Abstract
Immunoglobulin (Ig) genes naturally acquire frequent premature termination codons during the error-prone V(D)J recombination process. Although B cell differentiation is linked to the expression of productive Ig alleles, the transcriptional status of nonfunctionally recombined alleles remains unclear. Here, we tracked transcription and posttranscriptional regulation for both Ig heavy-chain (IgH) alleles in mice carrying a nonfunctional knock-in allele. We show that productively and nonproductively VDJ-rearranged alleles are transcribed throughout B cell development, carry similar active chromatin marks, and even display equivalent RNA polymerase II (RNAPII) loading after B cell stimulation. Hence, these results challenge the idea that the repositioning of one allele to heterochromatin could promote the silencing of nonproductive alleles. Interestingly, the efficiency of downstream RNA surveillance mechanisms fluctuates according to B cell activation and terminal differentiation: unspliced nonfunctional transcripts accumulate in primary B cells, while B cell activation promotes IgH transcription, RNA splicing, and nonsense-mediated mRNA decay (NMD). Altogether, IgH transcription and RNA splicing rates determine by which RNA surveillance mechanisms a B cell can get rid of nonproductive IgH mRNAs.
INTRODUCTION
Development of the primary immunoglobulin (Ig) repertoire involves DNA recombination between variable (V), diversity (D), and joining (J) segments, and this diversity is extended by the imprecision of VDJ junctions. A collateral effect of this random process is the occurrence of out-of-frame rearrangements inherently associated with premature termination codons (PTCs) in two-thirds of cases. Previous reports documented that around 40 to 50% of B cells carry VDJ rearrangements (VDJ+/VDJ−) on both Ig heavy-chain (IgH) alleles, while the remainder retains incomplete DJ rearrangements on the nonproductive allele (VDJ+/DJ) (18, 24, 49).
Although B cell receptor (BCR) signaling, and, hence, the expression of productively rearranged Ig alleles, governs B cell development and survival, the transcriptional status of nonfunctional alleles remains unclear. If translated, these PTC-containing alleles might encode potentially harmful truncated Ig proteins that could disrupt the normal assembly of the BCR or elicit the unfolded protein response (UPR) (13, 32). Recently, it was demonstrated that the stability of untranslated nonsense μH mRNAs that escape degradation by nonsense-mediated mRNA decay (NMD) impairs IgH allelic exclusion and pro-B cell differentiation (36). Although nonsense mutations (in the leader exon) generating stable and untranslated μH mRNAs should not exist in pro-B cells, this model suggest that the persistence of nonsense μH mRNAs per se could be detrimental in early B cell development, yet the underlying mechanisms are currently unknown.
For activated B cells, it was reported previously that one IgH allele was localized mainly in heterochromatin domains (47), leading to the assumption that an asymmetric nuclear location could help the silencing of VDJ− alleles (25). In addition, the presence of a PTC on nonfunctionally recombined IgH transcripts might induce transcriptional silencing and heterochromatin repositioning by a mechanism referred to as nonsense-mediated transcriptional gene silencing (NMTGS) (8, 48). However, the NMTGS process observed for non-B cell lines for PTC-containing Ig genes does not take place in a pro-B cell line in which VDJ+ and VDJ− alleles are transcribed at equal rates (23). Accordingly, using RNA-fluorescence in situ hybridization (FISH) experiments, Daly and colleagues previously demonstrated a predominant biallelic IgH transcription pattern in primary B cells (18). It was also reported previously that the nuclear accumulation of pre-mRNAs near the site of transcription (39) or degradation by NMD can reduce the level of nonproductive IgH mRNAs in B cell hybridomas (6, 17, 33).
The degradation of PTC-containing mRNAs by NMD is dependent on translation by ribosomes and involves a complex set of protein interactions allowing PTC recognition (14, 31). These proteins are able to distinguish between the correct termination codon and the PTC. The exon junction complex (EJC) plays an important role in facilitating this, and NMD usually requires termination to occur at least 50 to 55 nucleotides (nt) upstream of the last exon-exon junction (14, 31). Because out-of-frame VDJ junctions lead to the appearance of a PTC within the variable or the first constant exon, nonproductive IgH mRNAs obey the “50- to 55-nt boundary rule” of EJC-dependent NMD. However, exceptions to this rule have been reported for T cell receptor β (TCRβ) and Ig genes, with NMD triggering for mRNA containing a PTC close to (<50 nt) or even downstream of the last exon-exon junction (9, 11, 15, 22, 30).
Many studies have reported that the inhibition of NMD factors has strong physiological and developmental impacts (28). In the mouse, the abrogation of the Upf1 (up-frameshift 1), Upf2, and Smg1 (suppressor with morphological effect on genitalia 1) genes elicits embryonic lethality (37, 38, 50). Since Upf1 and other NMD factors play significant roles in the maintenance of genome integrity and in cell cycle progression (1, 4, 5), it can be difficult to attribute the embryonic lethality to the sole inhibition of NMD. However, in two independent studies, T cells lacking nonproductive TCRβ transcripts appeared to be weakly sensitive to Upf1 or Upf2 inhibition (26, 50). Therefore, it seems complicated to definitely attribute the developmental arrest upon the inhibition of NMD factors to NMD-dependent or -independent mechanisms (28).
Although previous analyses with cell lines provided some insights into the RNA surveillance of nonproductively rearranged IgH transcripts (6, 9, 17, 23, 33, 39, 51), the regulation of this machinery during normal B cell development remains poorly understood. Using heterozygous IgH knock-in mice harboring a PTC on the targeted allele, we tracked both IgH alleles with regard to transcription and posttranscriptional regulation. Altogether, both the magnitude of IgH transcription and the rate of RNA splicing indicate by which RNA surveillance mechanism nonproductively rearranged IgH mRNAs are downregulated.
MATERIALS AND METHODS
Mice.
IgHfrVκ knock-in mice were described elsewhere previously and were created by the targeted insertion of an additional frameshift-inducing Vκ exon between JH4 and Eμ (44). To identify allotypic differences, IgHa/b animals were obtained after crossing 129/Sv (129) and C57BL/6 (B6) mice (129/B6 F1). Mice were maintained in our animal facilities under specific-pathogen-free conditions. Two- to six-month-old mice were used in all experiments. All the protocols used were approved by our institutional review board for animal experimentation.
Purification and culture of B cells.
Erythrocyte-depleted bone marrow and spleen cells were cultured in RPMI medium supplemented with 10% fetal calf serum, sodium pyruvate, nonessential amino acids, β-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). B cell populations were sorted on a FACS-Vantage instrument (BD Biosciences) after staining with anti-B220 (RA3-6B2; e-Bioscience), anti-CD43 (S7; BD Biosciences), anti-CD138 (281-2; BD Biosciences), anti-IgM (eB121-15F9; e-Bioscience), and a allotype-specific anti-IgM (anti-IgMa) (DS-1; BD Biosciences) monoclonal antibodies (MAbs), as indicated in Fig. 3. Spleen resting B cells were purified by negative selection using anti-CD43 microbeads (Miltenyi Biotech) and stimulated (0.5 × 106 cells/ml) with lipopolysaccharide (LPS) (20 μg/ml; Sigma) for 2 to 4 days. To determine the magnitude of RNA surveillance with regard to the proliferative response, resting B cells were labeled before LPS stimulation with 5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) for 15 min at 37°C according to the manufacturer's instructions. For NMD analysis, cells (<106 cells/ml) were treated with 100 μg/ml of cycloheximide (CHX) (in dimethyl sulfoxide [DMSO]) (Sigma) or 20 μg/ml of wortmannin (Sigma) for 4 h. Histone deacetylase inhibitor (HDACi) treatments were performed by using trichostatin A (TSA) (25 ng/ml; Sigma) or sodium butyrate (SB) (3 mM) for 8 h. According to the drug used, control cells were left untreated or incubated with DMSO alone (dilution factor, 1/1,000).
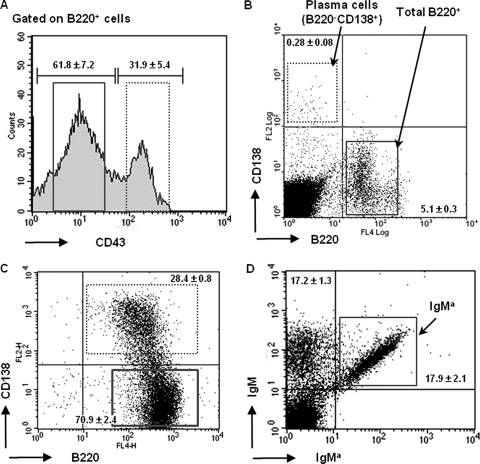
Fig 3.
FACS analysis of B cell populations. (A to C) Primary B cells were isolated from heterozygous IgHfrVk/wt mice and sorted after staining with anti-B220, anti-CD43, and anti-CD138 Abs. (A) Percentages of B220+ CD43+ and B220+ CD43− cells and gates used for cell sorting. The B220+ CD43+ population (solid rectangle) includes pro-B and large pre-B cells (precursor B cells before allelic exclusion), and the B220+ CD43− population (dotted rectangle) represents B cells after pre-BCR-mediated allelic exclusion (including small pre-B and immature and mature B cells). (B) Representative dot plot indicating the gates used for cell sorting and the percentages of total bone marrow B cells (solid rectangle) (B220+ CD138−) and plasma cells (dotted rectangle) (B220− CD138+). (C) Dot plot indicating the frequency of activated B cells (B220+ CD138−) and plasmablasts (B220+ CD138+) upon LPS stimulation (day 4) and gates used for cell sorting. (D) Spleen cells were isolated from B6/129 F1 mice (IgHa/b), and IgMa-positive cells (solid rectangle) were sorted after staining with total and a-allotype-specific anti-IgM Abs. Representative results from 5 to 7 (A to C) or 2 (D) independent cell sorting experiments are shown. The purity of sorted populations was above 90%.
Chromatin immunoprecipitation (ChIP) experiments.
Erythrocyte-depleted spleens cells from IgHfrVκ/wt mice were cultured (106 cells/ml) in the presence of LPS (20 μg/ml) for 4 days. On day 3, dead cells were eliminated by Ficoll centrifugation (Lympholyte-Mammal; Cedarlane), and living cells were stimulated for an additional 24-h period. Chromatin preparations were made with 5 × 107 to 1 × 108 LPS-stimulated B cells using standard fixation and sonication methods as described previously (29). Immunoprecipitations were performed with protein G-Sepharose (Sigma) and 1 μg of anti-RNA polymerase II (RNAPII) (CTD4H8; Santa Cruz) or anti-acetyl-histone H3 (Ac-H3) (Millipore) antibody (Ab) or nonimmune rabbit IgG (Millipore) as a control.
PCR and RT-PCR.
Genomic DNA and total RNA were prepared by using standard proteinase K (Eurogentec) and Tri reagent (Invitrogen) procedures, respectively. Reverse transcription-PCR (RT-PCR) was carried out with DNase I (Invitrogen)-treated RNA and was checked to be negative in the absence of reverse transcription, ruling out contamination by genomic DNA. Reverse transcription was performed by using a high-capacity cDNA reverse transcription kit (Applied Biosystems) on 1 to 3 μg of total RNA. Priming for reverse transcription was done with random hexamers. Real-time PCR using TaqMan Universal or SYBR green Mastermix (Applied Biosystems) was performed on DNA (50 ng) or cDNA (equivalent to 10 ng RNA) samples per reaction. Data were analyzed by comparing threshold cycle (CT) values according to the 2−(ΔΔCT) method. Unless otherwise stated, values obtained for resting B cells served as a reference, and their mean was set to 1. Primers used for analysis are depicted in Fig. 1A and Table S1 in the supplemental material.
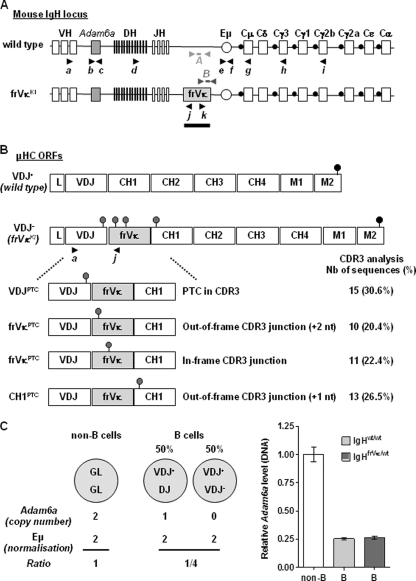
Fig 1.
Normal VDJ rearrangements in B cells from heterozygous IgHfrVκ/wt mice. (A) Schematic representation of wild-type and targeted IgH loci. Primers are represented by black arrows and referenced using letters in italics (a to k). Primers and probes used for allele-specific qPCR assays are represented by light gray (IgHwt) (set A) and gray (IgHfrVκ) (set B) arrows and short rectangles. The probe used for Northern blot experiments is shown below the frVκ exon (black rectangle). (B) Analysis of VDJ junctions, performed as described in Materials and Methods, after RT-PCR using the a/j PCR primer set. After VDJ recombination, PTCs appeared in VDJ, frVκ, and CH1μ exons with equal frequencies in splenic B cells from IgHfrVκ/wt mice. ORFs, open reading frames. (C) V(D)J recombination was analyzed by quantifying the copy number of Adam6a by qPCR (primer set b/c) after normalization to the DNA copy number of Eμ (primer set e/f). DNA amounts of Adam6a were determined for spleen B cells isolated from wild-type and IgHfrVκ/wt animals and compared to reference nonlymphoid DNA from tail tissues (empty bar). Results from 3 to 4 independent mice are shown.
Northern blots.
Northern blotting was conducted by migrating 10 μg of total RNA on a 1% agarose denaturing gel, followed by transfer onto nylon sheet membranes (MP Biomedicals). The blots were hybridized by using a 32P-labeled mouse Vκ6 segment probe allowing the simultaneous detection of IgHfrVκ and Vκ6-bearing Igκ transcripts, hence helping normalization. Blots were revealed with a Cyclone imager (Perkin-Elmer).
Analysis of VDJ junctions.
Sequencing of VDJ junctions was performed after the cloning of RT-PCR products (VH7183-for/frVκ-rev) into the pCR2.1-TOPO vector (Invitrogen). DNA sequences were obtained by using the M13 reverse primer (M13-rev) and a cycle sequencing kit together with an automated sequencer (Applied Biosystems). Sequences from VDJ junctions were analyzed by using V-QUEST software (International ImMunoGeneTics Information System [http://www.imgt.org/]).
Statistical analysis.
Results are expressed as means ± standard errors of the means (SEM), and overall differences between variables were evaluated by a two-tailed unpaired Student t test using GraphPad Prism software (GraphPad, San Diego, CA).
RESULTS
Features of VDJ rearrangements in B cells from heterozygous IgHfrVκ/wt mice.
To analyze both productive and nonproductive IgH alleles with regard to transcription and posttranscriptional regulation, we used an IgH knock-in mouse strain harboring a “frameshift-inducing V kappa exon” (frVκ) between JH and Cμ (Fig. 1A) (20, 44). After VDJ recombination, this inactivating exon was included in mature IgH transcripts between the VDJ and the CH1μ exons. Due to splicing frameshifts at both the 5′ and 3′ ends of the frVκ exon, any VDJ-rearranged transcript from the IgHfrVκ allele is nonfunctional. Hence, homozygous (IgHfrVκ/frVk) mutant animals are unable to produce any detectable μ IgH and have a developmental arrest at the pro-B cell stage. In heterozygous IgHfrVκ/wt mice, B cells develop normally through the expression of the wild-type (wt) allele (44). Because the mutation does not introduce any exogenous regulatory element that could modify the transcriptional status of the targeted IgH locus, this mouse strain stands as a good model to simultaneously track both productive and nonproductive IgH alleles.
Sequencing analysis of CDR3 junctions revealed that VDJ rearrangements on IgHfrVκ alleles involve all reading frames, leading to the appearance of PTCs in VDJ, frVκ, or CH1 exons (Fig. 1B). By quantifying the level of Adam6a, a gene located in the V-D intergenic region and lost upon a V-to-DJ rearrangement, we also determined the frequency of incomplete DJ rearrangements on IgHfrVκ alleles. Compared to non-B cell DNA, the relative level of Adam6a was similarly reduced by ∼4-fold in DNA from wt and IgHfrVκ/wt B cells, suggesting that in both cases, one-fourth of IgH alleles were DJ rearranged (Fig. 1C). Thus, VDJ recombination occurs normally in IgHfrVκ/wt mice, with an equivalent proportion of B cells harboring either biallelic (VDJ+/VDJ−) or monoallelic (VDJ+/DJ) VDJ rearrangements.
Active transcription and absence of heterochromatin silencing of nonproductive IgH alleles.
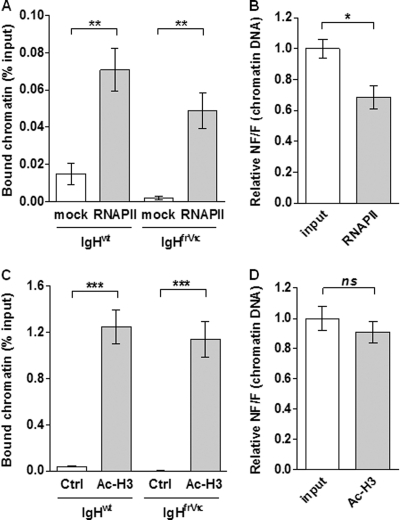
Conflicting results have been obtained with regard to the monoallelic or biallelic transcription of IgH alleles after B cell activation (18, 47). To address this discrepancy and determine the transcriptional accessibility of both productive and nonproductive alleles, we performed ChIP experiments with LPS-stimulated B cells from IgHfrVκ/wt mice using anti-RNA polymerase II (RNAPII) and anti-acetyl-histone H3 (Ac-H3) Abs (Fig. 2). We found a significant binding of RNA polymerase II (RNAPII) on both IgH alleles, suggesting an active transcription of nonproductive alleles after activation (Fig. 2A). From these data, we also determined nonfunctional/functional (NF/F) ratios in RNAPII-enriched chromatin fractions and found an NF/F ratio of 0.68 ± 0.07 (Fig. 2B). Although NF/F ratios were found to be decreased in RNAPII-bound chromatin fractions compared to input chromatin reference samples, these data are consistent with an equal RNAPII loading of both VDJ-rearranged alleles, with two times less VDJ− than VDJ+ alleles together with a low level of RNAPII binding on weakly transcribed DJ-recombined alleles (12, 18). In addition, we found that both IgH alleles exhibited hallmarks of active chromatin and similar Ac-H3 profiles (Fig. 2C). In these experiments, NF/F ratios were similar in input chromatin- and Ac-H3-immunoprecipitated fractions (NF/F ratio=0.91 ± 0.07), indicating an open chromatin structure for nonproductively VDJ- or DJ-rearranged alleles (Fig. 2D). Hence, this quantitative analysis argues against the heterochromatin repositioning and silencing of nonfunctional IgH alleles and suggests that both VDJ-rearranged alleles display equivalent RNAPII loading after B cell activation.
Fig 2.
Equivalent transcription and accessibility of functional and nonfunctional IgH alleles. ChIP assays were performed with spleen B cells isolated from heterozygous IgHfrVκ/wt mice and stimulated with LPS for 4 days. Immunoprecipitations were done by using antibodies to RNA polymerase II (RNAPII) (A and B) and acetylated histone H3 (Ac-H3) (C and D). (A and C) The relative enrichments (percent input) were analyzed by quantitative PCR using the allele-specific primers described in the legend of Fig. 1A (sets A and B) and were compared to those of negative controls obtained without Ab (mock) (A) or using a control IgG Ab (ctrl) (C). (B and D) Nonfunctional/functional (NF/F) ratios were obtained from these data, and the mean of ratios obtained for chromatin inputs (NF/Finput) was set to 1. Results from 5 (A and B) or 4 (C and D) independent ChIP experiments are shown. (ns [not significant], P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
From these experiments, we can also conclude that nonfunctional alleles are not silenced by NMTGS in activated B cells. Although histone deacetylase inhibitor (HDACi) treatments have been used previously to reverse the heterochromatin silencing of nonsense Ig minigenes (8), these drugs are potent inhibitors of the IgH enhancers Eμ and 3′ regulatory region (3′RR) in B cells (35). In agreement, we found that levels of IgHfrVκ mRNAs were strongly decreased in activated B cells treated with trichostatin A (TSA) and sodium butyrate (SB), whereas mRNA levels from a control gene (CD79a) were unaffected (see Fig. S1 in the supplemental material). Therefore, ChIP assays should be preferred over HDACi treatments to study NMTGS of endogenous nonproductive IgH alleles.
Accumulation of nonproductive IgH pre-mRNAs during B cell development.
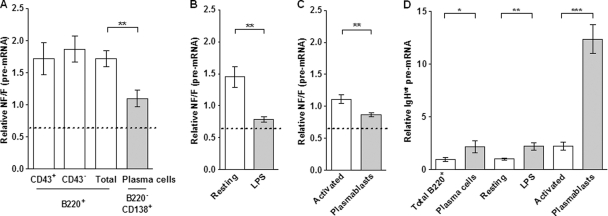
Next, we performed quantitative assessments of primary transcripts from both functional and nonfunctional IgH alleles in B cell populations sorted from IgHfrVκ/wt mice (Fig. 3A to C). We determined NF/F pre-mRNA ratios and compared these values to the threshold reference given by the mean of NF/F ratios (mean NF/F ratio=0.68) obtained from RNAPII binding assays (Fig. 2B). This mean was considered the reference because the analysis of RNAPII loading somehow reflects the transcription of both IgH alleles and accounts for the equivalent proportion of B cells harboring either biallelic (VDJ+/VDJ−) or monoallelic (VDJ+/DJ) VDJ rearrangements, together with the weak strength of DH promoters compared to VH (12, 18).
We found that NF/F pre-mRNA ratios were between 0.75 and 2 for all B cell populations (Fig. 4A to C). These values were higher than those obtained for RNAPII-bound chromatin fractions and hence reinforce the idea of a biallelic IgH transcription pattern during B cell development (47). High NF/F pre-mRNA ratios also indicate a slower processing of NF transcripts, resembling a splicing inhibition mechanism previously observed for PTC-containing Igκ or TCRβ transcripts (2, 15, 34, 43) and here referred to as nonsense-mediated upregulation of pre-mRNA (NMUP) (29). In B220+ bone marrow cells, sorted before (CD43+) or after (CD43−) the pre-BCR-mediated allelic exclusion (Fig. 3A), and in spleen resting B cells, NF/F values were between 1.5 and 2 (Fig. 4A and B, empty bars). Compared to the threshold reference (NF/F ratio=0.68) (Fig. 2B), these data suggest that NMUP increases the level of NF pre-mRNAs in primary B cells by ∼2- to 3-fold. Surprisingly, we observed significant drops in NF/F pre-mRNA ratios for bone marrow plasma cells (Fig. 3B and 4A) and upon B cell stimulation (Fig. 4B). According to data shown in Fig. 2B suggesting equivalent RNAPII loading on both VDJ-rearranged IgH alleles in LPS-stimulated B cells and, hence, no transcriptional silencing, the fluctuations of NF/F pre-mRNA ratios strongly indicate that NMUP displays variable efficiencies during B cell development.
Fig 4.
Correlation between IgH transcription and accumulation of nonfunctional pre-mRNAs. B cell populations were sorted from heterozygous IgHfrVκ/wt mice as described in the legend of Fig. 3. (A to C) NF/F IgH pre-mRNA ratios were determined for cDNAs by qPCR using allele-specific primers (sets A and B in Fig. 1A) after normalization to the NF/F ratio obtained by using DNA from IgHfrVκ/wt mice (NF/FDNA, set to 1). Dotted lines corresponds to the mean of NF/F values obtained for RNAPII-bound fractions (NF/F ratio=0.68) (Fig. 2B). This value was used as a threshold reference and reflects the RNAPII loading on both VDJ-rearranged IgH alleles in B cell populations harboring either biallelic (VDJ+/VDJ−) or monoallelic (VDJ+/DJ) VDJ rearrangements. (D) Relative levels of functional IgH pre-mRNAs (IgHwt) in B and plasma cells were determined by using allele-specific primers (set A in Fig. 1A) after normalization to β-actin gene transcripts. Values obtained for resting B cells were set to 1. Results from 5 to 7 independent cell sorting experiments are shown. (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
We also analyzed NF/F pre-mRNA ratios in activated B cells (B220+ CD138−) and plasmablasts (B220+ CD138+) sorted 4 days after LPS stimulation (Fig. 3C and 4C). We found a weak accumulation of NF pre-mRNAs in both activated B cells and plasmablasts (Fig. 4C) and NF/F ratios similar to those of the bulk of LPS-stimulated cells (Fig. 4B). Furthermore, NF/F pre-mRNA ratios were significantly lower for plasmablasts than for activated B cells (Fig. 4C), as previously observed for bone marrow plasma cells compared to B cells (Fig. 4A). Therefore, these data demonstrate that the efficiency of NMUP decreases after B cell activation and during plasma cell differentiation.
Previous studies have documented that IgH transcription rates are increased after LPS stimulation and in terminally differentiated plasma cells (16, 45). To confirm these findings, we determined pre-mRNA levels of functional IgH alleles that are PTC free and therefore not sensitive to NMUP. We found that these levels were significantly increased in bone marrow plasma cells compared to those in B cells, in LPS-stimulated cells compared to those in resting B cells, and, more strikingly, in plasmablasts compared to those in activated B cells (Fig. 3D). Altogether, our data indicate that the efficiency of NMUP decreases as the level of IgH transcription increases.
Levels of mature transcripts from nonproductively VDJ- and DJ-rearranged IgH alleles are efficiently lowered by NMD.
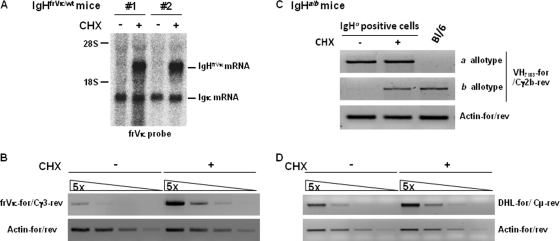
To determine whether mRNAs from nonproductively rearranged IgH alleles were sensitive to NMD, B cells were isolated from IgHfrVκ/wt and wt mice and were stimulated with LPS. Cells were then treated or not with cycloheximide (CHX), a protein synthesis inhibitor used to prevent degradation by NMD (10). In B cells isolated from IgHfrVκ/wt mice, Northern blot analysis revealed that levels of nonfunctional VDJ-rearranged mRNAs were strongly increased after CHX treatment and, hence, that a very efficient NMD process allowed an almost complete disappearance of nonproductive IgHfrVκ transcripts (Fig. 5A). Likewise, we found a robust NMD of IgHfrVκ mRNAs in B cells harboring class switch recombination (CSR) to Cγ3 (Fig. 5B). Next, we confirmed that NMD degrades almost all nonproductive VDJ-rearranged IgH mRNAs in wt (129/B6 F1) mice harboring allotypic differences on their IgH alleles (IgHa/b) (Fig. 5C). To this aim, IgMa-positive B cells were sorted by fluorescence-activated cell sorter (FACS) (Fig. 3D) and stimulated with LPS for 3 days. RT-PCR was then performed by using a reverse Cγ2b primer specific for the b allotype. As shown in Fig. 5C, levels of nonproductive VDJ-Cγ2bb mRNAs were strongly increased after CHX treatment. Regarding DJ-recombined mRNAs, they were extremely rare and undetectable by Northern blotting (Fig. 5A); as PTC-containing transcripts, they were also shown to be degraded by NMD (Fig. 5D). Thus, mRNAs arising from nonproductive VDJ- or DJ-rearranged IgH alleles are highly sensitive to NMD in LPS-stimulated B cells.
Fig 5.
Nonfunctional VDJ- and DJ-rearranged IgH mRNAs are efficiently degraded by NMD. mRNA levels from nonproductively rearranged IgHfrVκ or IgHwt alleles in LPS-stimulated B cells treated or not with CHX on day 4 were determined. (A) Northern blotting was performed by using the frVκ probe that is specific for Vκ6 segments and allows the simultaneous identification of IgHfrVκ and Igκ (normalization) mRNAs. The stimulation of B cells with LPS in the absence of additional cytokines induces preferential class switch recombination (CSR) to γ2b and γ3 isotypes. (B) Nonproductive mRNAs from switched IgHfrVκ alleles were assessed by semiquantitative PCR using primers frVκ-for and Cγ3-rev (primer set k/h in Fig. 1A). (C) IgMa-positive B cells were sorted from 129/B6 F1 (IgHa/b) mice (as described in the legend to Fig. 3D) and stimulated with LPS for 4 days. Nonproductive VDJ-rearranged mRNAs from switched IgHb alleles were then analyzed by using a consensus VH7183 forward primer (VH7183-for) and allotype-specific Cγ2b reverse primers (primer set a/i in Fig. 1A). (D) Degradation of DJ-Cμ mRNAs by NMD was assayed with wt mice by semiquantitative RT-PCR using a consensus forward primer (DHL-for) located at the 5′ end of most DH segments (primer set d/g in Fig. 1A). Representative results from 3 to 4 different mice (A, B, and D) or from 2 independent cell sorting experiments (C) are shown.
Levels of both RNA splicing and NMD of nonproductive IgH transcripts are increased upon B cell stimulation.
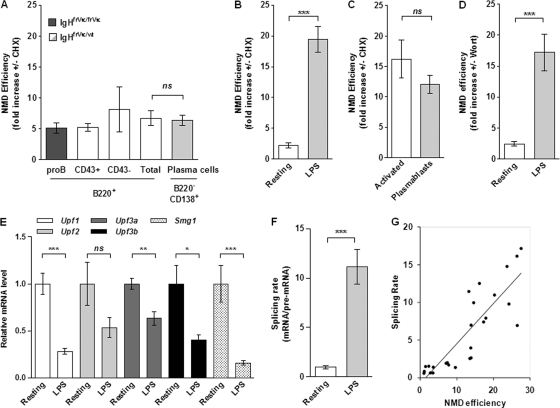
Next, we sought to determine whether variations in the NMD efficiency occur during B cell development. For this purpose, B cell populations were isolated from heterozygous (Fig. 3A to C) and homozygous (pro-B cells) IgHfrVκ mice and were treated or not with CHX. We measured nonfunctional Igμ mRNAs by quantitative PCR (qPCR), and the fold increase obtained after CHX treatment was used to determine the NMD efficiency (Fig. 6A to C and see Fig. S2A in the supplemental material). In bone marrow, sorted B and plasma cell populations exhibited similar NMD efficiencies, which ensured about a 5-fold degradation (i.e., ∼80%) of nonproductive Igμ mRNAs (Fig. 6A). However, the NMD efficiency dropped significantly in resting B cells, with only a 2-fold increase (i.e., ∼50% degradation) upon CHX treatment (Fig. 6B). Interestingly, a strong improvement of the NMD efficiency was observed upon LPS stimulation, leading to a 20-fold degradation (i.e., ∼95%) of nonproductive Igμ mRNAs (Fig. 6B). In addition, we found a strong and similar NMD efficiency for activated B cells and plasmablasts sorted 4 days after LPS stimulation (Fig. 3C and 6C). Therefore, the equivalent degradations by NMD observed for activated B cells and plasmablasts (Fig. 6C) and for bone marrow B and plasma cells (Fig. 6A) indicate that the NMD efficiency is not further enhanced during plasma cell differentiation.
Fig 6.
Correlation between the RNA splicing rate and the extent of degradation by NMD. The degradation of nonproductive Igμ mRNAs was assessed with pro-B cells (dark gray bar) isolated from homozygous IgHfrVκ/frVκ mice and with the B cell populations (empty and light gray bars) described in the legend for Fig. 3A to C. (A to D) RT-qPCRs were performed with cells treated or not with CHX (A to C) or wortmannin (Wort) (D) using primers frVκ-for and Cμ-rev (primer set k/g in Fig. 1A). Relative mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene transcripts, and the NMD efficiency corresponds to the fold increase in levels of nonproductive Igμ mRNAs upon treatment with inhibitors (see Fig. S2A in the supplemental material). (E) Relative expression levels of Upf1, Upf2, Upf3a, Upf3b, and Smg1 were determined for resting B cells (with a mean set to 1) and LPS-stimulated B cells after normalization to GAPDH gene transcripts. (F) Splicing rates correspond to the nonproductive mRNA/pre-mRNA ratio obtained for CHX-treated cells. Values obtained for resting B cells were set to 1. (G) The NMD efficiency was plotted against the splicing rate using data from resting and LPS-stimulated B cells (B and F) and those from activated B cells and plasmablasts (C; also see Fig. S2B in the supplemental material). Results from 3 (A, pro-B cells) or 5 to 7 independent cell sorting experiments are shown. (ns, P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
We also analyzed whether the degradation of nonproductive Igμ mRNAs was sensitive to wortmannin (Fig. 6D). This drug inhibits the phosphorylation of Upf1 by the phosphatidylinositol 3-kinase (PI3K)-related kinase Smg-1 and prevents the activation of the canonical Upf1-dependent NMD pathway (41). For resting and LPS-stimulated B cells, we found that the NMD efficiencies were similar upon CHX (Fig. 6B) and wortmannin (Fig. 6D) treatments. Therefore, we conclude that the degradation of nonproductive Igμ mRNAs involves the canonical Upf1-dependent NMD pathway. It was previously reported that the NMD efficiency does not correlate with the expression levels of genes encoding NMD factors (52). Likewise, we found that the level of expression of the Upf1, Upf2, Upf3, and Smg1 genes was decreased in LPS-stimulated B cells (Fig. 6E), whereas the degradation by NMD was strongly enhanced (Fig. 6B). Hence, the absence of a correlation between the expression of genes involved in NMD and the NMD efficiency indicates that these NMD factors (Upf1, Upf2, Upf3, and Smg1) are not rate limiting in B cells.
Next, we examined whether a correlation exists between the level of RNA splicing and the efficiency of NMD. We determined mRNA/pre-mRNA ratios as previously described to measure splicing rates (3, 27). Because almost all nonproductive Igμ mRNAs were degraded by NMD, splicing rates were quantified by using CDNAs from CHX-treated cells. We observed efficient RNA splicing in LPS-stimulated B cells compared to resting B cells (Fig. 6F). We also found similar splicing rates for activated B cells and plasmablasts (see Fig. S2B in the supplemental material), and these data are consistent with the equivalent NMD efficiencies observed for those cells (Fig. 6C). Altogether, the positive correlation retrieved after the plotting of the NMD efficiency against the splicing rate (Fig. 6G) indicates that the level of RNA splicing may influence the downstream degradation of nonproductive Igμ mRNAs by NMD.
Cooperation between RNA surveillance mechanisms.
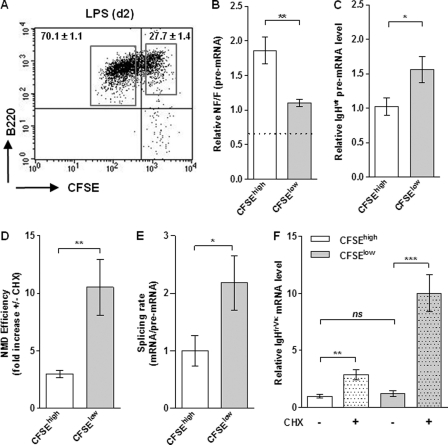
Compared to resting B cells, NMUP decreased (Fig. 4B) whereas the NMD efficiency was strongly enhanced (Fig. 6B) after LPS stimulation. These data strongly suggest that the magnitude of RNA surveillance mechanisms is linked to B cell activation. To model this issue with closely related cells, resting B cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE) before LPS stimulation, and the transcription and RNA surveillance of nonproductive IgH alleles in undivided (CFSEhigh) and divided (CFSElow) cells, sorted at day 2, were then analyzed (Fig. 7A). As expected, the vast majority of cells performed cellular divisions after stimulation (Fig. 7A and see Fig. S3 in the supplemental material). Compared to undivided (CFSEhigh) cells, we found a significant drop in NF/F pre-mRNA ratios (Fig. 7B) and an elevated IgH transcription level (Fig. 7C) in divided (CFSElow) cells. Likewise, the level of degradation by NMD was significantly increased in CFSElow cells compared to undivided cells (Fig. 7D), and this increase did not correlate with the expression of the Upf1, Upf2, Upf3, and Smg1 genes (see Fig. S4 in the supplemental material). RNA splicing rates were also higher in CFSElow than in CFSEhigh cells (Fig. 7E), and this is consistent with the link between RNA splicing and NMD (Fig. 6G). Lastly, we found that while CFSElow and CFSEhigh B cells exhibited strong variations in NMUP and NMD efficiencies, the levels of nonproductive Igμ mRNAs remained remarkably constant in those populations (without CHX) (Fig. 7F) and extremely low, as indicated by Northern blotting, in LPS-stimulated B cells (without CHX) (Fig. 5A). The same observation was made for activated B cells compared to plasma cells (without CHX) (see Fig. S2A in the supplemental material). Therefore, these data reveal compensatory effects of NMUP and NMD mechanisms and that B cell activation and/or proliferation triggers qualitative changes in the RNA surveillance machinery.
Fig 7.
Regulation of RNA surveillance mechanisms in undivided and divided cells. (A) Resting B cells from IgHfrVκ/wt mice were stained with CFSE and then stimulated with LPS for 2 days. A representative dot plot indicates the frequency of divided (CFSElow) and nondivided (CFSEhigh) B cells and the gates used for cell sorting. (B and C) In those sorted cells, NF/F pre-mRNA ratios (B) and relative levels of functional IgH pre-mRNA (C) were obtained as described in the legend to Fig. 4. (D) The NMD efficiency was assessed as described in the legend to Fig. 6 and corresponds to the fold increase in levels of nonproductive Igμ mRNAs upon treatment with CHX. (E) For each sorted population, the splicing efficiency of nonfunctional IgHfrVκ transcripts was calculated for CHX-treated cells by dividing mRNA by pre-mRNA levels, as described in the legend to Fig. 6F. Values obtained for CFSEhigh cells were set to 1. (F) Relative levels of nonproductive Igμ mRNAs were calculated for cells treated (+) or not treated (−) with CHX after normalization to GAPDH gene transcripts. Values obtained for CFSEhigh cells (without CHX) were set to 1. Results from 7 independent cell sorting experiments are shown. (ns, P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
DISCUSSION
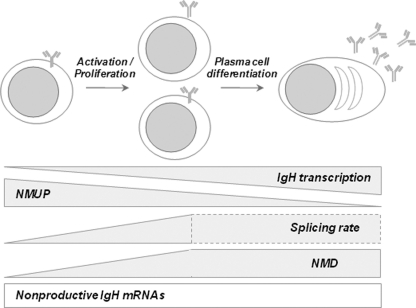
In the present study, we confirm that IgH transcription is mainly biallelic during B cell development (18), and we show that the RNA surveillance machinery ensures the monoallelic expression of functional IgH proteins by lowering the mRNA amount of nonproductively rearranged alleles. Furthermore, upon B cell activation and plasma cell differentiation, we observed a strong correlation between the magnitude of IgH transcription, the level of RNA splicing, and the degradation of nonproductive IgH mRNAs by NMD (Fig. 8).
Fig 8.
Model for the regulation of IgH transcription and RNA surveillance during B cell differentiation. Although the transcriptional pattern of IgH alleles is mainly biallelic during B cell development, RNA surveillance mechanisms strongly decrease the mRNA amount of nonproductively rearranged alleles (empty rectangle) and thus ensure the monoallelic expression of functional alleles. This model shows that the improvement in levels of IgH transcription, upon B cell activation and during plasma cell differentiation, is associated with a low level of NMUP and a strong degradation by NMD. Similar RNA splicing rates and NMD efficiencies in plasma cells and activated B cells are also depicted. Dotted lines indicate that our analysis accounts only for the splicing of full-length nonproductive Igμ mRNAs and hence might minimize the overall splicing rate by excluding alternatively spliced mRNAs (see also Discussion).
The IgHfrVκ/wt mouse model proves very useful to track both productive and nonproductive IgH alleles simultaneously. Our ChIP data show that both IgH alleles exhibit hallmarks of active chromatin and display equivalent RNAPII loading after B cell stimulation (Fig. 2). Likewise, we previously found a similar frequency of transcription-dependent somatic hypermutation (SHM) on both VDJ-rearranged alleles in germinal center B cells (21). This finding is in contrast to previously reported observations by Skok and coworkers showing that IgH alleles displayed a monoallelic expression pattern 4 days after B cell activation (47). For LPS-stimulated cells, we also found that pre-mRNA levels from both IgH alleles were strongly increased in plasmablasts compared to activated B cells, arguing against a transcriptional silencing of nonfunctional alleles in terminally differentiated plasma cells (Fig. 4D and see Fig. S2C in the supplemental material). Furthermore, the analysis of NF/F pre-mRNA ratios demonstrates that nonproductive IgH alleles are transcribed throughout B cell development and accumulate as unspliced transcripts (Fig. 4). Another major conclusion from our study is the lack of a physiological role of NMTGS during B cell development. These data are consistent with previously reported observations of NFS70 pro-B cells (23) and suggest that the transcriptional silencing by NMTGS might be restricted to nonproductive Ig transgenes in non-B cell lines (8, 48).
Previous reports, including ours, have documented NMUP for Igκ, Igμ, and TCRβ transcripts, showing that the presence of a PTC induces a specific accumulation of pre-mRNAs (2, 15, 30, 34, 39). The present study provides evidence for a decrease in NMUP as the level of IgH transcription increases. Accordingly, NMUP is downregulated upon B cell stimulation and during plasma cell differentiation. Compared to resting B cells, which exhibited high NF/F pre-mRNA ratios, a low level of accumulation of NF pre-mRNAs was found for LPS-stimulated B cells. This decrease of NF/F pre-mRNA ratios should help to reconcile previous findings, since it confirms that pre-mRNAs from one IgH allele decrease after B cell activation (47). However, our data strongly suggest that it is caused by variations of NMUP rather than the transcriptional silencing of nonproductively rearranged IgH alleles. Further studies using the IgHfrVκ/wt mouse strain should help to determine whether nonfunctional IgH pre-mRNAs accumulate near the site of transcription in primary B cells, as observed previously for hybridomas (39).
Although it has been demonstrated that the NMD efficiency displays variability among different mouse tissues (52), less is known about the importance of NMD during B cell differentiation. Here, we show that nonproductive VDJ- or DJ-rearranged alleles are degraded by NMD. Interestingly, we provide evidence that the degradation of nonproductive Igμ mRNAs by NMD fluctuates according to the proliferation and/or activation status of B cells. Since the level of degradation by NMD is high (>80%) in long-lived bone marrow plasma cells compared to resting B cells (∼50%), we posit that the NMD efficiency is not intrinsically linked to B cell proliferation and is rather influenced by the activation status. Although they deserve further investigations, the proliferative response and B cell activation are inherently connected, and it can be difficult to definitely distinguish between their respective influences.
Bruno et al. have recently shown that microRNA-128 (miR-128) represses NMD by decreasing the level of expression of Upf1 (by ∼5-fold) during brain development (7). However, we observed no correlation between the NMD efficiency and the expression of genes involved in NMD (Fig. 6E and see Fig. S4 in the supplemental material), suggesting that the regulation of NMD might be different in B cells. In addition, Upf1 mRNA levels decreased by ∼4-fold upon LPS stimulation, whereas we found that the NMD efficiency increased by ∼10-fold (Fig. 6B). Likewise, a strong expression of NMD factors and a low NMD efficiency in testis were found previously, and these factors obviously do not appear to be rate limiting (52).
Compared to resting B cells, we also found that the RNA splicing efficiency was strongly increased upon B cell activation and that the magnitude of NMD was correlated with RNA splicing rates (Fig. 6G). Hence, these results are consistent with data from Gudikote and coworkers showing that RNA splicing promotes translation and RNA surveillance of nonsense transcripts in HeLa cells (27).
It is now well accepted that transcription and splicing are connected (19, 42) and that the RNAPII elongation rate can influence the splicing efficiency (40, 46). Although our analysis is limited to one location (i.e., the JH-Cμ region) and is not representative of the activity of the whole expressed IgH gene, we speculate that cotranscriptional splicing occurs on highly transcribed IgH alleles and precludes strong NMUP in plasma cells and LPS-stimulated B cells. Since introns can vary in their rates of splicing, and the RNAPII density can vary across genes, other models are required to analyze whether transcription and splicing are similarly correlated all along the IgH locus.
It was reported previously that the rate of RNAPII elongation shows some increase in plasma cells (45). We indeed observed a boost of the IgH transcription level and a low level of NMUP in plasma cells compared to B cells (Fig. 4). However, upon the quantification of full-length nonproductive Igμ mRNAs, we found similar levels of RNA splicing in plasmablasts and activated B cells (see Fig. S2 in the supplemental material). Because our methodology excludes the analysis of alternatively spliced mRNAs with skipped frVκ or CH1μ exons, we hypothesize that high levels of nonsense-associated altered splicing (NAS) in plasma cells might underestimate the overall splicing rate of nonproductive Igμ transcripts. Although nonproductive transcripts that have skipped the frVκ exon are hardly distinguishable from productive Igμ mRNAs, this assumption is in agreement with our previous findings with cell lines showing that plasma cells exhibit a higher NAS efficiency than B cells (15).
In summary, our study provides evidence for an equivalent biallelic transcription of VDJ-rearranged IgH alleles rather than a transcriptional silencing of nonfunctional alleles and that cumulative RNA surveillance mechanisms ensure an almost complete disappearance of nonproductively rearranged IgH mRNAs throughout B cell development. In addition, our data indicate that the increased IgH transcription after B cell activation correlates with a low NMUP efficiency and also with strong RNA splicing and degradation of nonfunctional IgH mRNAs by NMD. Altogether, RNA surveillance mechanisms can somehow relay to each other to finally achieve the monoallelic expression of functional IgH proteins during B cell development.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the animal facility at the Institut Fédératif de Recherche 145 (IFR 145) (GEIST, Limoges, France) and C. Ouk-Martin (flow cytometric platform) for their technical assistance.
This work was supported by grants from La Ligue Nationale contre le Cancer (comité départemental de la Haute-Vienne) and Conseil Régional du Limousin. A.T. is funded by a French government fellowship.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abraham RT. 2004. The ATM-related kinase, hSMG-1, bridges genome and RNA surveillance pathways. DNA Repair (Amst.) 3:919–925 [DOI] [PubMed] [Google Scholar]
- 2. Aoufouchi S, Yelamos J, Milstein C. 1996. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell 85:415–422 [DOI] [PubMed] [Google Scholar]
- 3. Audibert A, Weil D, Dautry F. 2002. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 22:6706–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azzalin CM, Lingner J. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16:433–439 [DOI] [PubMed] [Google Scholar]
- 5. Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318:798–801 [DOI] [PubMed] [Google Scholar]
- 6. Baumann B, Potash MJ, Köhler G. 1985. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 4:351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bruno IG, et al. 2011. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42:500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bühler M, Mohn F, Stalder L, Mühlemann O. 2005. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Mol. Cell 18:307–317 [DOI] [PubMed] [Google Scholar]
- 9. Bühler M, Paillusson A, Mühlemann O. 2004. Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 32:3304–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carter MS, et al. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270:28995–29003 [DOI] [PubMed] [Google Scholar]
- 11. Carter MS, Li S, Wilkinson MF. 1996. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 15:5965–5975 [PMC free article] [PubMed] [Google Scholar]
- 12. Casola S, et al. 2004. B cell receptor signal strength determines B cell fate. Nat. Immunol. 5:317–327 [DOI] [PubMed] [Google Scholar]
- 13. Cenci S, van Anken E, Sitia R. 2011. Proteostenosis and plasma cell pathophysiology. Curr. Opin. Cell Biol. 23:216–222 [DOI] [PubMed] [Google Scholar]
- 14. Chang YF, Imam JS, Wilkinson MF. 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76:51–74 [DOI] [PubMed] [Google Scholar]
- 15. Chemin G, et al. 2010. Multiple RNA surveillance mechanisms cooperate to reduce the amount of nonfunctional Ig kappa transcripts. J. Immunol. 184:5009–5017 [DOI] [PubMed] [Google Scholar]
- 16. Chen-Bettecken U, Wecker E, Schimpl A. 1987. Transcriptional control of mu- and kappa-gene expression in resting and bacterial lipopolysaccharide-activated normal B cells. Immunobiology 174:162–176 [DOI] [PubMed] [Google Scholar]
- 17. Connor A, Wiersma E, Shulman MJ. 1994. On the linkage between RNA processing and RNA translatability. J. Biol. Chem. 269:25178–25184 [PubMed] [Google Scholar]
- 18. Daly J, Licence S, Nanou A, Morgan G, Mårtensson IL. 2007. Transcription of productive and nonproductive VDJ-recombined alleles after IgH allelic exclusion. EMBO J. 26:4273–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das R, et al. 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 20:1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delpy L, Le Bert M, Cogné M, Khamlichi AA. 2003. Germ-line transcription occurs on both the functional and the non-functional alleles of immunoglobulin constant heavy chain genes. Eur. J. Immunol. 33:2108–2113 [DOI] [PubMed] [Google Scholar]
- 21. Delpy L, Sirac C, Le Morvan C, Cogné M. 2004. Transcription-dependent somatic hypermutation occurs at similar levels on functional and nonfunctional rearranged IgH alleles. J. Immunol. 173:1842–1848 [DOI] [PubMed] [Google Scholar]
- 22. Delpy L, Sirac C, Magnoux E, Duchez S, Cogné M. 2004. RNA surveillance down-regulates expression of nonfunctional kappa alleles and detects premature termination within the last kappa exon. Proc. Natl. Acad. Sci. U. S. A. 101:7375–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eberle AB, Herrmann K, Jäck HM, Mühlemann O. 2009. Equal transcription rates of productively and nonproductively rearranged immunoglobulin mu heavy chain alleles in a pro-B cell line. RNA 15:1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehlich A, Martin V, Müller W, Rajewsky K. 1994. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4:573–583 [DOI] [PubMed] [Google Scholar]
- 25. Fisher AG. 2002. Cellular identity and lineage choice. Nat. Rev. Immunol. 2:977–982 [DOI] [PubMed] [Google Scholar]
- 26. Frischmeyer-Guerrerio PA, et al. 2011. Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 108:10638–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. 2005. RNA splicing promotes translation and RNA surveillance. Nat. Struct. Mol. Biol. 12:801–809 [DOI] [PubMed] [Google Scholar]
- 28. Hwang J, Maquat LE. 2011. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 4:422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iacovoni JS, et al. 2010. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29:1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imam JS, Gudikote JP, Chan WK, Wilkinson MF. 2011. Frame-disrupting mutations elicit pre-mRNA accumulation independently of frame disruption. Nucleic Acids Res. 38:1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Isken O, Maquat LE. 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9:699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwakoshi NN, et al. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329 [DOI] [PubMed] [Google Scholar]
- 33. Jäck HM, Berg J, Wabl M. 1989. Translation affects immunoglobulin mRNA stability. Eur. J. Immunol. 19:843–847 [DOI] [PubMed] [Google Scholar]
- 34. Lozano F, Maertzdorf B, Pannell R, Milstein C. 1994. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 13:4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu ZP, Ju ZL, Shi GY, Zhang JW, Sun J. 2005. Histone deacetylase inhibitor trichostatin A reduces anti-DNA autoantibody production and represses IgH gene transcription. Biochem. Biophys. Res. Commun. 330:204–209 [DOI] [PubMed] [Google Scholar]
- 36. Lutz J, et al. 2011. Pro-B cells sense productive immunoglobulin heavy chain rearrangement irrespective of polypeptide production. Proc. Natl. Acad. Sci. U. S. A. 108:10644–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McIlwain DR, et al. 2010. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. U. S. A. 107:12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Medghalchi SM, et al. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10:99–105 [DOI] [PubMed] [Google Scholar]
- 39. Mühlemann O, et al. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8:33–43 [DOI] [PubMed] [Google Scholar]
- 40. Nogués G, et al. 2003. Control of alternative pre-mRNA splicing by RNA Pol II elongation: faster is not always better. IUBMB Life 55:235–241 [DOI] [PubMed] [Google Scholar]
- 41. Pal M, Ishigaki Y, Nagy E, Maquat LE. 2001. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7:5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pandit S, Wang D, Fu XD. 2008. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell Biol. 20:260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qian L, et al. 1993. T cell receptor-beta mRNA splicing: regulation of unusual splicing intermediates. Mol. Cell. Biol. 13:1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reynaud S, et al. 2005. Interallelic class switch recombination contributes significantly to class switching in mouse B cells. J. Immunol. 174:6176–6183 [DOI] [PubMed] [Google Scholar]
- 45. Santos P, Arumemi F, Park KS, Borghesi L, Milcarek C. 2011. Transcriptional and epigenetic regulation of B cell development. Immunol. Res. 50:105–112 [DOI] [PubMed] [Google Scholar]
- 46. Schwartz S, Ast G. 2011. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 29:1629–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Skok JA, et al. 2001. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2:848–854 [DOI] [PubMed] [Google Scholar]
- 48. Stalder L, Mühlemann O. 2007. Transcriptional silencing of nonsense codon-containing immunoglobulin micro genes requires translation of its mRNA. J. Biol. Chem. 282:16079–16085 [DOI] [PubMed] [Google Scholar]
- 49. ten Boekel E, Melchers F, Rolink A. 1995. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int. Immunol. 7:1013–1019 [DOI] [PubMed] [Google Scholar]
- 50. Weischenfeldt J, et al. 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22:1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wittmann J, Hol EM, Jäck HM. 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 26:1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zetoune AB, et al. 2008. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.