Abstract
Sperm motility and hence male fertility strictly depends on proper development of the sperm tail and its tight anchorage to the head. The main protein of sperm tail outer dense fibers, ODF1/HSPB10, belongs to the family of small heat shock proteins that function as molecular chaperones. However, the impact of ODF1 on sperm tail formation and motility and on male fecundity is unknown. We therefore generated mutant mice in which the Odf1 gene was disrupted. Heterozygous mutant male mice are fertile while sperm motility is reduced, but Odf1-deficient male mice are infertile due to the detachment of the sperm head. Although headless tails are somehow motile, transmission electron microscopy revealed disturbed organization of the mitochondrial sheath, as well as of the outer dense fibers. Our results thus suggest that ODF1, besides being involved in the correct arrangement of mitochondrial sheath and outer dense fibers, is essential for rigid junction of sperm head and tail. Loss of function of ODF1, therefore, might account for some of the cases of human infertility with decapitated sperm heads. In addition, since sperm motility is already affected in heterozygous mice, impairment of ODF1 might even account for some cases of reduced fertility in male patients.
INTRODUCTION
Heat shock proteins play an essential role as molecular chaperones by preventing the aggregation of misfolded proteins and, therefore, execute strong cytoprotective effects. A structurally divergent group within the molecular chaperone family is constituted by the small heat shock proteins (sHSPs). The superfamily of sHSPs comprises 10 proteins with molecular masses ranging from 12 to 43 kDa. Their most pronounced structural feature is the conserved α-crystallin domain of 80 to 100 amino acids. sHSPs interact with a lot of essential cellular structures. Hence, sHSPs seem to be implicated in a wide variety of human diseases (23, 32, 54, 55).
The most recent protein assigned to sHSP family is ODF1, based on its overall structural features and especially on its conserved α-crystallin domain that are characteristic of sHSPs (19). ODF1 was, therefore, also called HSPB10 (19). ODF1/HSPB10 is the major protein of the mammalian sperm tail outer dense fibers (ODFs), which are cytoskeletal structures specifically found in the sperm tails of vertebrates but not in eukaryotic cilia and flagella in general (16). ODFs are accessory fibers accompanying the axonemal tubuli doublets on their outer site. They are composed of more than a dozen different proteins of which only a few have been identified (43, 46, 61). Although not involved in active motility, ODFs seem to be important for the stability and the elastic recoil of the sperm tail, as well as for support of the flagellar beat (3, 34). Impaired development of the ODFs has been described as a major cause of tail abnormalities in infertile men, indicating an important function in sperm motility and/or morphology (21).
ODF1/HSPB10 has a molecular mass of ∼27 kDa with a high content of cysteine (between 13.7 and 17%) and proline (up to 10%), capable of forming disulfides (8, 9, 20, 25, 40, 60). The high cysteine content of ODF1 may provide free sulfhydryl groups for the retention of zinc ions that are enriched in the sperm tail (11). Cysteine and proline are mainly found in the C-terminal end of ODF1 as a repetitive tripeptide motif of C-X-P. Although variability of C-X-P repeat frequencies has been reported, it did not affect male fertility in humans (24).
ODF1 is a self-interacting protein, albeit its self-association is weak (50). Moreover, several binding partners were identified over the past years, including ODF2. Interaction between ODF1 and ODF2 was proven in a yeast two-hybrid assay and was shown to depend on their leucine-zipper motifs (51). The scaffold of the ODF fiber may thus be structured by the two main ODF proteins, ODF1 and ODF2 (46). Other ODF1 interacting proteins are SPAG4, KLC3 (kinesin light chain 3), OIP1 (ODF1-interacting protein), and SPAG5 (4, 18, 52, 63). SPAG4 is located in the transient manchette and associated with the axoneme in elongating spermatids and epididymal sperm. It binds to ODF1, but not to ODF2 (52). SPAG5 is found in rat elongated spermatids and epididymal sperm. SPAG5 associates with ODF, but not with the axonemal microtubules (18). OIP1 is a member of the RING finger family that are often E3 ubiquitin protein ligases. OIP1 binds to the conserved C-X-P motif in the C-terminal end of ODF1 (63), and ODF1-OIP1 interaction is enhanced by Cdk5/p35-mediated ODF1 phosphorylation (48). In addition to being components of the ODFs, ODF1, ODF2, and ODF1 interacting protein SPAG4 have all been located to the manchette, a transient microtubular structure that seems to be important for the shaping of the sperm nucleus and the development of the sperm tail (29).
The expression of mouse Odf1 is restricted to testis and is first detectable at the round spermatid stage. In addition, immunoelectron microscopy revealed that ODF1 protein is specifically located to the ODFs (10, 40). The Odf1 gene, which is a single copy gene located in mice to chromosome 15 region B2-C, as well as its encoded protein, are highly conserved in evolution (20, 46), indicating an important function in sperm motility and/or morphology. To study the role of ODF1 in sperm function, we generated Odf1-deficient mice and show here that ODF1 is essential for male fertility. Homozygous ODF1-deficient male mice are infertile, whereas heterozygous male mice are fertile but show reduced sperm motility. Male infertility by loss of ODF1 resulted from a faulty sperm head-to-tail linkage.
MATERIALS AND METHODS
Generation of the targeting vector.
All mouse experiments were reviewed and approved by the local ethics commission. Licensing for animal experiments was obtained by the Institute of Human Genetics. A genomic Odf1 cosmid clone (121 J 1787Q3; 129/Ola) was obtained from the Resource Centre of the German Human Genome Project at the Max-Planck-Institute for Molecular Genetics (Berlin, Germany) and mapped for restriction enzyme recognition sites. To assemble the targeting vector, an XbaI fragment containing ∼4 kb of the 5′ region, including the promoter region, as well as exon1, was subcloned into pGEMT (Promega, Madison, WI), and exoIII deletions, starting from the 3′ region, were performed (ExoIII/S1 deletion kit; MBI Fermentas, St. Leon-Rot, France). A fragment of ∼3 kb consisting of the upstream genomic region of the Odf1 gene was first subcloned via XbaI/EcoRI into pBluescript (Stratagene, La Jolla, CA), followed by NotI/XhoI cloning into pPNT-M1 (59). The ∼3.8-kb PstI intron fragment was first subcloned into pBluescript (Stratagene), followed by XbaI/EcoRI directional cloning into pPNT-M1. The resulting targeting vector (Fig. 1A) was sequenced to verify the correct orientations of both fragments. The targeting construct was linearized at the unique NotI site before electroporation.
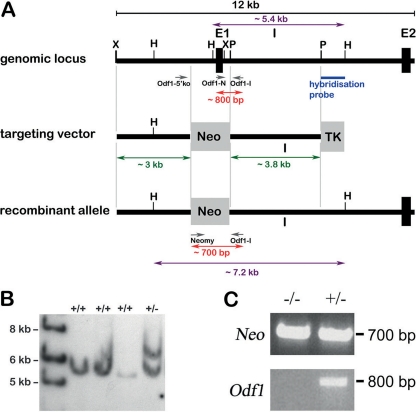
Fig 1.
Targeted disruption of the promoter region and of exon 1 of the Odf1 gene. (A) Structure of the wild-type allele, targeting vector, and recombinant allele are shown, together with relevant restriction sites, primer positions, and external hybridization probe position. The targeting vector consists of 5′ upstream region (∼3 kb) and intronic sequences (∼3.8 kb) flanking the pgk-neomycin selection cassette (Neo). TK, thymidine kinase cassette; X, XbaI; H, HindIII; P, PstI; E1, exon 1; E2, exon 2; I, intron; Odf1-5′ko, Odf1-I, Odf1-N, and Neomy, primers used for genotyping. Southern blot hybridization of HindIII-digested genomic DNA with the 3′ external probe resulted in an ∼5.4-kb fragment of the wild-type allele and a ∼7.2-kb fragment of the recombined allele. Genotyping with primer pair Odf1-N/Odf1-I resulted in an ∼800-bp fragment of the wild-type allele, and genotyping with the primer pair Neomy/Odf1-I resulted in an ∼700-bp fragment of the recombined allele. (B) Southern blot hybridization of HindIII-digested genomic DNA isolated from individual ES cell colonies generated by electroporation with the targeting construct, followed by selection with G418 and ganciclovir. Hybridization with the 3′ external probe to a fragment of ∼7.2 kb revealed homologous recombination. The fragment of the wild-type allele is shown at ∼5.4 kb. +/+, no homologous recombination; +/−, homologous recombination. (C) Genotyping using the primer pair Neomy/Odf1-I for detection of the recombined allele (Neo) and the primer pair Odf1-N/Odf1-I for detection of the wild-type allele (Odf1) in homozygous (−/−) and heterozygous (+/−) mice.
Generation of the 3′ external probe and Southern blot hybridization.
The 3′ external hybridization probe was generated by PCR amplification using the primer pair Odf1SPst (CTGCAGCTCAGAGTCTTGTCTGTG) and Odf1SHindIII (AAGCTTAGACTGTTCTCCACAG) out of a genomic Odf1 clone and cloned into pGEMT.
For Southern blot hybridization, genomic DNA was extracted from embryonic stem (ES) cells, digested with HindIII, electrophoretically separated on agarose gels, and transferred onto Hybond N membranes (Amersham, Freiburg, Germany) by capillary blotting. The 3′ external probe was labeled with [32P]dATP by the random hexanucleotide primer method (17), and hybridized overnight in 5× SSPE (20× SSPE is 3.6 M NaCl, 0.2 M NaH2PO4, and 0.02 M Na2EDTA at pH 7.7), 5× Denhardt's solution, 0.1% SDS (sodium dodecyl sulfate), and 100 μg of denatured salmon sperm DNA/ml (53) at 65°C. Filters were washed twice at room temperature in 2× SSC (20× SSC is 3 M NaCl plus 0.3 M trisodium citrate at pH 7.0), then in 1× SSC–0.1% SDS, and finally in 0.1× SSC–0.1% SDS at hybridization temperature.
ES cell culture and generation of chimeric mice.
ES cell line R1 (provided by A. Nagy, Toronto, Ontario, Canada) was cultured as described previously (27). Trypsinized and resuspended ES cells were mixed with 50 mg of linearized target vector and electroporated at 250 V and 500 μF using a Bio-Rad gene pulser apparatus (Bio-Rad, Munich, Germany). Cells were plated in nonselective medium containing leukemia inhibitory factor in the presence of G418-resistant embryonic mouse fibroblasts. Selection for homologous recombined ES cells was carried out with medium containing G418 at 350 μg/ml and ganciclovir at 2 μmol/liter. After 10 days of selection, individual drug-resistant clones were picked into 24-well trays. Three days later, individual recombinant ES clones were replicated into 24-well trays for freezing and for isolation of genomic DNA. A total of 10 to 15 compacted recombinant ES cells were microinjected into 3.5-day-old embryos of the C57BL/6J mouse strain (27). The chimeric male mice generated were mated with C57BL/6J females to produce offspring.
Genotyping.
Genomic DNA from tail tip biopsy specimens was isolated using Viagen DirectPCR-Tail (Viagen Biotech) and proteinase K digestion overnight. Genotyping was performed with the following primer pairs: Odf1-I (ATCAACTCTGCCTGAGAC) and Neomy (CCTTCTATCGCCTCCTTGACG) for detection of the neomycin cassette in the recombined allele, Odf1-5′ko (AGGAAGAAGGGACTAGAG) and Odf1-I for differentiation between the wild-type allele of ∼1.5 kb and the recombined allele of ∼ 2.4 kb, and Odf1-I and Odf1-N (GAGCTCAAGCTTTGGCCGCACTGAGTTGTC) for detection of the wild-type allele. We routinely performed a first PCR with the primer pair Odf1-I/Neomy for detection of the recombined allele and a second PCR with the primer pair Odf1-I/Odf1-N for detection of the wild-type allele and to discriminate between heterozygous and homozygous animals.
Fertility test.
The fertility of the Odf1-deficient males on a mixed background (C57BL/6J × 129/Sv) was investigated by mating with wild-type females. Mating tests were performed with 11 homozygous male mice (Odf1−/−) for at least 3 months. As a control group, six of their male littermates with a heterozygous genotype (Odf1+/−) were mated with wild-type females. Females were checked for the presence of vaginal plugs and/or pregnancy. Pregnant females were removed to holding cages to give birth. The numbers and size of litters sired by each group of males were determined.
Reverse transcriptase PCR.
Total RNA was extracted from testes of wild-type, heterozygous, and homozygous Odf1 knockout mice, respectively, using peqGOLD RNAPure (PEQLAB, Erlangen, Germany) and digested with RQ1 DNase. RNA was reverse transcribed using RevertAid H Minus First Strand cDNA synthesis kit (Fermentas) and oligo(dT)18 primer, followed by PCR. The sequences of primers used and their corresponding PCR parameters (annealing temperature, elongation time, and size of amplified fragments) are shown in Table 1. PCR amplification was performed with an initial denaturation step at 95°C for 3 min, followed by 36 cycles at 95°C for 30 s, annealing for 30 s, and elongation at 72°C, and a final extension step at 72°C for 8 min.
Table 1.
Primer sequences, reaction conditions, and product lengths
| Target gene | Primer | Sequence (5′–3′) | Annealing temp (°C) | Elongation time | Amplified fragment size (bp) |
|---|---|---|---|---|---|
| SYCP3 | MSYCP35′Kpn1 | GACGGTACCATGCTTCGAGGGTGTGGG | 52 | 1 min | 900 |
| MSYCP3r | TTGACACAATCGTGGAGAGAA | ||||
| Stra8 | Stra8 for | TCACAGCCTCAAAGTGGCAGG | 56 | 30 s | 444 |
| Stra8 rev | GCAACAGAGTGGAGGAGGAGT | ||||
| HANP1/H1T2 | Hanp1-H1T2-for | GCTGGCTACTTCAGGGTCT | 50 | 1 min | 800 |
| Hanp1-H1T2-rev | TGTATGCTGGGAGCGTTG | ||||
| MIS | MMISf | TTGGTGCTAACCGTGGACTT | 56 | 30 s | 315 |
| MMISr | GCAGAGCACGAACCAAGCGA | ||||
| Ins13 | mIns13f | TACTGATGCTCCTGGCTCTGG | 56 | 30 s | 543 |
| mIns13r | TTAGACTGTTTGGGACACAGG | ||||
| Odf2 | 138NCEcoRI | GGAATTCATGTCCGCCTCATCCTCAGGC | 59 | 1 min | 822/677 |
| DH1rev | CAGCTTCCCGATGGTATCCTTCAAG | ||||
| Odf1 | Odf1-N | GAGCTCAAGCTTTGGCCGCACTGAGTTGTC | 52 | 1 min | 800 |
| Odf1-C | CCGCGGTACCCAAGATCATCTTCCTACA |
Histology and immunocytology.
Testes were alternatively fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; 145 mM NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4) or in Bouin's solution and embedded in paraffin. Then, 4-μm sections were cut and placed onto Superfrost slides. After deparaffinization and rehydration the probes were stained with hematoxylin and eosin.
For immunocytology, epididymides were minced in PBS, and the cell suspension was transferred onto Superfrost plus slides (Thermo, Braunschweig, Germany) by centrifugation at 1,000 × g for 5 min. The cells were fixed in 3.7% paraformaldehyde, blocked in PBT (PBS, at pH 7.5, containing 1% bovine serum albumin and 0.5% Tween 20) for 30 min, and incubated with anti-α-tubulin antibody (Calbiochem, San Diego, CA) for 1 h. The primary antibody was detected with Cy3-labeled anti-mouse IgG (Sigma), the DNA was counterstained with DAPI (4′,6′-diamidino-2-phenylindole), and the acrosome was decorated with fluorescein isothiocyanate (FITC)-labeled peanut lectin (PL-FITC). Images were taken by confocal microscopy (LSM 510; Zeiss) and processed using Adobe Photoshop 7.0.
Electron microscopy.
Testes and epididymides were fixed in 5% glutaraldehyde in 0.2 M phosphate buffer, postfixed with 2% osmium tetroxide, and embedded in epoxy (Epon) resin. Selected areas were sectioned and examined by transmission electron microscopy.
Sperm analysis.
Heterozygous and homozygous Odf1-deficient male mice of the mixed background, as well as wild-type males of C57BL/6J strain, were used for sperm analyses. Epididymides were collected and dissected in Tyrode's medium. Sperm number in corpus and cauda epididymis was determined using the Neubauer cell chamber. For acrosome reaction, spermatozoa were capacitated for 1.5 h in Tyrode's medium and then incubated for 5 min at 37°C in 5% CO2 with Tyrode's medium plus 20 μM calcium ionophore A23187 (Sigma-Aldrich). For the determination of the percentage of sperm that had undergone an acrosome reaction, sperm were fixed and stained with Coomassie brilliant blue R250 as described previously (58). At least 200 spermatozoa from each male were examined for the presence or absence of the characteristic dark blue crescent.
Sperm motility analysis.
Epididymides of three wild-type, heterozygous, and homozygous Odf1-deficient males, respectively, were dissected in in vitro fertilization medium (Medi-Cult, Jyllinge, Denmark). Spermatozoa were allowed to swim out of the epididymides and incubated for 1.5 h at 37°C. A total of 13 μl of the sperm suspension was transferred to the incubation chamber, which was set at a temperature of 37°C. Sperm movement was quantified using the CEROS computer-assisted semen analysis system (version 10; Hamilton Thorne Research, Beverly, MA). Then, 1,000 to 3,000 spermatozoa from each individual were analyzed using the following parameters: negative phase-contrast optics; recording, 60 frames/s; minimum contrast, 60; minimum cell size, 6 pixels; straightness (STR) threshold, ≥50%; cutoff of the average path velocity (VAP) and straight line velocity (VSL), 25 and 30 μm/s, respectively; minimum progressive average path velocity (VAP), 75 μm/s; slow cells motile, no (this limit avoids counting sperm moved by others or Brownian motion and low-velocity nonprogressive cells); and minimum static contrast, 15 pixels.
For statistical analysis, the frequencies of the six sperm motility parameters VAP, VSL, VCL, ALH, BCF, and STR were examined. Because all analyzed parameters were significantly different from normal distribution (Shapiro-Wilk's W test) and could not be normalized by any transformation, the nonparametric alternative for the t test, the Mann-Whitney U test, was used. For statistical testing, sperm motility measurements of each parameter were pooled for mouse type. Statistical analyses were performed by Statistica 9 (StatSoft, Inc., Tulsa, OK).
RESULTS
Generation of mice with disruption of the Odf1 gene.
The Odf1 gene consists of two exons separated by one large intron. Transcription and translation start sites are both in exon 1 (8, 9, 20). The targeting vector used to disrupt Odf1 was designed with a neomycin cassette to replace exon 1 and the promoter region (Fig. 1A). After electroporation of the linearized targeting vector into mouse ES cells recombinant colonies were established by selection with G418 and ganciclovir. Next, Southern blot hybridization with HindIII digested genomic DNA of individual ES cell clones and 3′ external probe was performed. Out of 96 ES cell clones tested, 20 clones showed the expected hybridization pattern of a band of ∼5.4 kb that refers to the wild-type allele and a band of ∼7.2 kb that refers to the homologous recombined allele (Fig. 1B). One ES cell clone that showed the expected hybridization pattern after repeated Southern blot hybridization with the external 3′ probe was chosen for microinjection into blastomeres of 3.5-day-old embryos of C57BL/6 mice. After reimplantation of embryos, several chimeras were born, most of them with a high chimerism (>90%). Male chimeras mated with C57BL/6 females transmitted the targeted allele to their offspring. Offspring were genotyped by PCR on genomic DNA isolated from tail tip biopsy specimens using primer pair Neomy/Odf1-I (Fig. 1C). Heterozygous mice were crossed with C57BL/6 mice to establish the heterozygous Odf1+/− line. Eventually, heterozygous male and female mice were crossed to generate Odf1-deficient mice. Genotyping was generally performed on tail tip biopsy specimens using the primer pair Neomy/Odf1-I for detection of the recombined allele and with primer pair Odf1-N/Odf1-I for detection of the wild-type allele (Fig. 1C).
Odf1-deficient male mice are infertile.
Heterozygous and homozygous Odf1-deficient male mice were first inspected regarding their overall morphological features taking into consideration testes weight related to body length and weight. However, no significant difference between both groups could be found (not shown). Phenotypic differences mostly rely on individual deviations irrespective of the presence of ODF1. Testis weight especially was related to body length and weight but not to the presence or absence of ODF1. Odf1 deficiency, therefore, did not obviously influence these morphological criteria.
However, we observed infertility in Odf1-deficient male mice, whereas fecundity was not impaired in Odf1-deficient female mice or in heterozygous male mice. To further confirm the infertility of Odf1-deficient male mice, we mated 11 Odf1−/−-male mice with wild-type females. Over a time period of at least 3 months, none of these pairings gave birth to offspring or even resulted in pregnancy. In contrast, their heterozygous Odf1+/− male littermates (n = 6) gave rise to offspring generally after about 4 weeks of mating with wild-type females. In test matings, we found no sperm in the uterus of the female mouse when mated to Odf1-deficient male, although the vaginal plug inspection was positive. The litter size of matings of wild-type C57BL/6 females with heterozygous Odf1+/− males (n = 29 litters; average litter size of 6.7) was comparable to that of wild-type breeding (litter size of 6). Furthermore, offspring of heterozygous males have sex ratios of approximately Mendelian ratio (57 male and 43 female pups were born), suggesting that Odf1 deficiency in the heterozygote state did not impair embryonic development. In addition, Odf1-deficient female mice (Odf1−/−) are not impaired in fertility (Table 2). We thus established our Odf1-deficient mouse line by mating of Odf1-deficient females with heterozygous males. The male infertility prompted us to have a closer look at the testis of Odf1-deficient mice.
Table 2.
Fertility, fecundity, and testis weight of wild-type and Odf1 mutant micea
| Genotype | Male fertility | Avg litter size (males) | Mean testis/body wt (×10−3) ± SD | Female fertility | Avg litter size (females) |
|---|---|---|---|---|---|
| +/+ | 28/28 | 6.2 | ND | 28/28 | 6.2 |
| +/– | 29/29 | 6.7 | 3.1 ± 0.37 (n = 3) | 19/19 | 5.8 |
| –/– | 11/0 | 0 | 2.92 ± 0.31 (n = 6) | 10/10 | 5.2 |
To test fertility, sexually mature mice were bred for at least 3 months. Litter size was recorded for each mating. Testis weight and body weight were measured from mice older than 5 months of age. ND, not determined.
Spermatogenesis in Odf1-deficient mice.
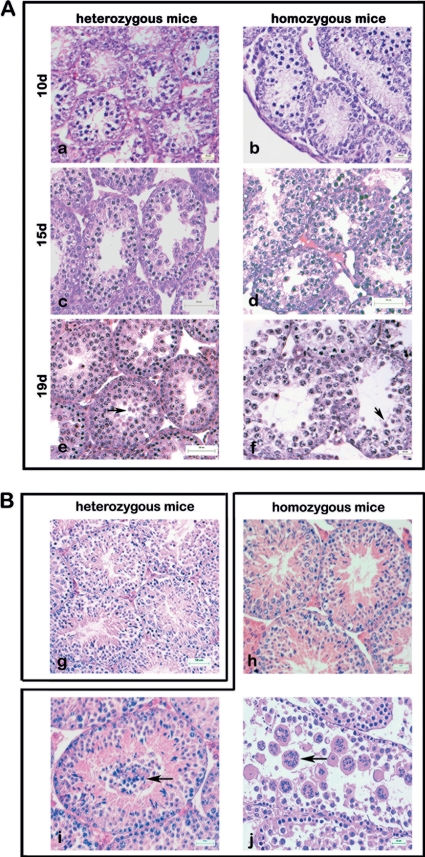
Histological analyses of testes from heterozygous and homozygous Odf1-deficient adult mice revealed that spermatogenesis proceeded normally in all heterozygous mice (n = 4) (Fig. 2B). However, three different phenotypes were observed in Odf1−/− mice. One of nine homozygous mice analyzed showed an obvious wild-type phenotype with elongating spermatids in the seminiferous tubules (Fig. 2Bh). Six mice had displaced germ cells in the lumen (Fig. 2B, arrow in panel i), and in testes from two mice we observed multinuclear cells in the lumen (Fig. 2B, arrow in panel j). Mature sperm are barely found.
Fig 2.
Testicular histology of heterozygous and homozygous Odf1-deficient mice. Testis sections were stained with hematoxylin and eosin. (A) Spermatogenesis in heterozygous and homozygous Odf1-deficient mice. Testes from 10-day-old mice (10d), 15-day-old mice (15d), and 19-day-old mice (19d) were examined. No differences in spermatogenic progression between heterozygous and homozygous mice were observed. Round spermatids are found in 19d testes of heterozygous and homozygous Odf1-deficient mice (arrows in panels e and f). For each developmental stage, the testes of one to three mice were prepared. Bars: 20 μm (a, b, and f), 50 μm (c to e). (B) Spermatogenesis proceeded normally in adult heterozygous mice (g). In adult homozygous mice three different phenotypes were observed. One in nine animals showed an obvious wild-type phenotype (h); six animals also had undifferentiated cells prematurely released in the lumen of the seminiferous epithelium (i; arrow), and in testes of two animals the seminiferous epithelium was flat and highly disorganized, and numerous multinuclear cells were found in the lumen (j; arrow). Bars: 50 μm (g to i), 25 μm (j).
In order to narrow down the developmental stage when spermatogenesis is affected, we inspected testis sections of one to three individuals each aged 10, 15, and 19 days post partum (dpp). Histological analyses of testis sections from 10 dpp onward revealed no obvious differences between heterozygous and homozygous Odf1-deficient mice (Fig. 2A). Spermatogenesis proceeded normally as previously described (42). Meiosis starts around day 10, and early round spermatids are present ∼10 days later. As demonstrated in Fig. 2A (b, d, and f), the spermatogenesis of Odf1-deficient males proceeded normally, and at 19-dpp round spermatids were observed in heterozygous, as well as in homozygous mutants (arrows in Fig. 2Ae and f). The early elongating spermatid stage then is reached at about day 26 and elongating spermatids were also detected in most of the Odf1-deficient testes (Fig. 2Bh and i).
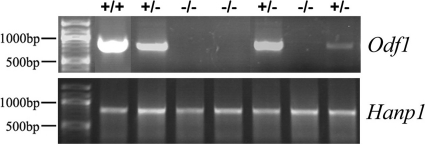
Furthermore, we monitored testis composition and spermatogenic progression by reverse transcription-PCR (RT-PCR) of marker genes. We inspected Insl3 as a marker gene for Leydig cells, Mis (Müllerian inhibiting substance) as a marker gene for Sertoli cells, Stra8 as a marker for spermatogonia, Sycp3 (Scp3, synaptonemal complex protein 3) as a marker gene for meiosis, Hanp1/H1T2 as a marker gene to monitor spermiogenic progression, and Odf2 as a ubiquitously expressed gene that is upregulated during spermiogenesis (1, 7, 13, 26, 33, 35, 38, 44, 45, 57). In addition, expression of Odf1 by RT-PCR was checked to confirm the correct genotyping and absence of Odf1 in homozygous Odf1-deficient mice (Fig. 3). We performed RT-PCR for all marker genes using cDNA generated from testes of two wild-type mice, nine heterozygous mice, and 17 homozygous Odf1-deficient mice. Besides Odf1, which was absent in homozygous Odf1-deficient mice, we found the expression of all marker genes in all cases, including mice that have undifferentiated or multinuclear cells present in their testis tubules (see Fig. 2B; PCR results not shown). To note, marker genes specific for the differentiation phase of spermatids are expressed as well, e.g., Hanp1 (Fig. 3).
Fig 3.
Marker gene expression revealed spermatogenic progression irrespective of the presence or absence of ODF1. RT-PCR for Odf1 or Hanp1 was performed on cDNA generated from the testes of wild-type (+/+), heterozygous (+/−), and homozygous Odf1-deficient (−/−) mice.
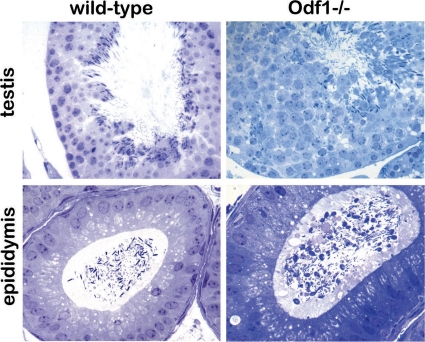
Although we found normal progression of spermatogenesis up to the spermatid stage, examination of semithin sections revealed a disorganized seminiferous epithelium, and strong reduction of mature spermatozoa in caput epididymides of Odf1-deficient mice (Fig. 4). Instead, we found an increase of nonmature germ cells and a high percentage of dysplastic sperm in the epididymes (Fig. 4).
Fig 4.
Semithin sections of wild-type and homozygous Odf1-deficient mouse testes and epididymides. Sections were stained with toluidine blue. Seminiferous tubules of wild-type and of Odf1-deficient mouse testis showed spermatogenic progression up to elongated spermatozoa and sperm tails. Sections of caput epididymides showed mature spermatozoa in the wild-type genotype, whereas in Odf1-deficient mice mature spermatozoa are barely found. Magnifications: testis, ×400; epididymis, ×630.
Sperm analyses of Odf1-deficient mice.
Histological analyses, as well as expression profiling, revealed that Odf1 deficiency did not affect the premeiotic, meiotic, and early postmeiotic phases of spermatogenesis. Even elongated spermatozoa, including the formed sperm tail, could be detected in the testes of homozygous mice. We therefore analyzed the epididymal spermatozoa more closely. Sperm motility was measured after capacitation in vitro, taking into consideration the sperm velocity parameters (curvilinear velocity [VCL], average path velocity [VAP], straight line velocity [VSL], and also the straightness of the movement [STR]) and parameters describing the head behavior (amplitude of the lateral head displacement [ALH] and the beat cross frequency [BCF]). We found that homozygous Odf1-deficient spermatozoa showed significantly reduced velocities (VAP, VCL, and VSL) and a reduction in lateral head displacement (ALH) compared to wild-type and heterozygous Odf1-deficient spermatozoa (see Fig. S1 in the supplemental material). BCF and STR were significantly increased in homozygous mutants. Heterozygous Odf1-deficient spermatozoa revealed properties generally in between wild-type and homozygous spermatozoa (see Fig. S1 in the supplemental material).
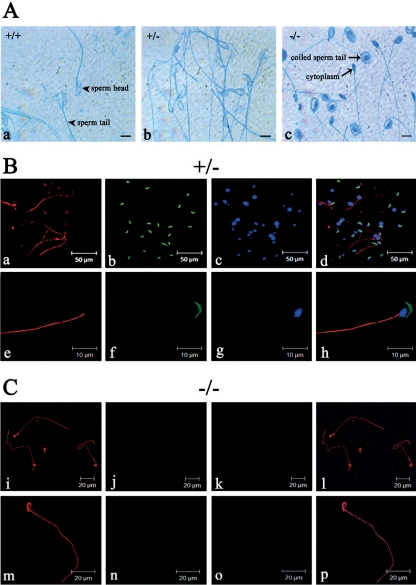
We then analyzed the capability of spermatozoa to perform the acrosome reaction. Whereas in wild-type sperm as well as in heterozygous Odf1-deficient sperm a clear acrosome reaction could be monitored, homozygous sperm did not show any acrosome reaction at all and were of a conspicuously different phenotype (Fig. 5A). Odf1-deficient spermatozoa were characterized by their coiling sperm tail (Fig. 5Ac). Immunocytology using anti-αtubulin antibodies revealed that the axoneme is formed in heterozygous (Fig. 5B), as well as in homozygous Odf1-deficient spermatozoa (Fig. 5C). PL-FITC staining showed the presence of the acrosome in heterozygous spermatozoa but not in probes of Odf1−/− mice. Furthermore, sperm heads were generally missing in the epididymal probes of Odf1−/− mice (Fig. 5B and C).
Fig 5.
Acrosome and axoneme in Odf1-mutant sperm. (A) Acrosome reaction in epididymal sperm of wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice. A characteristic light blue staining of the reacted acrosome is visible in wild-type (a; +/+) and heterozygous (b; +/−) sperm, whereas in Odf1−/− sperm (c; −/−) no acrosome reaction was found. In addition, a remarkable sperm coiling in Odf1−/− sperm is visible. Bars: 10 μm. (B and C) Detection of the axonemal microtubules in heterozygous (B) and homozygous (C) Odf1-deficient spermatozoa. Sperm were released from the cauda epididymides of heterozygous (B, +/−) and homozygous (C, −/−) mice and stained for the acrosome with PL-FITC (in green) and for tubulin with anti-α-tubulin antibodies (in red). Nuclear counterstain with DAPI is shown in panels c, g, k, and o (in blue). In heterozygous and homozygous sperm the sperm tail is decorated with anti-α-tubulin antibodies demonstrating the presence of the axonemal structure (a, e, i, and m). In homozygous sperm the tail showed a conspicuous coiling (i, m, l, and p). The acrosome is present in heterozygous sperm (b and f) but absent in homozygous sperm (j and n). In addition, characteristic DAPI-stained sperm heads are also barely visible in homozygous preparations (k and o).
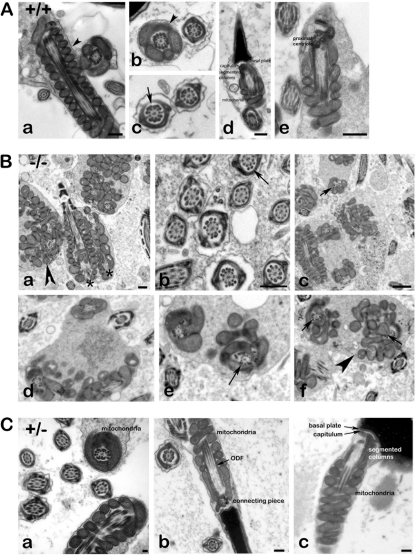
Loss of Odf1 affects the ultrastructure of the spermatozoon.
Inspection of transmission electron micrographs prepared from wild-type and homozygous Odf1-deficient mice, respectively, revealed a disturbed ultrastructure in homozygous animals (Fig. 6B). Most remarkably, the organization of the mitochondrial sheath in the midpiece of spermatozoa discernible in longitudinal sections and cross-sections is disturbed (Fig. 6B, arrowheads). In the midpiece of wild-type spermatozoa the mitochondria are elongated, crescent-shaped organelles that are aligned end-to-end to form helices and are enclosed by a rigid capsule, the mitochondrial sheath (44). This orderly mitochondrial array is visible in longitudinal sections and in cross-sections of wild-type spermatozoa (Fig. 6A, arrowheads). A disturbed mitochondrial organization is found in epididymal sperm (Fig. 6Ba to c, arrowhead) as well as in testicular sections of Odf1−/− mice (Fig. 6Bd to f, arrowhead). In addition, the well-structured pattern of the ODFs is disturbed. ODFs are no more tightly aligned to their corresponding microtubule doublet. The disturbed structural organization of the sperm tail is evident in cross-sections of the midpiece when the mitochondrial sheath surrounds the sperm tail cytoskeleton (Fig. 6Bc to f, arrows) but is also found in the principal piece of the sperm tail characterized by its surrounding fibrous sheath (Fig. 6Bb, arrow). In the principal piece, the ODFs are either not correctly aligned to their corresponding tubule doublets or are dislodged from them (Fig. 6Bb, arrow). In addition, in the cytoplasm of a single cell more than one axonemal cross-section could be found very often, sometimes together with a longitudinal section through the axoneme, or two longitudinal sections through axonemata in parallel (Fig. 6Ba, asterisks). In contrast, in wild-type sperm tail cross-sections a highly regular pattern is found with tight association of the ODFs to their corresponding tubule doublets (Fig. 6Ab and c, arrow). Wild-type sperm showed a tight linkage of the sperm head to the tail and the well-known ultrastructural organization comprising basal plate, capitulum, segmented columns, and even the proximal centriole (Fig. 6Ad and e). Sperm of heterozygous animals likewise did not reveal obvious disturbances. Mitochondria, ODF, and the connecting piece are well structured (Fig. 6Ca to c). However, in ODF1-deficient specimens no sperm heads could be found but instead only detached tails. It was thus not possible to analyze the ultrastructure of the neck region.
Fig 6.
Ultrastructure of spermatozoa. (A) Transmission electron microscopy of epididymal spermatozoa from wild-type mice. The well-structured organization of the sperm is shown, including the mitochondrial sheath (a, b, d, and e, arrowheads), the outer dense fibers (c; arrow), and the neck region (d and e). Bars: 250 nm (a and d), 500 nm (e). (B) Transmission electron microscopy of Odf1−/− sperm. Epididymides (a to c) and testes (d to f) from Odf1-deficient mice were prepared, and sections analyzed by electron microscopy. Sperm are highly disorganized including the mitochondrial sheath (a, c to f, arrowheads), as well as the outer dense fibers in the midpiece (c to f, arrows) and in the principal piece (b, arrow). In addition, very often in the cytoplasm of one cell more than one axonemal section was found (two longitudinal sections of axonemata in panel a, asterisks). Arrowheads, disturbed mitochondrial sheath; arrows, disturbed alignment of ODFs. Bars: 500 nm (a and b), 1,000 nm (c). (C) Transmission electron microscopy of sperm from heterozygous animals. No obvious disturbances are found but instead well-organized mitochondria (a to c), ODF (a and b), and connecting piece (b and c). Bars: 100 nm (a), 250 nm (b and c).
DISCUSSION
According to the World Health Organization up to 15% of reproductive-aged couples worldwide suffer from infertility. In 50% of cases male infertility has been found to be the cause (13). In male infertility diagnostics, the sperm tail cytoskeleton has received increasing attention over the years since sperm tail-associated defects may impair sperm motility that eventually results in infertility (35). Likewise, sperm tail defects have been considered as a prognostic factor in assisted conception in humans (33). Unfortunately, in most instances morphological defects of the sperm tail have not been traced back to their molecular cause, although more than 400 mutant mouse models with a reproductive phenotype have been generated (38) and the sperm proteome is available (14, 36, 37). Only ca. 10% of knockout mouse models that display male reproductive disorders have spermatozoa with impaired motility and/or sperm flagellum structure anomalies (15, 62).
The sperm tail generates progressive motility necessary to reach the oocyte. Its functional unit is the axoneme, a tubulin-based structure containing microtubule-associated proteins and motor proteins that convert chemically bound energy into progressive mechanical force by ATP hydrolysis (22, 30, 41, 47). The orderly array of axonemal nine doublet microtubules surrounding a pair of singlet microtubules is complemented by associated ODFs that accompany the microtubule doublets throughout most of the sperm tail. In addition, in the midpiece of the flagellum the mitochondrial sheath wraps around the ODFs, and in the principal piece the fibrous sheath (FS) surrounds the axoneme and the residual seven ODFs (16). Both the ODFs and the FS provide structural rigidity to the tail (3). The FS additionally is involved in the generation of ATP (39).
A main protein component of mammalian ODFs is ODF1. Moreover, ODF1 is exclusively expressed in spermatids and restricted to the ODF compartment (40). Not surprisingly, it was observed that Odf1 deficiency did not affect female mice. Loss of ODF1 exclusively affected male mice resulting in infertility. However, spermatogenesis in general was not impaired. We found expression of all marker genes characteristic for the various germ cell types, demonstrating that spermatogenesis proceeded from spermatogonia to spermatids. Expression of Hanp1 especially substantiated spermiogenic progression in Odf1-deficient mice because Hanp1 transcription is restricted to spermatids (57). Histological examinations of testes likewise revealed no hints of disturbed spermatogenesis up to at least 19 days after birth. Perturbations of the germinal epithelium were found not until sexual maturity when two-thirds of animals exhibited undifferentiated cells, and two of nine animals showed multinuclear cells in the lumen of their seminiferous tubules. However, in most cases elongated spermatids were present. Semithin sections of epididymides from Odf1−/− mice revealed an increase of nonmature germ cells and a high percentage of dysplastic sperm. Sperm heads were barely found, thus accounting for observed infertility. The presence of large multinucleate cells found in some tubules suggests that the lack of ODF1 also might affect germ cell separation, leading eventually to the coalescence of adjacent germ cells.
Measurement of sperm parameters revealed that Odf1-deficient epididymal sperm are motile, although with strongly reduced velocities but increased beat cross frequencies and straightness, both of which might be caused by the absence of the normally built sperm head. In addition, half-reduction of the amount of ODF1 in sperm of heterozygous mice, originating from the syncytial nature of the germinal epithelium (6), affected their motility but eventually did not impact fecundity. At the light microscopic level, Odf1-deficient sperm are characterized by their remarkable tail coiling, as well as by their missing heads. At the ultrastructural level, we observed disturbed mitochondrial organization in the midpiece of the sperm tail and disturbed organization of ODFs, both in the midpiece and in the principal piece. Most remarkably, however, is the absence of intact spermatozoa. Our results thus strongly support the view that ODF1 is essential for the tight connection of the sperm head to the tail and for the orderly arrangement of the mitochondria and the ODFs in the sperm tail.
The observed disorganization of the mitochondrial sheath in the midpiece of Odf1-deficient sperm tails is reminiscent to loss of Nectin-2, a Ca2+-independent immunoglobulin-like cell-cell adhesion molecule, or of Gopc (Golgi-associated PDZ- and coiled-coil motif-containing protein), respectively. Loss of either Nectin-2 or Gopc showed impairment of tight packaging of mitochondria into a helical sheath. Male mice lacking both alleles of Nectin-2 are infertile, whereas heterozygous males and females, as well as homozygous Nectin-2-deficient females, are fertile and display no obvious phenotype (5). In Gopc−/− spermatozoa the distribution of GPX4/PHGPx, a major element of the mitochondrial capsule, is disturbed, whereas SPATA19, an important factor for mitochondrial aggregation, is not affected (56). However, neither Nectin-2 nor Gopc have previously been identified as ODF1 binding partners. Instead, a promising candidate that could be responsible for mediating the tight packaging of mitochondria and their association to the ODFs in the midpiece of the sperm tail is the kinesin light chain, KLC3. KLC3 has not only been shown to interact with ODF1 but also to associate with mitochondria in the sperm tail (4, 64). Lack of its binding partner ODF1 could thus affect the tight mitochondrial association to the ODFs eventually resulting in its disturbed arrangement.
In addition, we found a disorganization of the ODF themselves in that their tight association to the axonemal microtubule doublets is loosened, becoming apparent in the midpiece with its disturbed mitochondrial arrangement and in the principal piece that is characterized by its surrounding FS. Moreover, lack of ODF1 seems to result in reduction or even loss of the central part of the ODF, the medulla, and eventually in hollow fibers (see, for example, Fig. 6Bb). Beyond that, the axonemal structure itself seems not to be affected. The most striking phenotype, however, was the detachment of the tail from the nucleus, thus resembling a type of oligoteratoasthenospermia in humans with sperm tail fragility and decapitated sperm heads (2, 12, 28). Detachment might occur during sperm passage from testis to epididymis due to the loosening of the sperm head-to-tail linkage by missing the ODF1 that eventually could not withstand mechanical forces.
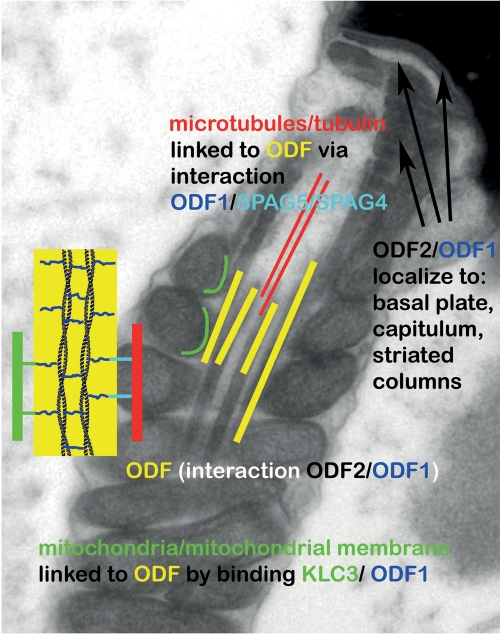
The different domains and regions characteristic of sHSPs are present in ODF1. Noteworthy, however, is the primary structure of the variable C-terminal tail of ODF1 consisting of repeats of the motif C-X-P. Similar repeat motifs have only been found in D. melanogaster male-specific proteins encoded by the Mst84 gene cluster (31), and several mammalian intermediate filament proteins (19). Whether these repeat motifs are important for mediating protein-protein interaction is not known. The impact of ODF1 on the structural organization of the spermatozoon is illustrated in Fig. 7, emphasizing known interaction partners. Linking ODF to microtubules might occur via ODF1/SPAG5/SPAG4 interaction and to mitochondria via ODF1/KLC3 interaction. ODF1/ODF2 binding might account for rigidity of ODF, basal plate, capitulum, and striated columns (49). However, proteins that mediate the linkage of the sperm tail to the head via ODF1 have yet to be defined.
Fig 7.
Illustration depicting the localization of ODF1 and its reported interaction partners that might account for observed disturbances in ODF1-deficient sperm. ODF1 interacts with ODF2, and both are located to the ODF, basal plate, capitulum, and striated columns (49). Linking of ODF to axonemal microtubules might be mediated via binding of ODF1 to SPAG5/SPAG4 and of ODF to mitochondria via KLC3 binding. Proteins that mediate the binding of the connecting piece to the sperm head are not known at present.
To sum up, our results suggest that ODF1, besides being involved in the correct arrangement of mitochondrial sheath and ODF, is essential for the rigid junction of sperm head and tail and that a loss of function of ODF1 might account for some of the cases of human infertility with decapitated sperm heads. In addition, since half-reduction of ODF1 in heterozygous mice already affected sperm motility, impairment of ODF1 might account for some cases of reduced fertility in male patients.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge scientific support by Wolfgang Engel and the technical assistance of Gerd Kripp.
K.Y., B.Z., P.G., and I.M.A. performed analyses, and A.M. performed the electron microscopy. S.H.-F. led this study and wrote the manuscript, with input from the other authors. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Adham IM, Burkhardt E, Benahmed M, Engel W. 1993. Cloning of a cDNA for a novel insulin-like peptide of the testicular Leydig cells. J. Biol. Chem. 268:26668–26672 [PubMed] [Google Scholar]
- 2. Baccetti B, et al. 2001. Genetic sperm defects and consanguinity. Hum. Reprod. 16:1365–1371 [DOI] [PubMed] [Google Scholar]
- 3. Baltz JM, Williams PO, Cone RA. 1990. Dense fibers protect mammalian sperm against damage. Biol. Reprod. 43:485–491 [DOI] [PubMed] [Google Scholar]
- 4. Bhullar B, Zhang Y, Junco A, Oko R, van der Hoorn FA. 2003. Association of kinesin light chain with outer dense fibers in a microtubule-independent fashion. J. Biol. Chem. 278:16159–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bouchard MJ, et al. 2000. Defects in nuclear and cytoskeletal morphology and mitochondrial localization in spermatozoa of mice lacking Nectin-2, a component of cell-cell adherens junctions. Mol. Cell. Biol. 20:2865–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braun RE, Behringer RR, Peschon JJ, Brinster RL, Palmiter RD. 1989. Genetically haploid spermatids are phenotypically diploid. Nature 337:373–376 [DOI] [PubMed] [Google Scholar]
- 7. Brohmann H, Pinnecke S, Hoyer-Fender S. 1997. Identification and characterization of new cDNAs encoding outer dense fiber proteins of rat sperm. J. Biol. Chem. 272:10327–10332 [DOI] [PubMed] [Google Scholar]
- 8. Burfeind P, Hoyer-Fender S. 1991. Sequence and developmental expression of a mRNA encoding a putative protein of rat sperm outer dense fibers. Dev. Biol. 148:195–204 [DOI] [PubMed] [Google Scholar]
- 9. Burfeind P, Belgardt B, Szpirer C, Hoyer-Fender S. 1993. Structure and chromosomal assignment of a gene encoding the major protein of rat sperm outer dense fibres. Eur. J. Biochem. 216:497–505 [DOI] [PubMed] [Google Scholar]
- 10. Burmester S, Hoyer-Fender S. 1996. Transcription and translation of the outer dense fiber gene (Odf1) during spermiogenesis in the rat. A study by in situ analyses and polysome fractionation. Mol. Reprod. Dev. 45:10–20 [DOI] [PubMed] [Google Scholar]
- 11. Calvin HI, Hwang FH-F, Wohlrab H. 1975. Localization of zinc in a dense fiber-connecting piece fraction of rat sperm tails analogous chemically to hair keratin. Biol. Reprod. 13:228–239 [DOI] [PubMed] [Google Scholar]
- 12. Chemes HE, et al. 1999. Acephalic spermatozoa and abnormal development of the head-neck attachment: a human syndrome of genetic origin. Hum. Reprod. 14:1811–1818 [DOI] [PubMed] [Google Scholar]
- 13. Cui W. Mother or nothing: the agony of infertility. 2010. Bull. World Health Organ. 88:881–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Mateo S, et al. 2007. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 7:4264–4277 [DOI] [PubMed] [Google Scholar]
- 15. Escalier D. 2006. Knockout mouse models of sperm flagellum anomalies. Hum. Reprod. Update 12:449–461 [DOI] [PubMed] [Google Scholar]
- 16. Fawcett DW. 1975. The mammalian spermatozoon. Dev. Biol. 44:394–436 [DOI] [PubMed] [Google Scholar]
- 17. Feinberg AP, Vogelstein B. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6–13 [DOI] [PubMed] [Google Scholar]
- 18. Fitzgerald CJ, Oko RJ, van der Hoorn FA. 2005. Rat Spag5 associates in somatic cells with endoplasmic reticulum and microtubules but in spermatozoa with outer dense fibers. Mol. Reprod. Dev. 73:92–100 [DOI] [PubMed] [Google Scholar]
- 19. Fontaine J-M, Rest JS, Welsh MJ, Benndorf R. 2003. The sperm outer dense fiber protein is the 10th member of the superfamily of mammalian small stress proteins. Cell Stress Chaperones 8:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gastmann O, et al. 1993. Sequence, expression, and chromosomal assignment of a human sperm outer dense fiber gene. Mol. Reprod. Dev. 36:407–418 [DOI] [PubMed] [Google Scholar]
- 21. Haidl G, Becker A, Henkel R. 1991. Poor development of outer dense fibers as a major cause of tail abnormalities in the spermatozoa of asthenoteratozoospermic men. Hum. Reprod. 6:1431–1438 [DOI] [PubMed] [Google Scholar]
- 22. Haimo LT, Rosenbaum JL. 1981. Cilia, flagella, and microtubules. J. Cell Biol. 91:125s–130s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hishiya A, Takayama S. 2008. Molecular chaperones as regulators of cell death. Oncogene 27:6489–6506 [DOI] [PubMed] [Google Scholar]
- 24. Hofferbert S, et al. 1993. A homozygous deletion of 27 basepairs in the coding region of the human outer dense fiber protein gene does not result in a pathologic phenotype. Hum. Mol. Genet. 2:2167–2170 [DOI] [PubMed] [Google Scholar]
- 25. Hoyer-Fender S, Burfeind P, Hameister H. 1995. Sequence of mouse Odf1 cDNA and its chromosomal localization: extension of the linkage group between human chromosome 8 and mouse chromosome 15. Cytogenet. Cell Genet. 70:200–204 [DOI] [PubMed] [Google Scholar]
- 26. Hüber D, Hoyer-Fender S. 2007. Alternative splicing of exon 3b gives rise to ODF2 and Cenexin. Cytogenet. Genome Res. 119:68–73 doi:10.1159/000109621 [DOI] [PubMed] [Google Scholar]
- 27. Joyner AD. (ed.) 1993. Gene targeting: a practical approach, p 230 IRL Press, Oxford, United Kingdom [Google Scholar]
- 28. Kamal A, et al. 1999. Easily decapitated spermatozoa defect: a possible cause of unexplained infertility. Hum. Reprod. 14:2791–2795 [DOI] [PubMed] [Google Scholar]
- 29. Kierszenbaum AL, Tres LL. 2002. Bypassing natural sperm selection during fertilization: the azh mutant offspring experience and the alternative of spermiogenesis in vitro. Mol. Cell Endocrinol. 187:133–138 [DOI] [PubMed] [Google Scholar]
- 30. King SM. 2000. The dynein microtubule motor. Biochim. Biophys. Acta 1496:60–75 [DOI] [PubMed] [Google Scholar]
- 31. Kuhn R, Schäfer U, Schäfer M. 1988. cis-Acting regions sufficient for spermatocyte-specific transcriptional and spermatid-specific translational control of the Drosophila melanogaster gene mst(3)gl-9. EMBO J. 7:447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lanneau D, et al. 2008. Heat shock proteins: essential proteins for apoptosis regulation. J. Cell. Mol. Med. 12:743–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim CC, Lewis SEM, Kennedy M, Donnelly ET, Thompson W. 1998. Human sperm morphology and in vitro fertilization: sperm tail defects are prognostic for fertilization failure. Andrologia 30:43–47 [DOI] [PubMed] [Google Scholar]
- 34. Lindemann CB. 1996. Functional significance of the outer dense fibers of mammalian sperm examined by computer simulation with the geometric clutch model. Cell Motil. Cytoskeleton 34:258–270 [DOI] [PubMed] [Google Scholar]
- 35. Marmor D, Grob-Menendez F. 1991. Male infertility due to asthenozoospermia and flagellar anomaly: detection in routine semen analysis. Int. J. Androl. 14:108–116 [DOI] [PubMed] [Google Scholar]
- 36. Martinez-Heredia J, Estanyol JM, Ballesca JL, Oliva R. 2006. Proteomic identification of human sperm proteins. Proteomics 6:4356–4369 [DOI] [PubMed] [Google Scholar]
- 37. Martinez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballesca JL, Oliva R. 2008. Identification of proteomic differences in asthenozoospermic sperm samples. Hum. Reprod. 23:783–791 [DOI] [PubMed] [Google Scholar]
- 38. Matzuk MM, Lamb DJ. 2008. The biology of infertility: research advances and clinical challenges. Nat. Med. 14:1197–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miki K, et al. 2004. Glyceraldehyde 3-phosphatase dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc. Natl. Acad. Sci. U. S. A. 101:16501–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morales CR, Oko R, Clermont Y. 1994. Molecular cloning and developmental expression of an mRNA encoding the 27-kDa outer dense fiber protein of rat spermatozoa. Mol. Reprod. Dev. 37:229–240 [DOI] [PubMed] [Google Scholar]
- 41. Mortimer ST. 1997. A critical review of the physiological importance and analysis of sperm movement in mammals. Hum. Reprod. Update 3:403–439 [DOI] [PubMed] [Google Scholar]
- 42. Nebel BR, Amarose AP, Hackett EM. 1961. Calendar of gametogenic development in the prepuberal male mouse. Science 134:832–833 [DOI] [PubMed] [Google Scholar]
- 43. Oko R. 1988. Comparative analysis of proteins from the fibrous sheath and outer dense fibers of rat spermatozoa. Biol. Reprod. 39:69–182 [DOI] [PubMed] [Google Scholar]
- 44. Otani H, Tanaka O, Kasai K, Yoshioka T. 1988. Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: regular dispositions and synchronized changes. Anat. Rec. 222:26–33 [DOI] [PubMed] [Google Scholar]
- 45. Oulad-Abdelghani M, et al. 1996. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J. Cell Biol. 135:469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petersen C, Füzesi L, Hoyer-Fender S. 1999. Outer dense fibre proteins from human sperm tail: molecular cloning and expression analyses of two cDNA transcripts encoding proteins of ∼70 kDa. Mol. Hum. Reprod. 5:627–635 [DOI] [PubMed] [Google Scholar]
- 47. Porter ME, Sale WS. 2000. The 9+2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37–F42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosales JL, et al. 2007. ODF1 phosphorylation by Cdk5/p35 enhances ODF1-OIP1 interaction. Cell Physiol. Biochem. 20:311–318 [DOI] [PubMed] [Google Scholar]
- 49. Schalles U, Shao X, van der Hoorn F, Oko R. 1998. Developmental expression of the 84-kDa ODF sperm protein: localization to both the cortex and medulla of outer dense fibers and to the connecting piece. Dev. Biol. 199:250–260 [DOI] [PubMed] [Google Scholar]
- 50. Shao X, van der Hoorn FA. 1996. Self-interaction of the major 27-kilodalton outer dense fiber protein is in part mediated by a leucine zipper domain in the rat. Biol. Reprod. 55:1343–1350 [DOI] [PubMed] [Google Scholar]
- 51. Shao X, Tarnasky HA, Schalles U, Oko R, van der Hoorn FA. 1997. Interactional cloning of the 84-kDa major outer dense fiber protein Odf84. J. Biol. Chem. 272:6105–6113 [DOI] [PubMed] [Google Scholar]
- 52. Shao X, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. 1999. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 211:109–123 [DOI] [PubMed] [Google Scholar]
- 53. Southern EM. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 54. Sun Y, MacRae TH. 2005. The small heat shock proteins and their role in human disease. FEBS J. 272:2613–2627 [DOI] [PubMed] [Google Scholar]
- 55. Sun Y, MacRae TH. 2005. Small heat shock proteins: molecular structure and chaperone function. Cell. Mol. Life Sci. 62:2460–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Suzuki-Toyota F, et al. 2007. Factors maintaining normal sperm tail structure during epididymal maturation studied in Gopc−/− mice. Biol. Reprod. 77:71–82 [DOI] [PubMed] [Google Scholar]
- 57. Tanaka H, et al. 2005. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol. Cell. Biol. 25:7107–7119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thaler CD, Cardullo RA. 1995. Biochemical characterization of a glycosyl-phosphatidylinositol-linked hyaluronidase on mouse sperm. Biochem. 34:7788–7795 [DOI] [PubMed] [Google Scholar]
- 59. Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 28:1153–1163 [DOI] [PubMed] [Google Scholar]
- 60. Van der Hoorn FA, Tarnasky HA, Nordeen SK. 1990. A new rat gene RT7 is specifically expressed during spermatogenesis. Dev. Biol. 142:147–154 [DOI] [PubMed] [Google Scholar]
- 61. Vera JE, Brito M, Zuvic T, Burzio LO. 1984. Polypeptide composition of rat sperm outer dense fibers: a simple procedure to isolate the fibrillar complex, J. Biol. Chem. 259:5970–5977 [PubMed] [Google Scholar]
- 62. Yan W. 2009. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol. Cell Endocrinol. 306:24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zarsky HA, Cheng M, van der Hoorn FA. 2003. Novel RING finger protein OIP1 binds to conserved amino acid repeats in sperm tail protein ODF1. Biol. Reprod. 68:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y, Oko R, van der Hoorn FA. 2004. Rat kinesin light chain 3 associates with spermatid mitochondria. Dev. Biol. 275:23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.