and data not shown).
Fig 2.
Expression patterns of Notch signaling-related genes and activation of Notch signaling in the developing mouse testis. (A to C) Expression patterns of Notch1 in the testes at P1, P10, and P20. (D) Expression pattern of Jagged1 in the testis at P30. (E to G) Expression patterns of Jagged2 in the testes at P5, P10, and P20. (H) Distribution of cleaved-N1ICD in P5 testis. (I to K) Immunostaining of Hes1 and N1ICD and their merged signal in P20 testis. Nuclei were stained with DAPI (blue). Scale bars, 80 μm (A to G) and 40 μm (H and I).
Notch activation was next examined using sections from P1, P5, and P20 testes. In the P1 testes, there was no evident N1ICD signal (data not shown), and N1ICD became faintly detectable in immature Sertoli cells at P5 when the nuclei of these cells were still located in the center of the tubules (Fig. 2H). This was further found to be coincident with the onset of Jagged2 expression at around P5; however, Hes1 expression was not detectable, probably due to the low activation level of Notch signaling (data not shown). At P20, when Sertoli cells become mature and Jagged2 expression increases in germ cells, N1ICD signals were detected in a proportion of Sertoli cell nuclei located on the periphery of the tubules, and those cells also expressed Hes1 (Fig. 2I to K). These results thus suggest that Notch1 activation and Hes1 expression in Sertoli cells are temporally and spatially regulated by the expression of Notch1 in Sertoli cells and Jagged2 in germ cells.
Notch signaling regulates stage-dependent expression of Hes1.
To clarify the role of Notch signaling in stage-dependent gene expression and mouse spermatogenesis, we used a conditional knockout (cKO) mouse carrying a loxP-flanked allele of Pofut1. Pofut1 encodes O-fucosyltransferase which has an essential function in the proper folding and/or trafficking of all Notch receptors and therefore is required for activation of Notch signaling (30, 31). Hence, all Notch signaling will be abolished in this mutant mouse, independent of any other Notch receptor that may be expressed in Sertoli cells below the limit of detection by in situ hybridization.
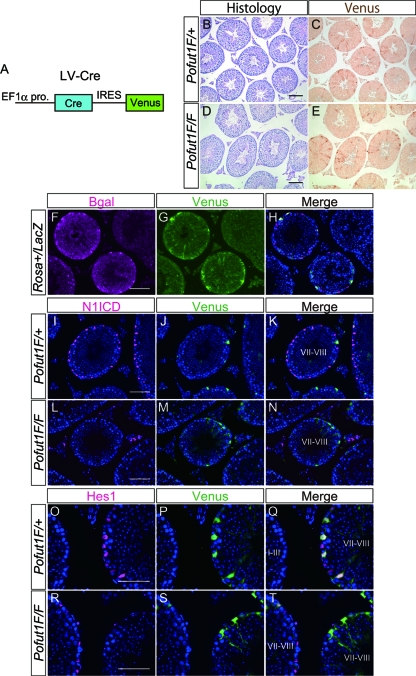
It was reported that injection of lentivirus into seminiferous tubules resulted in Sertoli cell-specific transfection (10). Therefore, to remove the floxed allele specifically in Sertoli cells, we utilized lentivirus containing Cre recombinase followed by IRES Venus (LV-Cre) (Fig. 3A) to identify lentivirus-infected Sertoli cells. LV-Cre was injected into P7 to P10 testes of Rosa-LacZ mice, in which LacZ reporter expression would be initiated following Cre-dependent recombination. We found that Venus was specifically expressed in Sertoli cells that were also positive for LacZ, indicating that recombination is specifically induced in Sertoli cells (Fig. 3F to H). To inactivate Pofut1, we next injected LV-Cre into Pofut1flox/flox testes at P7 to P10 and analyzed the testes at the age of 8 weeks. In those testes, expression of N1ICD in Venus-positive Sertoli cells became undetectable (Fig. 3I to N), suggesting that Notch signaling was inactivated in Sertoli cells. Furthermore, expression of Hes1 in stage VII to VIII Sertoli cells was also diminished (Fig. 3O to T). These results indicate that the stage-dependent expression of Hes1 is regulated by the cyclical activation of Notch signaling.
Fig 3.
Inactivation of Notch signaling in mouse Sertoli cells induced by using lentivirus. (A) Construction of LV-Cre. Cre and the following IRES-Venus are under the control of EF1α promoter (pro). (B to E) Histological sections stained with PAS and hematoxylin (B and D) and their serial sections stained with anti-GFP antibody (C and E) in adult testes from Pofut1flox/+ (Pofut1F/+) or Pofut1flox/flox (Pofut1F/F) mice injected with LV-Cre. (F to H) Immunostaining of β-galactosidase (β-Gal) and Venus and their merged signal in a testis from a 6-week-old Rosa-LacZ reporter mouse injected with LV-Cre. (I to N) Immunostaining of N1ICD and Venus and their merged signal in adult testes from Pofut1flox/+ or Pofut1flox/flox mice injected with LV-Cre. (O to T) Immunostaining of Hes1 and Venus and their merged signal in adult testes from Pofut1flox/+ or Pofut1flox/flox mice injected with LV-Cre. Nuclei were stained with DAPI (blue). Scale bar, 80 μm.
Amh-cre mouse successfully deletes Pofut1 and inactivates Notch signaling in Sertoli cells.
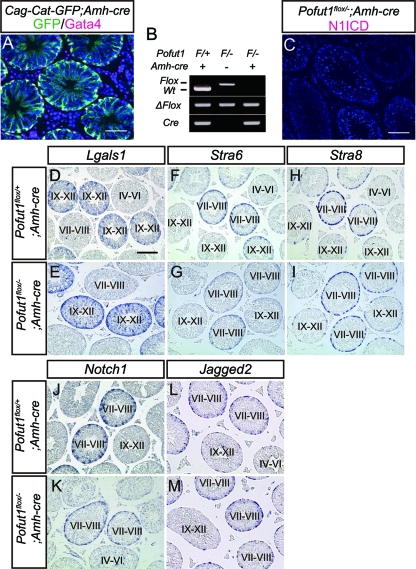
Despite the inactivation of Notch signaling and the loss of Hes1 expression in Sertoli cells, spermatogenesis in the Pofut1 cKO testes injected with LV-Cre was not affected (Fig. 3B to E). We hypothesized that the absence of any noticeable abnormality in the Pofut1 cKO testes injected with LV-Cre might be due to nonautonomous rescue of Notch-deficient cells by the Notch-expressing population. Therefore, to inactivate Pofut1 in most Sertoli cells, we generated an Amh-cre mouse, which induces Cre recombinase in embryonic Sertoli cells (9). The specificity of Amh-cre was confirmed by crossing with Cag-Cat-GFP reporter mice in which GFP is expressed after cre-mediated recombination. In 6-week-old Amh-cre; Cag-Cat-GFP mice, more than 95% of Sertoli cells became GFP positive (Fig. 4A). In addition, a small number of cells in other tissues, including liver, heart, and kidney, also expressed this reporter gene, suggesting that some leaky Cre expression takes place (data not shown). When Pofut1flox/flox mice were crossed with Amh-cre; Pofut1+/− mice, the resulting Pofut1flox/−; Amh-cre mice were not born at the expected Mendelian frequency. This suggested that leaky deletion of Pofut1 led to embryonic lethality. However, we were able to obtain enough viable, adult Pofut1flox/−; Amh-cre mice to be analyzed.
Fig 4.
Inactivation of Notch signaling in mouse Sertoli cells by Amh-cre and expression pattern of stage-dependent genes in Pofut1flox/−; Amh-cre mice. (A) Immunostaining of GFP (green) and Gata4 (magenta) in a testis obtained from an adult Cag-Cat-GFP; Amh-cre mouse. (B) Genotyping PCR of cultured Sertoli cells isolated from the indicated mice. (C) Immunostaining of N1ICD (magenta) in a Pofut1flox/−; Amh-cre testis. (D to M) Expression of Lgals1, Stra6, Stra8, Notch1, and Jagged2 in Pofut1flox/+; Amh-cre (D, F, H, J, and L) and Pofut1flox/−; Amh-cre (E, G, I, K, and M) mice detected by in situ hybridization. Seminiferous epithelial stages were determined by staining of serial sections with PAS and hematoxylin. Scale bar, 80 μm. Wt, wild type.
To confirm that Pofut1 was actually deleted in Sertoli cells by Amh-cre, we analyzed the Pofut1 locus in cultured Sertoli cells isolated from Pofut1flox/−; Amh-cre mice by PCR and found that deletion of the Pofut1 locus was complete, as expected (Fig. 4B). Consistent with this, adult Pofut1flox/−; Amh-cre testes lost N1ICD immunoreactivity in Sertoli cells (Fig. 4C). These results confirmed the successful inactivation of Notch signaling.
The majority of the stage-dependent gene expression remains unchanged in Sertoli cell-specific Pofut1 cKO mice.
We assessed the possibility that Notch signaling is involved in the regulation of stage-dependent gene expression other than Hes1 in mouse Sertoli cells. We first examined the expression pattern of Lgals1, whose expression peaked in stage IX to XII Sertoli cells (40), and we found that Lgals1 was normally expressed in Pofut1flox/−; Amh-cre mice (Fig. 4D and E). We next analyzed the expression of Stra6 and Stra8, which are originally identified as targets of RA signaling in embryonic carcinoma cells and highly expressed in stage VII to VIII Sertoli cells and stage VII to VIII germ cells, respectively (44). In Pofut1flox/−; Amh-cre mice, the expression pattern of these genes was also comparable to that of the control mice (Fig. 4F to I). Finally, to investigate a feedback regulation of Notch signaling, we examined Notch1 and Jagged2 expression in the Pofut1flox/−; Amh-cre mice, and we found that their stage-dependent expression levels were also normal (Fig. 4J to M). These results suggest that the stage-dependent gene expression in seminiferous tubules except for Hes1 would be independent of Notch signaling and that the stage-dependent expression of Notch1 and Jagged2 is not regulated by Notch signaling itself.
Activation of Notch signaling in Sertoli cells is dispensable for mouse spermatogenesis.
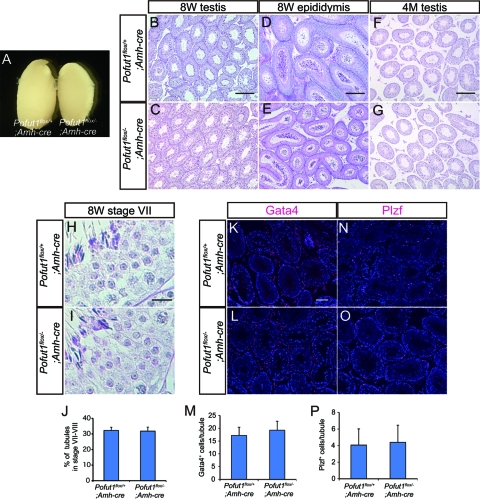
We next examined fertility of Pofut1flox/−; Amh-cre mice and found that 8-week-old mutant males were fertile and produced normal litter sizes, comparable to those of the control males mated with wild-type females (Table 1). We confirmed that testis size (Fig. 5A), the histology of the testis (Fig. 5B and C) and epididymis (Fig. 5D and E), and the number of spermatozoa in the epididymides (Table 1) in Pofut1flox/−; Amh-cre testis were comparable to those of the control. We also checked a possibility of any changes in the seminiferous epithelial cycle in Pofut1flox/−; Amh-cre mice due to the lack of Notch activation in stages VII to VIII. However, we detected no significant change in germ cell group observed in each seminiferous stage (Fig. 5H and I and data not shown) and proportion of stage VII to VIII tubules (Fig. 5J). Moreover, the numbers of Gata4-positive Sertoli cells (Fig. 5K to M) and Plzf-positive undifferentiated spermatogonia (Fig. 5N to P) were unchanged in these mice. To examine the role of Notch signaling in mouse spermatogonial stem cell maintenance, we analyzed 4-month-old Pofut1flox/−; Amh-cre males. The older mutant testes were comparable to those of the control mice histologically (Fig. 5F and G), and the older males generated normal litter sizes (Table 1). These results indicate that Notch signaling in Sertoli cells is dispensable for normal differentiation of spermatogenic cells and for stem cell maintenance in the mouse testes during the periods we examined.
Table 1.
Fertility of mutant mice
| Genotype | Sperm count (106) | Litter size (no. of pups) of mutant mice mated at: |
|
|---|---|---|---|
| 8 wk | 4 mo | ||
| Pofut1flox/+; Amh-cre | 19.0 ± 6.3 | 11.1 ± 3.0 | 11.3 ± 3.1 |
| Pofut1flox/−; Amh-cre | 20.0 ± 3.6 | 12.0 ± 3.0 | 10.0 ± 1.3 |
| Pofur1flox/+; Nanos3-cre | 15.6 ± 5.0 | 11.2 ± 4.4 | NDa |
| Pofut1flox/−; Nanos3-cre | 18.7 ± 4.4 | 10.9 ± 2.7 | ND |
ND, not determined.
Fig 5.
Spermatogenesis in Pofut1flox/−; Amh-cre mice. (A) Testes dissected from Pofut1flox/+; Amh-cre (left) and Pofut1flox/−; Amh-cre mice (right). (B to G) Histological sections of 8-week-old (8W) testes and epididymides and from 4-month-old (4M) testes obtained from Pofut1flox/+; Amh-cre and Pofut1flox/−; Amh-cre mice. (H and I) High-magnification image of spermatogenic cells in stage VII tubules obtained from Pofut1flox/+; Amh-cre and Pofut1flox/−; Amh-cre mice. (J) Proportion of stage VII to VIII tubules. (K to M) Distributions of Gata4-positive Sertoli cells (magenta) in Pofut1flox/+; Amh-cre (K) and Pofut1flox/−; Amh-cre (L) mice. The average numbers of Gata4-positive Sertoli cells in a seminiferous tubule are indicated in panel M. (N to P) Distributions of Plzf-positive spermatogonia (magenta) in Pofut1flox/+; Amh-cre (N) and Pofut1flox/−; Amh-cre (O) mice. The average numbers of Plzf-positive spermatogonia in a seminiferous tubule are indicated in panel P. Nuclei were stained with DAPI (blue). Error bars, standard deviations. Scale bars, 200 μm (B, D, and F), 20 μm (H), and 80 μm (K).
Inactivation of Notch signaling in mouse germ cells does not affect mouse spermatogenesis.
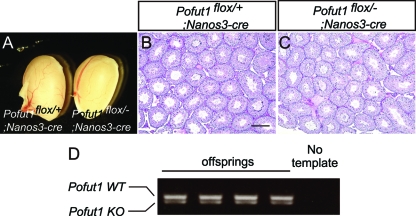
Since Notch signaling is required for the maintenance of germ line stem cells in C. elegans, we examined the possibility that Notch signaling may be activated and function in mouse germ cells in a cell-autonomous manner. For this purpose, Pofut1 cKO mice were crossed with Nanos3-cre mice which induce Cre in almost 100% of male germ cells from embryonic day 13.5 (E13.5) (38). In the 12-week-old Pofut1flox/−; Nanos3-cre mice, the external morphology of the testes (Fig. 6A), the histology of the seminiferous tubules (Fig. 6B and C), and the number of sperm in epididymides (Table 1) were normal. Furthermore, when 8-week-old Pofut1flox/−; Nanos3-cre male mice were crossed with wild-type female mice, fecundity was again similar to that of the control mice (Table 1), and all the pups derived from three different fathers were heterozygous for the Pofut1 allele (Fig. 6D and data not shown), indicating that both alleles of Pofut1 were inactivated in male sperm and that rare stem cells with an intact Pofut1 locus did not have an advantage. These results indicate that Notch signaling in germ cells is also dispensable for mouse spermatogenesis.
Fig 6.
Inactivation of Notch signaling in germ cells. (A) Testes dissected from Pofut1flox/+; Nanos3-cre and Pofut1flox/−; Nanos3-cre mice. (B and C) Histological sections prepared from Pofut1flox/+; Nanos3-cre and Pofut1flox/−; Nanos3-cre mice. (D) PCR genotyping of the offspring from a female wild-type mouse crossed with a male Pofut1flox/−; Nanos3-cre mouse. The DNA was amplified by specific primer pairs for wild-type (WT) and knockout (KO) alleles. Scale bar, 200 μm.
DISCUSSION
We report that Notch signaling is cyclically activated to regulate stage-dependent expression of Hes1 in mouse Sertoli cells during stages VII to VIII of spermatogenesis. Specifically, we found that Sertoli cells express Notch1 receptor in a stage-dependent manner that coincides with the stage-dependent expression of Jagged2 ligand in the adjacent germ cells. We addressed the question of how the cyclical expression of the Jagged2 and Notch1 is regulated and, more importantly, whether it is functionally important for spermatogenesis. It has been reported that Notch1 expression is regulated through feedback regulation of Notch signaling itself (47); however, Sertoli cell-specific Pofut1 deletion did not affect expression of either Notch1 or Jagged2, suggesting that cyclical activation is independent of Notch signaling. The cyclical expression patterns are thus regulated by another signaling pathway(s), for example, RA signaling. RA signaling has been shown to be important for the progression of germ cell differentiation as well as the regulation of cycling in Sertoli cells. It was suggested that activation of RA signaling is required for the transition from stage VII to stage VIII spermatogonia (42, 43). In the Pofut1 cKO mice, the expression levels of two RA downstream target genes, Stra6 and Stra8, were unchanged, suggesting that RA signaling in the mutant testis was unaffected. These findings indicate that RA signaling could work upstream from, or independent of, Jagged2 and Notch1 expression. Another possibility is that the spermatogenic cycle is robust, is independent of any perturbation in a single signaling axis, and utilizes species-specific mechanisms to ensure robustness. This idea is supported by the observation that when rat germ cells are transplanted to a mouse testis, spermatogenesis proceeds with the cycle characteristics of a rat (7). However, Sertoli cell-specific RA receptor α (RARα) deletion, a major RA receptor expressed in Sertoli cells, showed disruption of the stage-dependent expression of many genes without affecting the cycle of germ cells (44). This suggests that Notch signaling might be activated in the Sertoli cells by adjacent germ cells, which leads, in turn, to the induction of Hes1 in a stage-dependent manner. It remains to be tested if this is true and if other signaling pathways could play a role in the coordination between Sertoli and germ cell cycles.
To understand the function of Notch1 receptor in Sertoli cells, we performed conditional KO experiments of Pofut1 in Sertoli cells. Since the conventional Pofut1 KO results in embryonic lethality by E10, similar to other global Notch mutants (e.g., the Presenilin1 and -2 double KO [6] and RBP-Jk KO [27]), it is believed that a Pofut1 deletion leads to inactivation of Notch signaling mediated by all Notch receptors. We found that Sertoli cell-specific Pofut1 deletion did not produced any abnormality in spermatogenesis. Consistent with this report, another group reported that hyperactivation and inactivation of Notch signaling in somatic cells of mouse embryonic testes affected the development of Leydig cells but not Sertoli cells (39). Collectively, our results indicate that Notch signaling is dispensable for normal Sertoli cell development and mouse spermatogenesis. To our knowledge, this is the first report about a tissue in which Notch signaling is activated but is dispensable for its normal function. Hes1 expression was activated in a stage-dependent manner in Sertoli cells after birth. Hes1 expression is known to be regulated through other signaling pathways such as fibroblast growth factor (FGF) and transforming growth factor β (TGF-β) (24, 48), and therefore our current study cannot rule out the possibility that Hes1 in Sertoli cells play an important role during mouse spermatogenesis.
In Drosophila testis, Notch signaling was reported to be activated in the precursor of hub cells and to be important for development of hub cells (14). This is in striking contrast to our result that Notch signaling is dispensable for the function and development of Sertoli cells although activation of Notch signaling in niche cells is similar. In C. elegans, Notch signaling is activated in the germ line stem cells and prevents them from entering meiosis. Other groups have previously reported by immunostaining that the protein expression of Notch receptors occurs in spermatogonia, spermatocytes, and round spermatids in mammalian testes (5, 8, 45). We examined the possible involvement of Notch signaling in germ cell functions using Pofut1flox/−; Nanos3-cre mice. These mutant mice retained normal spermatogenesis and fertility over a long period, suggesting that Notch signaling in the germ cells is also dispensable for spermatogenesis in the mouse. Altogether, these differences suggest that mammals have changed the strategy to maintain spermatogenesis and evolved another signaling pathway(s) to compensate the function of Notch signaling.
Our results here demonstrate that Notch signaling is involved in the establishment of the stage-dependent expression of Hes1 in Sertoli cells and provide an insight into how the cyclicity of germ cells and Sertoli cells is coordinated. Our genetic approach revealed, however, that Notch signaling is dispensable for Sertoli cell function and germ cell maintenance and for mouse spermatogenesis. This is distinctly different from findings in Drosophila and C. elegans, indicating that not all aspects of germ cell development are evolutionarily conserved.
ACKNOWLEDGMENTS
We thank Rapahel Kopan for commenting on the manuscript and for the N1IP::Cre mouse, and Aya Satoh and Yuka Satoh (National Institute of Genetics) for animal care and technical assistance. We are also deeply grateful to A. Miyawaki (RIKEN BSI, Japan) for the Venus clone.
This study was supported in part by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists to K.H.
Footnotes
Published ahead of print 28 October 2011
REFERENCES
- 1. Assa-Kunik E, Torres IL, Schejter ED, Johnston DS, Shilo BZ. 2007. Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development 134:1161–1169 [DOI] [PubMed] [Google Scholar]
- 2. Bray SJ. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689 [DOI] [PubMed] [Google Scholar]
- 3. Chang C, et al. 2004. Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc. Natl. Acad. Sci. U. S. A. 101:6876–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Block M, Debrouwer D. 1993. RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal. Biochem. 215:86–89 [DOI] [PubMed] [Google Scholar]
- 5. Dirami G, Ravindranath N, Achi MV, Dym M. 2001. Expression of Notch pathway components in spermatogonia and Sertoli cells of neonatal mice. J. Androl. 22:944–952 [DOI] [PubMed] [Google Scholar]
- 6. Donoviel DB, et al. 1999. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13:2801–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franca LR, Ogawa T, Avarbock MR, Brinster RL, Russell LD. 1998. Germ cell genotype controls cell cycle during spermatogenesis in the rat. Biol. Reprod. 59:1371–1377 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi T, et al. 2001. Requirement of Notch 1 and its ligand jagged 2 expressions for spermatogenesis in rat and human testes. J. Androl. 22:999–1011 [DOI] [PubMed] [Google Scholar]
- 9. Holdcraft RW, Braun RE. 2004. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 131:459–467 [DOI] [PubMed] [Google Scholar]
- 10. Ikawa M, et al. 2002. Restoration of spermatogenesis by lentiviral gene transfer: offspring from infertile mice. Proc. Natl. Acad. Sci. U. S. A. 99:7524–7529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. 2008. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11:1247–1251 [DOI] [PubMed] [Google Scholar]
- 12. Karl AF, Griswold MD. 1990. Sertoli cells of the testis: preparation of cell cultures and effects of retinoids. Methods Enzymol. 190:71–75 [DOI] [PubMed] [Google Scholar]
- 13. Kimble J, Crittenden SL. 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23:405–433 [DOI] [PubMed] [Google Scholar]
- 14. Kitadate Y, Kobayashi S. 2010. Notch and Egfr signaling act antagonistically to regulate germ-line stem cell niche formation in Drosophila male embryonic gonads. Proc. Natl. Acad. Sci. U. S. A. 107:14241–14246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kotaja N, et al. 2004. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat. Methods 1:249–254 [DOI] [PubMed] [Google Scholar]
- 17. Kuroda H, Kutner RH, Bazan NG, Reiser J. 2009. Simplified lentivirus vector production in protein-free media using polyethylenimine-mediated transfection. J. Virol. Methods 157:113–121 [DOI] [PubMed] [Google Scholar]
- 18. Kusumi K, Dunwoodie SL, Krumlauf R. 2001. Dynamic expression patterns of the pudgy/spondylocostal dysostosis gene Dll3 in the developing nervous system. Mech. Dev. 100:141–144 [DOI] [PubMed] [Google Scholar]
- 19. Liu Z, et al. 2011. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J. Clin. Invest. 121:800–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Louvi A, Artavanis-Tsakonas S. 2006. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7:93–102 [DOI] [PubMed] [Google Scholar]
- 21. Maillard I, Adler SH, Pear WS. 2003. Notch and the immune system. Immunity 19:781–791 [DOI] [PubMed] [Google Scholar]
- 22. Meng X, et al. 2000. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287:1489–1493 [DOI] [PubMed] [Google Scholar]
- 23. Mori S, Kadokawa Y, Hoshinaga K, Marunouchi T. 2003. Sequential activation of Notch family receptors during mouse spermatogenesis. Dev. Growth Differ. 45:7–13 [DOI] [PubMed] [Google Scholar]
- 24. Nakayama K, Satoh T, Igari A, Kageyama R, Nishida E. 2008. FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr. Biol. 18:R332–R334 [DOI] [PubMed] [Google Scholar]
- 25. Oatley JM, Brinster RL. 2008. Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24:263–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oginuma M, Niwa Y, Chapman DL, Saga Y. 2008. Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development 135:2555–2562 [DOI] [PubMed] [Google Scholar]
- 27. Oka C, et al. 1995. Disruption of the mouse RBP-Jκ gene results in early embryonic death. Development 121:3291–3301 [DOI] [PubMed] [Google Scholar]
- 28. Okajima T, Irvine KD. 2002. Regulation of notch signaling by O-linked fucose. Cell 111:893–904 [DOI] [PubMed] [Google Scholar]
- 29. Okajima T, Xu A, Lei L, Irvine KD. 2005. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science 307:1599–1603 [DOI] [PubMed] [Google Scholar]
- 30. Okamura Y, Saga Y. 2008. Notch signaling is required for the maintenance of enteric neural crest progenitors. Development 135:3555–3565 [DOI] [PubMed] [Google Scholar]
- 31. Okamura Y, Saga Y. 2008. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mech. Dev. 125:663–673 [DOI] [PubMed] [Google Scholar]
- 32. Rao MK, et al. 2006. Tissue-specific RNAi reveals that WT1 expression in nurse cells controls germ cell survival and spermatogenesis. Genes Dev. 20:147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Russell L, Ettlin R, SinhaHikim A, Clegg E. 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater, FL [Google Scholar]
- 34. Sada A, Suzuki A, Suzuki H, Saga Y. 2009. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325:1394–1398 [DOI] [PubMed] [Google Scholar]
- 35. Sasaki N, Kiso M, Kitagawa M, Saga Y. 2011. The repression of Notch signaling occurs via the destabilization of mastermind-like 1 by Mesp2 and is essential for somitogenesis. Development 138:55–64 [DOI] [PubMed] [Google Scholar]
- 36. Sasaki N, et al. 2007. Polarized exocytosis and transcytosis of Notch during its apical localization in Drosophila epithelial cells. Genes Cells 12:89–103 [DOI] [PubMed] [Google Scholar]
- 37. Song X, Call GB, Kirilly D, Xie T. 2007. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 134:1071–1080 [DOI] [PubMed] [Google Scholar]
- 38. Suzuki H, Tsuda M, Kiso M, Saga Y. 2008. Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev. Biol. 318:133–142 [DOI] [PubMed] [Google Scholar]
- 39. Tang H, et al. 2008. Notch signaling maintains Leydig progenitor cells in the mouse testis. Development 135:3745–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Timmons PM, Rigby PW, Poirier F. 2002. The murine seminiferous epithelial cycle is pre-figured in the Sertoli cells of the embryonic testis. Development 129:635–647 [DOI] [PubMed] [Google Scholar]
- 41. Tiscornia G, Singer O, Verma IM. 2006. Production and purification of lentiviral vectors. Nat. Protoc. 1:241–245 [DOI] [PubMed] [Google Scholar]
- 42. Van Pelt AM, De Rooij DG. 1990. The origin of the synchronization of the seminiferous epithelium in vitamin A-deficient rats after vitamin A replacement. Biol. Reprod. 42:677–682 [DOI] [PubMed] [Google Scholar]
- 43. Van Pelt AM, de Rooij DG. 1990. Synchronization of the seminiferous epithelium after vitamin A replacement in vitamin A-deficient mice. Biol. Reprod. 43:363–367 [DOI] [PubMed] [Google Scholar]
- 44. Vernet N, et al. 2006. Prepubertal testis development relies on retinoic acid but not rexinoid receptors in Sertoli cells. EMBO J. 25:5816–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Schonfeldt V, Wistuba J, Schlatt S. 2004. Notch-1, c-kit and GFRalpha-1 are developmentally regulated markers for premeiotic germ cells. Cytogenet. Genome Res. 105:235–239 [DOI] [PubMed] [Google Scholar]
- 46. Ward EJ, et al. 2006. Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Curr. Biol. 16:2352–2358 [DOI] [PubMed] [Google Scholar]
- 47. Weinmaster G. 1997. The ins and outs of notch signaling. Mol. Cell Neurosci. 9:91–102 [DOI] [PubMed] [Google Scholar]
- 48. Xing Y, et al. 2010. Signaling via Alk5 controls the ontogeny of lung Clara cells. Development 137:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshida S, et al. 2001. Sgn1, a basic helix-loop-helix transcription factor delineates the salivary gland duct cell lineage in mice. Dev. Biol. 240:517–530 [DOI] [PubMed] [Google Scholar]
- 50. Yoshida S, et al. 2006. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 133:1495–1505 [DOI] [PubMed] [Google Scholar]