Abstract
In 2006, a large outbreak of Escherichia coli O157:H7 was linked to the consumption of ready-to-eat bagged baby spinach in the United States. The likely sources of preharvest spinach contamination were soil and water that became contaminated via cattle or feral pigs in the proximity of the spinach fields. In this study, we compared the transcriptional profiles of 12 E. coli O157:H7 isolates that possess the same two-enzyme pulsed-field gel electrophoresis (PFGE) profile and are related temporally or geographically to the above outbreak. These E. coli O157:H7 isolates included three clinical isolates, five isolates from separate bags of spinach, and single isolates from pasture soil, river water, cow feces, and a feral pig. The three clinical isolates and two spinach bag isolates grown in cultures to stationary phase showed decreased expression of many σS-regulated genes, including gadA, osmE, osmY, and katE, compared with the soil, water, cow, feral pig, and the other three spinach bag isolates. The decreased expression of these σS-regulated genes was correlated with the decreased resistance of the isolates to acid stress, osmotic stress, and oxidative stress but increases in scavenging ability. We also observed that intraisolate variability was much more pronounced among the clinical and spinach isolates than among the environmental isolates. Together, the transcriptional and phenotypic differences of the spinach outbreak isolates of E. coli O157:H7 support the hypothesis that some variants within the spinach bag retained characteristics of the preharvest isolates, whereas other variants with altered gene expression and phenotypes infected the human host.
INTRODUCTION
Escherichia coli O157:H7 infections accounted for 27 outbreaks linked to leafy produce between 1995 and 2008 (29), including large outbreaks related to contaminated bagged spinach and bagged Iceberg and Romaine lettuce (8, 10, 11). E. coli O157:H7 infections are a major concern due to the potential to cause severe symptoms, including hemolytic-uremic syndrome (HUS) and even death, especially among children (40). Indeed, 15% of the 205 humans infected during the 2006 spinach-associated outbreak developed HUS (8) and 6 of 11 food-borne-illness-related deaths in 2006 were linked to E. coli O157:H7, although only 2% of all reported food-borne illnesses were attributed to E. coli O157:H7 that year (11).
The ability of E. coli O157:H7 to survive and/or grow on leafy vegetables is due to its metabolic versatility and the ability to adapt to growth under a variety of environments. Previously, we have demonstrated that E. coli O157:H7 has the ability to multiply in the phyllosphere of whole lettuce plants under warm and humid conditions in the laboratory and to increase 11- and 2-fold in population size in only 4 h on shredded and intact harvested lettuce leaves, respectively (7). Plant injury is inherent to the harvesting and processing of fresh-cut leafy vegetables. We have reported that E. coli O157:H7 responded to the chemical environment in injured leaf tissue by upregulating genes involved in oxidative stress, osmotic stress, and antimicrobial resistance (26). These transcriptional changes affected an enhanced resistance of E. coli O157:H7 cells to hydrogen peroxide and calcium hypochlorite, which is used as a sanitizer for leafy vegetables in the fresh-cut produce industry. Hence, changes in transcriptional activity in this pathogen may play a key role in its adaptation to the stress encountered in processed and bagged produce, such as baby spinach.
In addition to an ability to transiently adapt to diverse environments, E. coli strains also adapt to environments through mutation. Recently, Zdziarski et al. (44) demonstrated patient-specific mutations of E. coli strain 83972 resulting in variants with unique adaptive phenotypes. Also, rpoS mutants and variants with reduced levels of RpoS (σS) are observed commonly among E. coli isolates (5, 22, 42). The rpoS gene encodes the stationary-phase sigma factor, σS, a global regulator that coordinates the expression of a large network of genes upon cells entering stationary phase or during exposure of cells to stresses such as acidity or high osmolarity (43). The appearance of rpoS mutants seems to be driven by the increased ability of such mutants to scavenge for scarce nutrients (22, 33); for example, rpoS mutants have been selected for by growth on nonpreferred carbon sources (14). Thus, rpoS mutants and strains with reduced σS levels sacrifice their ability to adapt to certain stresses in favor of nutritional scavenging in some environments.
This competition between stress protection and nutritional competence (SPANC) (22, 33, 39) predicts that certain environments encountered by E. coli O157:H7 will select for isolates with altered phenotypes. E. coli O157:H7 strains with distinct acid resistance (34) and differential expression of virulence and stress genes (20) have been observed. However, in these studies the relationship between the E. coli O157:H7 strains is unknown (34) or the strains are known to be from distinct genotypes (20). In this study, we explored the SPANC hypothesis by examining a set of E. coli O157:H7 isolates from different environmental sources that possess indistinguishable two-enzyme pulsed-field gel electrophoresis (PFGE) profiles and highly related multilocus variable-number tandem repeat (MLVA) types, in addition to being temporally and/or geographically associated with the 2006 outbreak attributed to fresh spinach in the United States (13). Thus, we designated these as “outbreak strains.” We compared the transcriptional profiles and stress phenotypes of stationary-phase cultures of E. coli O157:H7, including three clinical isolates, five isolates from separate bags of spinach linked to the outbreak, single isolates from pasture soil, river water, and cow feces, and a feral pig colon sample collected on the ranch in the vicinity of the spinach fields associated with the outbreak (13). We provide evidence herein that genetic variants occurring in the spinach bag and from human patients display reduced stress responses but exhibit a corresponding increased nutritional competence due to rpoS mutations and/or reduced σS levels.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli O157:H7 strains (Table 1) were cultured on Luria-Bertani (LB) agar plates (BD, Franklin Lakes, NJ) incubated at 37°C. It should be noted that this formulation of Luria-Bertani contains 10 g/liter NaCl. All 12 isolates had an indistinguishable PFGE profile (CDC PulseNet designation EXHX01.0124-EXHA26.0045) and were highly related by an 11-locus MLVA differing by only two repeats at one locus (13). The isolates had similar growth curves in LB broth (data not shown); thus, growth in LB broth was chosen over growth in clinical or environmental modeling conditions for ease of RNA extraction. Liquid cultures were obtained by inoculation of colonies into LB broth and growth at 37°C with shaking (150 rpm) to stationary phase.
Table 1.
E. coli O157:H7 strains used in this study
| Strain no. | Sourcea | State | MLVAd | Reference or provider |
|---|---|---|---|---|
| RM6067 | Spinach | PA | 163 | 13 |
| RM6069 | Clinical | PA | 163 | 13 |
| RM6101 | Pig (feral) | CAb | 176 | 13 |
| RM6102 | River water | CAb | 176 | 13 |
| RM6103 | Cow | CAb | 163 | 13 |
| RM6149 | Pasture soil | CAb | 176 | 13 |
| RM6655 | Clinical | NY | 163 | CDCc |
| RM6658 | Clinical | OR | 163 | CDC |
| RM9992 | Spinach | NM | 163 | CDC |
| RM9997 | Spinach | WI | 163 | CDC |
| RM9998 | Spinach | NV | 163 | CDC |
| RM10002 | Spinach | IL | 163 | CDC |
Strains are all associated with the 2006 spinach outbreak. Spinach, isolated from spinach in bag; clinical, human strain associated with the 2006 outbreak. The 12 strains had an indistinguishable PFGE profile (EXHX01.0124 to EXHA26.0045) and were highly related by an 11-locus MLVA.
Isolated from samples from the ranch (ranch A) linked to the spinach field associated with the 2006 outbreak.
CDC, provided by E. Hÿttia-Trees at the Centers for Disease Control and Prevention.
MLVA numbers were assigned according to Cooley et al. (13).
RNA extraction and microarray procedures.
Ice-cold phenol-ethanol (5%:95%) solution was added to bacterial liquid cultures, and the mixture was incubated on ice for 60 min. The bacteria were centrifuged, and the pellet was stored at −80°C. The RNA extraction was performed using the Promega SV total RNA kit (Madison, WI) according to the manufacturer's instructions, except that bacterial pellets were first treated with 50 mg/ml of lysozyme (Fisherbrand) and 1 U/μl of anti-RNase (Applied Biosystems, Ambion, Austin, TX). Total RNA was quantified with a Nanodrop ND 1000 spectrophotometer (Thermo Scientific), examined for quality on an Agilent bioanalyzer, and stored at −80°C until used for microarray analysis.
Transcriptional profiling experiments employing genomic DNA as a common reference were based on methods described previously (17, 26). The use of a common reference allows the direct comparison of multiple isolates. Microarray analysis was based on competitive hybridization between Cy5-cDNA (test cDNA) and Cy3-gDNA (the common reference). Genomic DNA (2 μg) was labeled via dye incorporation of Cy3-dCTP (GE Healthcare, Piscataway, NJ) using Klenow fragment (New England BioLabs, Ipswich, MA). Total RNA (20 μg) from each experimental condition and each biological replicate was labeled via incorporation of Cy5-dCTP (GE Healthcare) into a cDNA product with the Fairplay III microarray labeling kit (Agilent, Santa Clara, CA) as previously described (26). Hybridization of Cy5-cDNA and Cy3-gDNA was carried out overnight at 42°C. Each microarray contained 4,262 open reading frames (ORFs) from E. coli K-12 strain MG1655 supplemented with the 1,125 ORFs from E. coli O157:H7 strain EDL933 (including 25 from plasmid pO157) that are not present on the E. coli K-12 genome, as described previously (26). After hybridization, slides were scanned on a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA), and individual spots were analyzed with the GenePix Pro 6.0 Software (Molecular Devices). The coding portion of the E. coli O157:H7 chromosome, including the 25 protein-coding genes found on the pO157 plasmid, was examined (4,745 ORFs). Data normalization was conducted as described previously (35). Briefly, spots with a reference signal lower than background plus 2 standard deviations or spots covered by an obvious blemish were excluded. An average local background value was subtracted from all spots, and a Cy3/Cy5 ratio was calculated. Further normalization to account for differences in dye incorporation included data centering by setting the median natural logarithm to zero for each group of spots in one sector (printed by one microarray pin).
Two biological replicates were performed for each isolate, with a biological replicate defined as the growth and extraction of total RNA from an isolate. The cDNA/gDNA mixtures for each biological replicate were hybridized on three separate microarrays, providing three technical replicates for each biological replicate. The data from all replicates were tested by an unpaired t test with unequal variance using Genespring 7.3 (Agilent). Genes of the clinical isolate, RM6069, compared to the cow fecal isolate, RM6103, expressing a greater than 2-fold upregulation or downregulation and a Benjamini-Hochberg false discovery rate (FDR)-adjusted P value of ≤0.05 were considered to be regulated differentially. An average-linkage hierarchical clustering of the expression data (data sets of 4,669 genes) for each of the E. coli O157:H7 isolates was compiled in GeneSpring version 7.3 using the Pearson correlation similarity measure (16) and bootstrapping of 1,000 replications.
DNA sequencing, assembly, and analysis of rpoS.
PCR reagents were supplied by Epicentre (Madison, WI). Each PCR consisted of 1× MasterAmp Taq PCR buffer, 1× MasterAmp Taq Enhancer, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates, forward and reverse primers at 0.2 μM each, 0.2 U of MasterAmp Taq DNA polymerase (Epicentre) or Taq DNA polymerase (NEB, Beverly, MA), and approximately 50 ng of genomic DNA (final reaction volume, 25 μl). The nlpD-rpoS gene-specific forward and reverse primers (forward, 5′ TGTTCAGTATGGTGCCTTGC; reverse, 5′ GGTAGGACGCTGACGTGTCT) were designed using Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA) and purchased from Eurofins MWG Operon (Huntsville, AL). Amplification occurred under the following parameters: 30 cycles of 30 s at 94°C, 30 s at 58°C, and 2 min at 72°C, with a final extension at 72°C for 5 min using a Tetrad thermocycler (Bio-Rad, Hercules, CA).
The sequencing reactions were performed on a Tetrad thermocycler using the ABI Prism BigDye Terminator cycle sequencing kit (version 3.1) (Life Technologies, Foster City, CA) and standard protocols as recommended by the manufacturer. All labeled products were purified on DyeEx spin columns (Qiagen, Valencia, CA). DNA sequencing was performed on an ABI Prism 3130xl genetic analyzer using the POP-7 polymer and ABI Prism genetic analyzer data collection and ABI Prism genetic analyzer sequencing analysis software. The DNA primers used for sequencing were designed using Primer Premier 5.0. Sequencing reads were trimmed and assembled using Lasergene Seqman II (version 8.0; DNAstar, Madison, WI). Nucleotide sequences were compared also against the sequences of E. coli origin of the nonredundant DNA sequence NCBI database using the Basic Local Alignment Search Tool (BLAST) programs BLASTN and BLASTX analysis (2, 36) through the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/).
Western blots.
Bacterial samples for protein analysis were prepared from cultures grown in LB broth for >24 h into stationary phase. The equivalent of 1 optical density (OD) of cells (at 600 nm) was stored as a pellet at −80°C. The 1 OD equivalent of cells was resuspended in 50 μl of lysis buffer consisting of a final concentration of 50 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, and 0.01% sodium azide (6). Samples were placed in boiling water for 5 min, and then 2-mercaptoethanol and bromophenol blue were added to final concentrations of 2.5% and 0.1%, respectively. Two Novex Tris-glycine gels (8 to 16% gradient) (Invitrogen, Carlsbad, CA) were run at 125 V for 90 min. Proteins were transferred to nitrocellulose paper using a semidry system (Bio-Rad) for 1 h at 18 V. The two blots were probed with primary mouse antibodies against either σ70 or σS at 2 μg/ml (Neoclone, Madison, WI). A horseradish peroxidase-conjugated goat anti-mouse antibody was used as the secondary antibody (Invitrogen) at 0.5 μg/ml, and Super Signal West Pico (Thermo Fisher Scientific Inc., Rockford, IL) was used as the substrate according to the manufacturer's directions. The chemiluminescent signal was recorded using an Alpha Innotech Imager (Cell Biosciences, Inc., Santa Clara, CA).
Functional/stress challenge studies.
Isolates of E. coli O157:H7 used in challenge studies were grown from single colonies into stationary phase (>16 h) in LB broth (Difco) at 37°C with shaking at 150 rpm. Acid challenge protocols were developed from methods established previously (3, 18, 37, 42), in which the stress was pH 2.5 during 2 h of incubation to mimic the pH and digestion time of food in the stomach. In our study, a second collection at 6 h was added in order to further define acid resistance under a longer exposure time. For the acid challenge test, stationary cultures were diluted 1:1,000 into LB broth acidified to pH 2.5 using 1 N HCl (Fisherbrand). Osmotic stress challenge protocols were modified from methods established previously (12, 19). For the osmotic stress challenge, cultures were diluted 1:1,000 into LB broth containing 2.5 M NaCl. For both assay types, time zero samples were taken immediately and cultures were returned to 37°C at 150 rpm until later time points. Studies regarding oxidative (H2O2) stress were modified from Kyle et al. (26). Overnight stationary cultures for the H2O2 challenge studies were prepared as described above. Next, approximately 109 cells were washed once in 10 mM potassium phosphate (KP) buffer and then resuspended into 5 ml 0.5 mM KP. Time zero samples were collected immediately before the addition of a quantity of 3% H2O2 (Fisherbrand) that produced a final concentration of 12.5 mM H2O2. Three to six biological replicates (from individual colonies) were tested for each isolate in each assay type. All samples for quantitation were recovered by plating onto LB agar (directly or after dilution in 10 mM KP buffer) using an automated plater (Autoplate 4000; Spiral Biotech) and incubated overnight at 37°C.
Biolog GN2 microplates and bacterial growth kinetics.
Selected E. coli O157:H7 isolates were assayed for respiratory responses to substrates in GN2 microplates (Biolog Inc., Hayward, CA) according to the protocol provided by the vendor. The GN2 microplates were incubated for 24 h at 37°C. Color changes were measured at 600 nm using a Biolog microplate reader and were corrected for the no-substrate control. Three independent replicate assays were performed for each isolate. The cutoff point between negative results and positive results was an optical density at 600 nm of 0.2 as described previously (28). Student's t test was used to determine if data sets of E. coli O157:H7 isolates from the preharvest environment and clinical environment were significantly different as defined by P values of ≤0.01.
Screening for E. coli O157:H7 curli variants.
The population structure of curli variants in each E. coli O157:H7 isolate used in this study was examined as described previously (9). Briefly, the frozen glycerol stock of each isolate was scraped using an inoculating loop and resuspended in 1 ml LB with no salt (LBNS) broth. Ten-fold serial dilutions were prepared from these cell suspensions, and 50 μl of each dilution was plated onto Congo red indicator (CRI) plates (LBNS plates supplemented with 40 μg/ml of Congo red dye and 10 μg/ml of Coomassie brilliant blue). The plates were incubated at 28°C for 2 to 3 days. Colonies with various colors, e.g., red and brown (curli+) or white (curli−), were counted. The relative proportion of curli variants in each isolate was expressed as the percentage of red/brown (curli-producing) colonies over the total number of E. coli O157:H7 colonies examined.
Microarray data and nucleotide sequence accession numbers.
Microarray data were deposited in the NCBI GEO omnibus database and assigned the accession number GSE33131. The nucleotide sequences of the rpoS gene from individual colonies of E. coli O157:H7 isolates were deposited with GenBank and assigned accession numbers as follows: JN862236 to JN862243 for RM6103, RM6069-1, RM6069-11, RM6655, RM6658-1, RM6658-2, RM9998, and RM10002, respectively.
RESULTS
Global transcriptional profile of stationary-phase E. coli O157:H7.
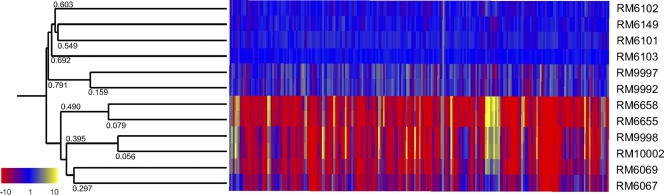
The transcriptomes of 12 E. coli O157:H7 strains (Table 1) grown to stationary phase were compared in two different biological experiments. Three isolates were from human clinical cases associated with the spinach outbreak, four (cow, pig, soil, and water) were collected from the preharvest environment, the ranch adjacent to the spinach fields, and five were cultured from separate packages of bagged spinach during the outbreak traceback investigation. Using hierarchical clustering, the relationship between the transcriptional profiles of 4,745 genes for each isolate was assessed. The smaller the distance scores shown in Fig. 1, the larger the correlation between the isolates' transcriptional profiles (Fig. 1). The cluster analysis suggests that the three clinical isolates (RM6069, RM6655, RM6658) have distinctly different expression patterns compared to the four environmental isolates (RM6101, RM6102, RM6103, RM6149). Furthermore, two of the spinach isolates (RM9998 and RM10002) clustered more closely with the clinical isolates, while two other spinach isolates (RM9992 and RM9997) clustered with the environmental/animal isolates. The fifth spinach isolate, RM6067, clustered away from the other isolates, indicating a distinct expression pattern compared to the other isolates.
Fig 1.
Hierarchical cluster analysis of E. coli O157:H7. The figure shows 230 E. coli O157:H7 genes with expression levels more than or less than 2-fold between clinical strains (RM6069, RM6655, RM6658), bagged spinach strains (RM6067, RM9992, RM9997, RM9998, RM10002), and environmental/animal strains (RM6101 to RM6103, RM6149). Each gene is color-coded according to gene expression relative to RM6103 (blue). Genes exhibiting a 10-fold increase in expression compared with RM6103 are labeled yellow, while those with a 10-fold decrease compared with RM6103 are labeled red. The gene list is ordered according to position on the chromosome. An average-linkage hierarchical clustering of the expression data (data sets of 4,669 genes) for each of the E. coli O157:H7 strains was compiled in GeneSpring version 7.3 using the Pearson correlation similarity measure and bootstrapping of 1,000 replications.
We compared the transcriptomic profiles from the three clinical isolates (RM6069, RM6655, RM6658) with the four environmental isolates (RM6101 to RM6103, RM6149) to identify gene expression that was significantly different between the groups (2-fold difference and P values of <0.01). In total, the expression of 189 genes was downregulated and the expression of 41 genes was upregulated among all of the clinical isolates (see Table S1 in the supplemental material). In comparing the 189 downregulated genes, we noted that 146, 141, and 140 genes were downregulated at least 2-fold in spinach isolates RM10002, RM9998, and RM6067, respectively. Also, of the 41 upregulated genes, we determined that 37, 36, and 20 genes were upregulated at least 2-fold in spinach isolates RM10002, RM9998, and RM6067, respectively. Despite the large number of genes shared between spinach isolate RM6067 and the clinical isolates, the average fold-change of the 189 genes for the clinical isolates and spinach isolates RM9998 and RM10002 was 8.3-fold lower than environmental isolate RM6103, while only 3.4-fold lower for RM6067 (Fig. 1).
Reduced expression of σS-controlled stress-responsive genes in clinical isolates.
Analysis of the transcriptional profiles of the clinical isolates revealed that many of the 189 downregulated genes were involved in stress responses (see Table S1 in the supplemental material and Fig. 1). These stress-responsive genes included those involved in acid resistance (gadA, gadE, hdeABD) (31), osmotic stress (osmY, osmC, osmE, otsAB), and oxidative stress (katE, ygiW). Moreover, most of these genes, including gadA, osmY, otsAB, and katE, have been shown to be σS-dependent in various E. coli backgrounds, including E. coli K-12 and E. coli O157:H7 (15, 43). Additionally, σS-dependent genes involved in starvation, including hya (4, 21), dps (38), and psiF (32), were also downregulated in the clinical isolates. The spinach isolates (RM9998 and RM10002) that clustered more closely with the clinical isolates exhibited decreased expression of these same σS-dependent genes, while two of the spinach isolates (RM9992 and RM9997) expressed these genes at a level similar to that of the environmental isolates (see Table S1). Again, spinach isolate RM6067 possessed an expression level for these genes that was intermediate between the clinical isolates and environmental isolates (see Table S1). These gene expression results suggest that all the clinical isolates and a subset of the spinach isolates (RM9998 and RM10002) have a defect in the RpoS regulon.
As mentioned above, transcriptional comparison of a wild type and an rpoS mutant of E. coli O157:H7 identified many of the same stress-responsive genes identified in E. coli K-12; however, many O-island (not found in E. coli K-12) and metabolic genes also were expressed differentially only in E. coli O157:H7 (15). This suggests that in E. coli O157:H7 there are additional genes within the RpoS regulon. In particular, the rpoS mutant of E. coli O157:H7 strain EDL933 exhibited reduced expression of genes involved in virulence (ler, espI, and cesF), arginine degradation (astCADBE), putrescine degradation (puuABCD), and fatty acid oxidation (fadBA and fadE) and many involved in transport (including oppABCDF, potFGH, and malEFGK). None of these genes were among the 189 genes that were downregulated in the clinical isolates, which may indicate that the RpoS regulon is distinct between different strains of E. coli O157:H7. However, we did observe decreased expression of genes involved in dipeptide transport (dppADF) and arginine transport (artMPQ). Interestingly, tcdC encoding a threonine/serine transporter and cstA encoding a peptide transporter showed decreased expression in the EDL933 rpoS mutant (15) but had increased transcription in the clinical isolates in this study.
Verification of rpoS deficiencies in the clinical isolates.
To determine if the σS-dependent transcriptional differences corresponded to the direct effects of defective or reduced levels of σS, we sequenced rpoS and measured the levels of σS from the various isolates by Western blot analysis. The sequences of the rpoS gene and 5′ flanking region from all four environmental isolates and the two spinach isolates (RM9992 and RM9997) that had similar expression patterns revealed that they all possessed the same wild-type rpoS allele. Also, spinach isolate RM6067 possessed the same rpoS sequence as the environmental isolates. In contrast, the clinical isolates (RM6069, RM6655, RM6658) and the two other spinach isolates (RM9998, RM10002) possessed a variety of mutations within the rpoS gene, including deletions, insertions, and nonsense mutations (Table 2). Moreover, clinical isolates RM6069 and RM6658 possessed more than one detectable allele among different colony picks, including one wild-type rpoS allele among 12 colonies of RM6069 sequenced. Western blot analysis of stationary-phase cultures (equivalent of 1 OD of cells at 600 nm) demonstrated that the environmental isolates possessed consistently high levels of σS, as exhibited by RM6149 (Fig. 2). Three spinach isolates, RM6067, RM9992, and RM9997 (data not shown for this isolate), showed similarly high levels of σS, whereas clinical isolates RM6655 and RM6658 and spinach isolate RM9998 exhibited lower levels of σS than the environmental isolates (Fig. 2). Clinical isolate RM6069 (Fig. 2), clinical isolate RM6658, and spinach isolate RM9998 (data not shown for this isolate) also possessed colonies that exhibited levels of σS similar to those of the environmental isolates. The amount of σ70 for each isolate was determined to be consistent among all samples. Together, these results suggest that microheterogeneity exists for rpoS and the levels of σS within the populations of cells of the clinical and spinach isolates, but this heterogeneity was not apparent among the environmental isolates.
Table 2.
Mutations in the rpoS genes of clinical and spinach isolates
| Isolatea | Sourceb | Position in the rpoS gene | Mutation |
|---|---|---|---|
| RM6069-1 to -10 | HC | 999–1009 | Deletion of GCTGAATATCG, frameshift |
| RM6069-11 | HC | 159 | Deletion of C, frameshift |
| RM6069-12 | HC | Wild-type as compared to RM6103 | |
| RM6655 | HC | 388 | G-to-A transition, missense (D to N) |
| RM6658-1 | HC | 125 | T-to-G transversion, nonsense |
| RM6658-2 | HC | 473 | Insertion of A, frameshift |
| RM9998 | SB | 976 | Insertion of A, frameshift |
| RM10002 | SB | 43 | C-to-T transition, nonsense |
Numbers after strain number correspond to separate single colony picks.
HC, human clinical strain associated with 2006 spinach outbreak; SB, bagged spinach strain associated with the 2006 spinach outbreak.
Fig 2.
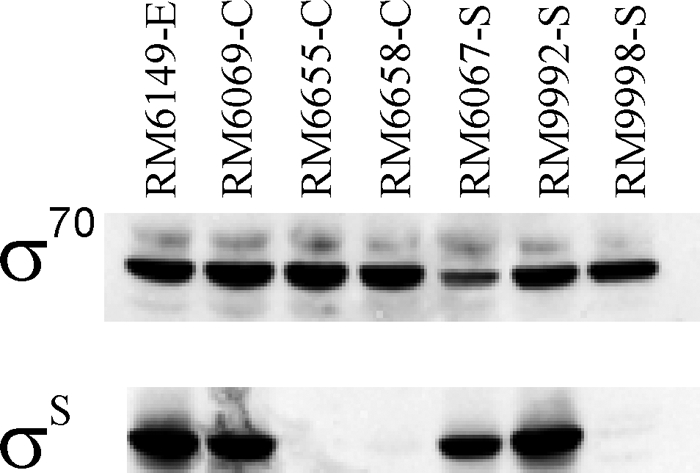
Production of σS in E. coli O157:H7 assessed by Western analysis using monoclonal anti-RpoS (σS) antiserum. The quantity of σS in stationary-phase cells at a concentration equivalent to 1 OD (at 600 nm) is shown for various clinical (C) and spinach (S) strains in comparison with an environmental strain from soil (E). Anti-RpoD (σ70) antiserum was used as an internal control.
Resistance of isolates to stress challenge corresponds to expression levels of stress-responsive genes.
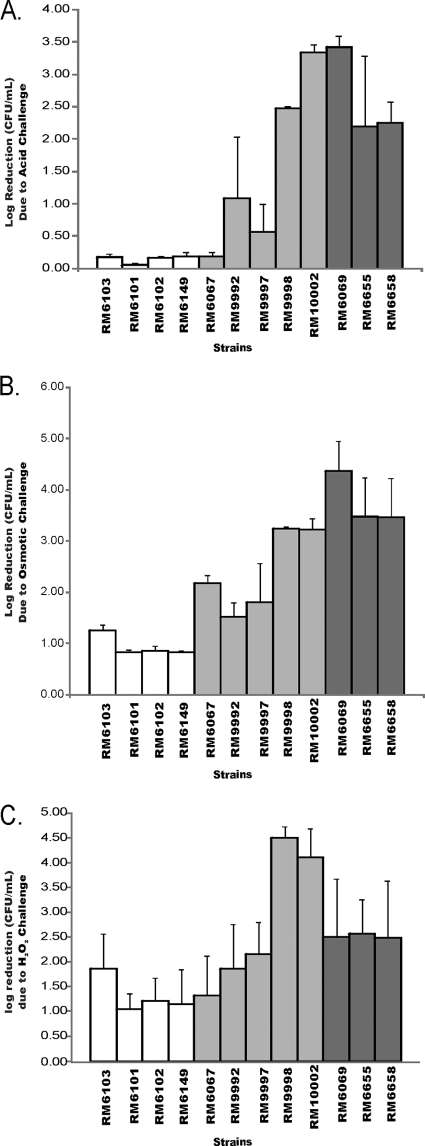
We performed several stress challenge experiments to determine if the differences in gene expression were relevant phenotypically. In response to acidic conditions, the E. coli O157:H7 isolates could be divided into two groups. The resistant group decreased by approximately one log or less over the 6-h exposure to pH 2.5 at 37°C (Fig. 3A). This group included all environmental isolates, as well as three spinach-derived isolates (RM6067, RM9992, RM9997). The second group comprised sensitive isolates, consisting of the three clinical isolates and two spinach isolates (RM9998, RM10002); approximately a 2- to 3-log decrease in survival was observed at the 6-h time point for these isolates (Fig. 3A). The E. coli O157:H7 isolates grouped with similar sensitivity patterns in response to osmotic or salt challenge conditions using a concentration of 2.5 M sodium chloride (Fig. 3B). Over the 25-h time course, the environmental isolates were observed to be the most fit isolates under these conditions, with spinach-derived isolates showing intermediate survival and clinical isolates least capable of surviving osmotic challenge. The sensitivity patterns exhibited during oxidative stress challenge using 12.5 mM H2O2 for 30 min were less obvious. Again, the environmental isolates as well as three spinach-derived isolates (RM6067, RM9992, RM9997) showed the smallest reduction in cell count (Fig. 3C). The two other spinach-derived isolates (RM9998, RM10002) were the most sensitive to oxidative stress (Fig. 3C). The clinical isolates were also more sensitive to oxidative challenge than the environmental isolates; however, approximately one-third of clinical isolate colonies were as resistant as the environmental isolates. It is noteworthy that in response to these functional challenges, the least robust isolates corresponded to those spinach-derived and clinical isolates showing decreased σS-dependent stress-responsive gene expression by microarray analysis.
Fig 3.
The reduction of E. coli O157:H7 cell numbers during stress challenge studies. (A) Exposure of E. coli O157:H7 cells to pH 2.5 for 6 h; (B) exposure of E. coli O157:H7 cells to 2.5 M NaCl for 25 h; (C) exposure of E. coli O157:H7 cells to 12.5 mM H2O2 for 30 min. The data represent the reduction of mean log cell concentration of E. coli O157:H7 in three replicate suspensions from time 0 h to the endpoint time for each condition. The replicates were each prepared from different stationary-phase inoculum cultures. The data bars are color-coded the same for each panel as follows: E. coli O157:H7 environmental strains, white; spinach bag strains, light gray; and clinical strains, dark gray. Error bars are one standard deviation of the mean.
Evidence for increased scavenging.
To address the SPANC hypothesis that rpoS mutants with reduced stress response have increased nutrient acquisition, we compared the metabolic differences in a Biolog assay with GN2 microplates of a clinical isolate (RM6069), two spinach bag isolates (RM6067 and RM9998), and the four environmental isolates (RM6101 to RM6103, RM6149). The two stress-sensitive isolates (RM6069 and RM9998) had similar metabolic fingerprints that were distinct compared with the environmental isolates and spinach bag isolate RM6067. Indeed, the two stress-sensitive isolates exhibited higher respiratory responses with 15 of 95 different substrates tested by the Biolog assay (Table 3). Of these 15 substrates inducing responses measured by the Biolog assay, the increased responses with l-serine and l-threonine correspond well with the increase in expression of tdcCA, genes involved in both l-threonine and l-serine uptake. Increased utilization of succinic acid (succinate) by RM6069 and RM9998 also is consistent with the SPANC hypothesis and the ability to select rpoS mutants on succinate minimal medium (14).
Table 3.
Substrates of Biolog GN2 microplates with distinct differences in activities between clinical and environmental isolates
| GN2 plate substrate | Respiration of E. coli O157:H7 isolatesa |
||||||
|---|---|---|---|---|---|---|---|
| Spinach RM6067 | Environmental |
Spinach clinical |
|||||
| RM6101 | RM6102 | RM6103 | RM6149 | RM9998 | RM6069 | ||
| Dextrin | 178 | 111 | 150 | 173 | 108 | 283 | 275 |
| l-Rhamnose | 61 | 20 | 48 | 92 | 23 | 197 | 192 |
| Methyl pyruvate | 204 | 198 | 211 | 282 | 162 | 416 | 477 |
| Mono-methylsuccinate | 176 | 217 | 242 | 226 | 134 | 486 | 442 |
| d-Glucuronic acid | 241 | 350 | 381 | 394 | 310 | 585 | 520 |
| d,l-Lactic acid | 152 | 159 | 197 | 367 | 192 | 539 | 433 |
| Succinic acid | 209 | 206 | 288 | 251 | 200 | 413 | 395 |
| d-Alanine | 6 | 3 | 7 | 190 | 7 | 318 | 214 |
| l-Alanine | 82 | 94 | 115 | 262 | 103 | 401 | 330 |
| l-Alanyl-glycine | 49 | 21 | 72 | 114 | 62 | 292 | 227 |
| Glycyl-l-aspartic acid | 51 | 17 | 22 | 42 | 21 | 251 | 265 |
| l-Serine | 93 | 14 | 40 | 48 | 21 | 333 | 465 |
| l-Threonine | 13 | 13 | 25 | 20 | 14 | 115 | 125 |
| Glycerol | 185 | 143 | 243 | 241 | 152 | 458 | 433 |
| d,l-α-Glycerolphosphate | 147 | 134 | 199 | 219 | 119 | 443 | 449 |
Values are a unitless measure of respiration based on the reduction of tetrazolium dye. Values are optical density units reflecting color formation due to oxidation of the compounds by bacterial cells. Optical density for each substrate was normalized to that in control wells (no substrate).
Interisolate and intraisolate E. coli O157:H7 curli variants.
We recently observed interisolate and intraisolate variation in curli expression in E. coli O157:H7 isolates (9). To investigate the incidence in curli variation among the E. coli O157:H7 isolates in this study, we determined the proportion of curli+ and curli− colonies on LBNS containing Congo red (CR). Curli bind CR; therefore, curli+ bacteria form red colonies on this medium. The interisolate variability was quite pronounced, with over 99% of the colonies formed by the environmental isolates and the spinach isolate RM6067 being curli− (Table 4). In contrast, the colonies from the E. coli O157:H7 clinical isolates were mostly curli+ (75% and 88%) (Table 4). The two other spinach isolates used in this study (RM9992 and RM9997) produced 45% and 62% curli+ colonies. These data reveal considerably more intraisolate variation among the spinach and from clinical isolates than among the environmental isolates of E. coli O157:H7 used in our study.
Table 4.
Curli variants in E. coli O157:H7 isolates used in this study
| Strain | Source | % curli variants ± SDa |
|---|---|---|
| RM6101 | Pig feces | 0.06 ± 0.06 |
| RM6102 | Water | 0.00 ± 0.00 |
| RM6103 | Cow feces | 0.06 ± 0.05 |
| RM6149 | Soil | 0.09 ± 0.07 |
| RM6067 | Spinach | 0.09 ± 0.10 |
| RM9992 | Spinach | 44.85 ± 5.30 |
| RM9997 | Spinach | 61.90 ± 9.87 |
| RM9998 | Spinach | 47.28 ± 8.53 |
| RM10002 | Spinach | 51.70 ± 10.85 |
| RM6069 | Clinical | 78.90 ± 2.35 |
| RM6655 | Clinical | 75.30 ± 5.58 |
| RM6658 | Clinical | 87.73 ± 8.11 |
Percentage of red/brown (curli-producing) colonies over the total number of E. coli O157:H7 colonies examined on CR medium.
DISCUSSION
In this study, we have demonstrated that during the passage of E. coli O157:H7 from “field to fork,” genetic mutations can occur that clearly affect global transcriptional patterns and survival under stress conditions that the pathogen likely will encounter during its contamination cycle. Cluster analysis of global transcriptional profiles from stationary-phase LB cultures revealed that genotypically related isolates (“outbreak strains”) from the spinach production environment (cow, feral pig, soil, and water) had expression patterns similar to each other but distinct from those in clinical isolates associated with the 2006 spinach outbreak. Furthermore, among isolates recovered from the spinach bags, some isolates exhibited transcriptional profiles similar to those of environmental isolates, others were similar to clinical isolate profiles, and one spinach bag isolate exhibited expression levels that were intermediate between the environmental and clinical isolate profiles. Among the genes that were differentially expressed (2-fold; P < 0.01) between the environmental and clinical isolates, we identified 189 genes that were downregulated in all of the clinical isolates compared with all of the environmental isolates (see Table S1 in the supplemental material). Many of these 189 genes were involved in adaptation to stresses, including acid resistance, osmotic shock, and oxidation, and have been reported previously by others to be part of the rpoS regulon of E. coli (43) and specifically in E. coli O157:H7 strain EDL933 (15). Indeed, we identified that isolates with a downregulated rpoS regulon possessed mutations within rpoS and/or had reduced levels of σS.
The appearance of isolates with reduced levels of σS corresponded to differential stress responses and metabolic patterns supporting the SPANC hypothesis, which predicts that certain environments encountered by E. coli O157:H7 will select for cells with an increased ability to scavenge for scarce nutrients at the expense of stress protection (22, 33). A relevant part of the hypothesis specifically suggests that competition between σS and σ70 defines the gene expression and phenotypes exhibited by the bacteria (22). In fact, rpoS mutants have been selected for by growth on nonpreferred carbon sources (14). Here, we demonstrated that the clinical and spinach isolates with mutations within rpoS and/or with decreased levels of σS exhibited decreased expression of stress-related genes (see Table S1 in the supplemental material) and lower resistance to acid, high-salt conditions, and oxidation (Fig. 2). Moreover, a clinical isolate and spinach isolate with mutated rpoS genes exhibited increased metabolic responses to 15 carbon sources compared to the responses of the four rpoS+ environmental isolates, a result supporting the SPANC hypothesis.
At this point, we can only speculate about the environment leading to the selection of the rpoS mutants. Considering that all of the environmental isolates examined in this study were rpoS+ and exhibited higher levels of σS compared with clinical isolates and some spinach isolates, it is not likely that the rpoS-selective niche was among these varied preharvest environments (cow, feral pig, pasture soil, and river water), although future studies may identify an environmental source that provides the rpoS mutant-selective niche. Conversely, E. coli O157:H7 isolates possessing rpoS mutant alleles were isolated from the spinach bag and the clinical samples. These clinical isolates were also acid sensitive, a result consistent with a report demonstrating the acid sensitivity of multiple clinical E. coli O157:H7 isolates compared with bovine isolates (34). Also, mouse colonization experiments using E. coli strain BJ4 showed that an rpoS mutant outcompeted the wild-type strain in the colon, perhaps due to the mutant's ability to scavenge limiting nutrients (24) and consistent with the concept that some intestinal environments might select for rpoS mutants. It is also possible that spinach outbreak rpoS+ E. coli O157:H7 strains were present from other clinical isolations but not represented in this study. The bagged spinach was the initial environment from “field to fork” in which the rpoS mutants were isolated. It is therefore possible that bagged spinach possesses the niche for selecting rpoS mutants and that these mutants were then merely maintained during passage through the human host. Considering that rpoS mutants have been selected for by growth on nonpreferred carbon sources (14), we speculate that bagged spinach provides such a niche. We recently demonstrated that E. coli O157:H7 cells in injured lettuce tissue exhibited upregulation of oxidative, osmotic, and antimicrobial stress genes and were correspondingly more resistant to oxidative compounds (26). Therefore, we also venture that these two different types of leafy greens, lettuce and spinach, likely provide distinctive environmental cues to the population of pathogenic bacteria that colonize them. To further understand the importance of the SPANC hypothesis, we are attempting to confirm the ability of bagged spinach to select for nutrient-competent but stress-sensitive mutants and to determine the rpoS state of clinical isolates related to the 2006 shredded lettuce outbreak.
It should be noted also that many of the E. coli O157:H7 isolates characterized in this study exhibited intraisolate variability. We observed very little intraisolate variation among the environmental isolates (RM6101, RM6102, RM6103, RM6149), where only wild-type rpoS was observed and less than 1% of colonies were curli+. However, we identified considerable intraisolate variation among spinach and clinical isolates. Specifically, E. coli O157:H7 isolates from spinach bags and clinical isolation possessed variability in rpoS alleles (Table 2), σS expression, differential sensitivity to oxidative stress, and curli expression (Table 4). Indeed, we previously reported that intraisolate curli+ and curli− variants occurred among E. coli O157:H7, and this coincided with differences in acid resistance (9). The higher incidence of curli− variants among environmental isolates and their greater resistance to acid stress, therefore, correlate well with our previous observation that curli− variants survive low-pH conditions better than curli+ variants. Such intraisolate variability of rpoS and associated phenotypes could allow E. coli O157:H7 to maintain a population of cells to ensure the presence of phenotypes that facilitate survival and rapid adaptation to a given niche and may be related to hypermutator isolates described previously in various pathogenic E. coli and Salmonella enterica isolates (27).
The isolation of rpoS mutants from clinical samples indicates that these spinach-associated E. coli O157:H7 strains were not impaired in survival through the human digestive tract and did not exhibit reduced virulence. In fact, the 2006 spinach outbreak resulted in levels of HUS that were significantly higher than in previous outbreaks (10). We did not observe any significant effect (2-fold difference and P < 0.01) on virulence genes when comparing the environmental (rpoS+) and clinical (rpoS mutant) isolates in this study. However, our rich medium growth conditions may have prevented differential expression of virulence genes. In contrast, the expression of over 50 O-island and virulence genes was decreased at least 2-fold in an rpoS mutant of E. coli O157:H7 EDL933 compared with that of the wild type (15). The transcriptional discrepancies between those data (15) and our results may be related to the potential differences in the rpoS regulon in different E. coli O157:H7 strains and may also be linked to the observed variations in virulence among E. coli O157:H7 strains in different clades (1, 30, 41). In fact, the E. coli O157:H7 isolates associated with the spinach outbreak are genotypically distinct from the commonly studied E. coli O157:H7 strains, EDL933 and Sakai. Genomic sequencing of a strain associated with the 2006 outbreak identified several genes that may be involved in its increased virulence (25). These included the Shiga toxin 2 gene variant (stx2c) that is associated with clade 8, a group of E. coli O157:H7 strains significantly associated with HUS patients (30). Sequencing and optical mapping of additional isolates from the spinach outbreak demonstrated prophage changes between isolates during the course of the outbreak (23). Our results reveal that changes occurred between isolates isolated from the preharvest environment and those isolated from human patients. Pressures hypothesized by SPANC (22, 33, 39) resulted in the selection of rpoS mutants of E. coli O157:H7 possessing increased nutrient scavenging ability but decreased stress resistance in the bagged spinach and/or within the patient. Ultimately, rpoS mutants were isolated by different public health laboratories from ill humans, suggesting they are virulent and may facilitate survival and rapid adaptation of E. coli O157:H7 at critical points of its contamination cycle from “field to fork.”
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Jay Hinton, Sacha Lucchini, and Arthur Thompson at the Institute for Food Research, Norwich, United Kingdom, for providing PCR products for E. coli K-12. We thank Anna Bates for technical assistance in this study. We thank Michael Cooley and Diana Carychao for providing the environmental isolates and W. Chmielecki (PA Department Public Health) and E. Hÿttia-Trees (CDC) for providing other outbreak isolates.
This work was supported also by the United States Department of Agriculture, Agricultural Research Service CRIS projects 5325-42000-044 and 5325-42000-045.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abu-Ali GS, Ouellette LM, Henderson ST, Whittam TS, Manning SD. 2010. Differences in adherence and virulence gene expression between two outbreak strains of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 156:408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold KW, Kaspar CW. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:2037–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atlung T, Knudsen K, Heerfordt L, Brondsted L. 1997. Effects of sigmaS and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J. Bacteriol. 179:2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhagwat AA, et al. 2005. Characterization of enterohemorrhagic Escherichia coli strains based on acid resistance phenotypes. Infect. Immun. 73:4993–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhagwat AA, et al. 2006. Functional heterogeneity of RpoS in stress tolerance of enterohemorrhagic Escherichia coli strains. Appl. Environ. Microbiol. 72:4978–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandl MT. 2008. Plant lesions promote the rapid multiplication of Escherichia coli O157:H7 on postharvest lettuce. Appl. Environ. Microbiol. 74:5285–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. California Food Emergency Response Team 2007. Investigation of an Escherichia coli O157:H7 outbreak associated with Dole prepackaged spinach. California Department of Public Health, Sacramento, CA: http://www.dhs.ca.gov/fdb/local/PDF/2006%20Spinach%20Report%20Final%20redacted%20no%20photosfigures.PDF.6 [Google Scholar]
- 9. Carter MQ, et al. 2011. Distinct acid resistance and survival fitness displayed by curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl. Environ. Microbiol. 77:3685–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. CDC 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb. Mortal. Wkly. Rep. 55:1–2 [PubMed] [Google Scholar]
- 11. CDC 2009. Surveillance for foodborne disease outbreaks—United States 2006. MMWR Morb. Mortal. Wkly. Rep. 58:609–615 [PubMed] [Google Scholar]
- 12. Cheville AM, Arnold KW, Buchrieser C, Cheng CM, Kaspar CW. 1996. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 62:1822–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cooley M, et al. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS One 2:e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong T, Chiang SM, Joyce C, Yu R, Schellhorn HE. 2009. Polymorphism and selection of rpoS in pathogenic Escherichia coli. BMC Microbiol. 9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dong T, Schellhorn HE. 2009. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics 10:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unraveling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 18. Gorden J, Small PL. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jenkins DE, Chaisson SA, Matin A. 1990. Starvation-induced cross protection against osmotic challenge in Escherichia coli. J. Bacteriol. 172:2779–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kailasan Vanaja S, Bergholz TM, Whittam TS. 2009. Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J. Bacteriol. 191:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. King PW, Przybyla AE. 1999. Response of hya expression to external pH in Escherichia coli. J. Bacteriol. 181:5250–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King T, Ishihama A, Kori A, Ferenci T. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kotewicz ML, Mammel MK, LeClerc JE, Cebula TA. 2008. Optical mapping and 454 sequencing of Escherichia coli O157:H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology 154:3518–3528 [DOI] [PubMed] [Google Scholar]
- 24. Krogfelt KA, Hjulgaard M, Sorensen K, Cohen PS, Givskov M. 2000. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect. Immun. 68:2518–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kulasekara BR, et al. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 77:3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kyle JL, Parker CT, Goudeau D, Brandl MT. 2010. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl. Environ. Microbiol. 76:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. LeClerc JE, Li B, Payne WL, Cebula TA. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211 [DOI] [PubMed] [Google Scholar]
- 28. Maharjan RP, Seeto S, Ferenci T. 2007. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J. Bacteriol. 189:2350–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mandrell RE. 2009. Enteric human pathogens associated with fresh produce: sources, transport and ecology, p 3–42 In Fan X, Niemira B, Doona CJ, Feeherry F, Gravani RB. (ed), Microbial safety of fresh produce: challenges, perspectives and strategies. IFT/Blackwell Publishing, Ames, IA [Google Scholar]
- 30. Manning SD, et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 189:2759–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Metcalf WW, Steed PM, Wanner BL. 1990. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J. Bacteriol. 172:3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Notley-McRobb L, King T, Ferenci T. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oh DH, et al. 2009. Escherichia coli O157:H7 strains isolated from environmental sources differ significantly in acetic acid resistance compared with human outbreak strains. J. Food Prot. 72:503–509 [DOI] [PubMed] [Google Scholar]
- 35. Rehfuss MY, Parker CT, Brandl MT. 2011. Salmonella transcriptional signature in Tetrahymena phagosomes and role of acid tolerance in passage through the protist. ISME J. 5:262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schäffer AA, et al. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29:2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stephani K, Weichart D, Hengge R. 2003. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol. Microbiol. 49:1605–1614 [DOI] [PubMed] [Google Scholar]
- 39. Stoebel DM, Hokamp K, Last MS, Dorman CJ. 2009. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 5:e1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tserenpuntsag B, Chang HG, Smith PF, Morse DL. 2005. Hemolytic uremic syndrome risk and Escherichia coli O157:H7. Emerg. Infect. Dis. 11:1955–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vanaja SK, Springman AC, Besser TE, Whittam TS, Manning SD. 2010. Differential expression of virulence and stress fitness genes between Escherichia coli O157:H7 strains with clinical or bovine-biased genotypes. Appl. Environ. Microbiol. 76:60–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Waterman SR, Small PL. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect. Immun. 64:2808–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zdziarski J, et al. 2010. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog. 6:e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.