Abstract
The genus Euduboscquella is one of a few described genera within the syndinean dinoflagellates, an enigmatic lineage with abundant diversity in marine environmental clone libraries based on small subunit (SSU) rRNA. The region composed of the SSU through to the partial large subunit (LSU) rRNA was determined from 40 individual tintinnid ciliate loricae infected with Euduboscquella sampled from eight surface water sites in the Northern Hemisphere, producing seven distinct SSU sequences. The corresponding host SSU rRNA region was also amplified from eight host species. The SSU tree of Euduboscquella and syndinean group I sequences from environmental clones had seven well-supported clades and one poorly supported clade across data sets from 57 to 692 total sequences. The genus Euduboscquella consistently formed a supported monophyletic clade within a single subclade of group I sequences. For most parasites with identical SSU sequences, the more variable internal transcribed spacer (ITS) to LSU rRNA regions were polymorphic at 3 to 10 sites. However, in E. cachoni there was variation between ITS to LSU copies at up to 20 sites within an individual, while in a parasite of Tintinnopsis spp., variation between different individuals ranged up to 19 polymorphic sites. However, applying the compensatory base change model to the ITS2 sequences suggested no compensatory changes within or between individuals with the same SSU sequence, while one to four compensatory changes between individuals with similar but not identical SSU sequences were found. Comparisons between host and parasite phylogenies do not suggest a simple pattern of host or parasite specificity.
INTRODUCTION
The dinoflagellate lineage can be roughly divided into two major groups, one that includes most familiar dinoflagellates with the typical dinokaryotic nucleus, or the dinokaryotes, and a second, the syndineans, that includes parasitic species lacking a dinokaryon (15). Syndinean dinoflagellates are a diverse and poorly understood marine group and may not represent a monophyletic lineage. Known syndinean hosts range from single-celled plankton, including other dinoflagellates, ciliates, and radiolarians, to parasites of fish eggs, copepods, and commercially important crustaceans such as lobsters and blue crabs (5, 10).
Interest in syndinean dinoflagellates was rekindled when marine environmental clone libraries dramatically expanded the breadth of the syndinean clade in small subunit (SSU) rRNA trees from just a few sequences for the genus Amoebophrya (18) to a series of clades that ultimately included thousands of sequences, although very few can be attributed to described species (17, 33). Originally described as belonging to marine alveolate groups I and II, sequences attributed to syndineans can now be classified into seven major groups (called clades I to VII), with groups I and II containing the bulk of the sequences.
In the past several years, progress has been made connecting syndinean rRNA sequences with the data from environmental clone libraries. Based on data currently available in GenBank, there are 204 rRNA sequences from syndinean taxa described to at least the genus level, mostly from the genera Hematodinium (176 sequences) (35, 42) and Amoebophrya (22 sequences) (18, 25), with a few sequences attributed to Duboscquella (2 sequences) (12a, 20), Ichthyodinium (12 sequences) (41, 48), and Syndinium (40). Syndinean group I was shown to contain species of both Ichthyodinium, parasites of fish eggs, and Duboscquella, parasites most often described as isolated from tintinnid ciliates.
Recently, several typical dinokaryotes such as Tintinnophagus and Duboscquodinium have also been shown to parasitize tintinnid ciliates (11), and the type species for the genus Duboscquella was also moved into the dinokaryotes, requiring the creation of a new genus, Euduboscquella, for the true syndinean parasites formerly in the genus Duboscquella (12a). Based on the SSU phylogeny, the two previously reported sequences for Duboscquella would represent syndineans assigned to Euduboscquella (12a, 20). Here we focus on the syndinean genus Euduboscquella, which, based on SSU rRNA phylogeny, belongs within clade I of the seven previously defined large-scale syndinean or marine alveolate clades (17, 20, 33).
The Euduboscquella life cycle involves a trophic intracellular growth phase emerging from the host cell and forming an elongating chain of rapidly dividing cells. After division is complete, different types of spores, including both motile and nonmotile spores, may be produced, although individual infections only produce one type of spore (4, 12a). For example, the recently described species E. crenulata produces two differently sized nonmotile spores and one motile spore, while E. anisospora produces two differently sized motile spores.
For the eight described Euduboscquella species, the most common hosts are tintinnid ciliates, although two species infect dinoflagellates and other protists may also act as hosts (4, 5, 10, 12a, 47). The tintinnid ciliates are attached to and surrounded by a lorica or shell, often with a wineglass- or bell-shaped outline (27). The lorica has distinctive features useful for host genus- or species-level taxonomy and also prevents the extracellular stages of the parasite life cycle from dispersing until late stages of infection. Here we describe the diversity and phylogeny of the genus Euduboscquella found within tintinnid ciliates as determined by the use of single-cell PCR for rRNA on field-collected material. A host phylogeny was simultaneously developed to compare host and parasite diversity characteristics.
MATERIALS AND METHODS
Sampling.
Plankton samples were collected using a 35-μm-pore-size plankton net at the water surface and were kept cool (∼15°C) during transport (see Table S1 in the supplemental material for site locations and dates). The samples were observed live with a dissecting or compound microscope for infected hosts and also fixed with Lugol's iodine to final concentrations of 0.04% (wt/vol) iodine and 0.06% (wt/vol) potassium iodide. Individual cells were picked with drawn glass capillaries and photographed with a digital camera mounted on a compound microscope at ×200 to ×400 magnification. Whenever possible, multiple replicate individual cells of the same morphotype were selected for amplification and sequencing. Host loricae were measured using Zeiss Axiocam and Axiovision software (Carl Zeiss Inc., Thornwood, NY). Whenever possible, 30 replicate host loricae from each ciliate morphotype were measured. Hosts were identified to the genus or species level, using the primary literature and assistance of S. Agatha (Fachbereich Organismische Biologie, Universität Salzburg).
Single-cell isolation.
For ciliates with apparent Euduboscquella infections, the hosts could often be dislodged from the lorica by repeated pipetting, with the developing parasite spores remaining behind in the lorica. Individual ciliate loricae were prepared by transferring each lorica through at least five drops of 0.45-μm-pore-filtered water under a dissecting microscope using drawn glass pipettes. For live cells, filtered site water was used, and for Lugol's iodine-fixed cells, filtered dionized water was used. The individual loricae containing either the parasite or host were then placed into a 1.5-ml centrifuge tube in a minimal volume (1 to 3 μl) of water and kept frozen.
Cell disruption.
Tubes containing individual host loricae were centrifuged at 10,000 × g for 1 min, and 40 μl of deionized UV-treated water was added to each tube. The cells were disrupted using a probe-tipped sonicator set to a 30% duty cycle and a power level of three with 3 pulses over 5 s (model W-225R; Heat Systems Ultrasonics Inc., Plainview, NY). Between samplings, the probe was rinsed with a 10% bleach solution for 30 s, rinsed with UV-treated deionized water, and dried with a lint-free tissue. Blank samples containing only water were interleaved after every third experimental sample, sonicated, and carried through the PCR in parallel with the experimental samples as negative controls. The sonicate was used as a template for PCR without further purification and contained sufficient volume for ∼8 individual reactions.
PCR methods.
The amplification reaction mixtures contained bovine serum albumin (Sigma A2053) (500 mg/ml), 50 mM Tris-HCl (Sigma T5128) at a pH of 8.3, 3 mM MgCl2, 10 μM deoxynucleoside triphosphates (dNTPs) (Promega, Madison, WI) (2.5 μM each nucleotide), 0.2 μm of each primer, and 0.12 U of GoTaq DNA polymerase (Promega) in a total volume of 20 μl, of which 4 μl was template. After initial denaturation at 94°C for 2 min, the reactions were run for 40 cycles of 94°C denaturing for 15 s, 55°C annealing for 15 s, and 90 s of extension at 72°C. For rRNA amplification, three partially overlapping primer pairs were used (see Table S2 in the supplemental material) as follows: EukA was paired with EukB for the SSU; a modified dinoflagellate-specific primer, DinoQ, from the end of the SSU was paired with a general large subunit (LSU) primer (25R1) for the region composed of the internal transcribed spacer (ITS) to the LSU; and, finally, a third general primer pair, 25F1 and LSUR2, was used to amplify the remaining LSU region. In several cases, taxon-specific primers were designed based on preliminary sequence data in order to obtain good-quality sequences (see Table S2 in the supplemental material). No nested PCR was done. PCR products were subjected to electrophoresis using 1% (wt/vol) agarose gels and Tris-acetate EDTA buffer, stained with ethidium bromide, and visualized using a GelDoc UV transilluminator and imager (Bio-Rad, Hercules, CA). Successful reactions were purified using polyethylene glycol precipitation (34). Sequencing reactions were run using a 10-μl volume and BigDye Terminator master mix (version 3.1; Life Technologies, Carlsbad, CA) and an Applied Biosystems 3730XL DNA analyzer according to the manufacturer's directions. For SSU rRNA sequencing, 6 primers were used, and for the remaining regions, an additional 6 primers were used (see Table S2 in the supplemental material).
Cloning.
For cloning of parasite ITS and other rRNA regions, high-fidelity Pfu Turbo (Stratagene, Hercules, CA) or Phusion (Fermentas, Glen Burnie, MD) polymerase was used with the reaction buffer supplied with the enzyme; the dNTP, primer, template, reaction volume, and thermocycling conditions were as described above. Cloning and transformation were done using ClonJET vector (Fermentas) and JM109 (Promega)- or Invitrogen TOP10 (Life Technologies, Carlsbad, CA)-competent cells according to manufacturer directions. The resulting individual bacterial colonies were added into 50 μl of 10 mM Tris (pH 8.0)–1 mM EDTA buffer, which was heated to 94°C for 5 min, followed by 5 min of centrifugation at 3,000 × g. Bacterial lysate (2 μl) was used as a template with pJET primers in a 20-μl PCR reaction mixture (Fermentas) for 30 cycles, with master mix, cycling conditions, PCR cleanup, and sequencing as described above.
The sequences were processed with Sequencher 4.10 software (Gene Codes, Ann Arbor, MI) to assemble and edit individual sequence reads into contiguous double-stranded sequences. The edited sequences were exported from Sequencher in FASTA format and manipulated in MacClade software (32). The nonredundant rRNA sequences from directly sequenced Euduboscquella spp., cloned Euduboscquella rRNA sequences, and novel ciliate SSU rRNA sequences have been deposited in GenBank (see below for accession numbers).
Host cell phylogeny.
For host sequences, all 61 SSU rRNA sequences for 41 different “tintinnida” (as circumscribed by GenBank) were downloaded. The sequences were aligned with Clustal W software (29), and the alignment was further refined manually with MacClade software. Positions were removed using Gblocks at different stringency levels to observe the overall effect on the number of useable sites and support for specific branches (6) (data not shown). Only SSU rRNA sequences were used for analysis of host relationships. For phylogenetic analysis, RAxML in the general time-reversible model with gamma correction and 100 bootstrap replicates was used (44).
Parasite phylogeny determined using SSU rRNA.
The large number of variable-quality environmental clone library sequences required a subsampling strategy. Analyses were performed using data sets with from 57 to 692 sequences from a starting group I syndinean SSU rRNA data set of 1,258 sequences (17). Pairwise comparisons of all the group I sequences were calculated using BLASTn. The BLAST results were parsed using a PERL script based on two criteria. First, only sequences that were redundant over 90% of the length were kept. Sequences were considered redundant at two stringency levels, allowing either two or five bases of difference. Second, three different size cutoffs (700, 1,000, and 1,400 bases) were used to exclude sequences. The two parameters created a matrix of six different nested subsampled data sets containing only sequences found at least twice in environmental clone libraries (3 sequence lengths × 2 identity cutoffs). The individual data sets were then aligned using MUSCLE software (14) and analyzed with RAxML using 100 bootstrap replicates and the general time-reversible model with gamma rate correction (44).
A second set of analyses with a more inclusive approach included singleton (unique) sequences. In this larger data set, duplicate sequences within two bases were eliminated as redundant and all sequences greater than 690 bases in length were kept, producing a data set with 700 nonredundant sequences. For alignment calculation of this large data set, the sequences were initially adjusted for length at the 5′ end. First, BLAST was used to align all sequences to the longest sequence (EU304548) as a reference and a PERL script was used to introduce the 5′ gap needed to make an initial alignment of the 5′ end to the reference sequence. Alignment without this adjustment was otherwise hampered, probably due to large differences in sequence sizes (from 690 bases to 6,000 bases) as well as large internal gaps in some of the sequences. After the initial 5′ alignment to the reference sequence, MUSCLE software was used to refine the alignment with 4 iterations, repeating the process three times and checking the alignment for changes after each repeat. The alignment was then iteratively checked using the RAxML tree as a guide to examine long branches for alignment artifacts. This was repeated four times. Eight sequences were excluded during this process due to extreme branch length, poor coverage across the alignment, or multiple unique insertions in the alignment. After the first alignment iteration, the data set was trimmed to include only the SSU region. The final data set was ultimately subjected to RAxML analysis with the general time-reversible model with gamma correction as described above but with a total of 500 bootstrap replicates.
ITS2 secondary structure for Euduboscquella.
Comparing the relatively conserved 5.8S, SSU, and LSU regions on either side of the more divergent ITS regions provided a rough estimate of the beginning and end of ITS1 and -2. Secondary structures were generated using mfold software with default parameters and putative ITS2 regions (49). The exact borders of the ITS2 region were then identified based on the base pairing between the end of the 5.8S and the start of the LSU region flanking the ITS2 (16, 23). When several structures were generated by the use of mfold software, comparisons were made between structures from closely related taxa to determine a common structure for the ITS2 in that clade. A query against the ITS2 database using the hidden markov model from the ITS2 database was also used but did not return a match (23). Compensatory base changes were identified based on predicted structures, and the alignment was adjusted to reflect the secondary structure.
Nucleotide sequence accession numbers.
The nonredundant rRNA sequences from directly sequenced Euduboscquella spp. were deposited in GenBank under accession no. JN934984 to JN934997, while cloned Euduboscquella rRNA sequences had the following accession numbers: JN934998 to JN935003 and JN966947 to JN966980. Novel ciliate SSU rRNA sequences were deposited under GenBank accession no. JN871720 to JN871726.
RESULTS
Host morphometry.
Morphological data used to identify host species are presented in Table S1 in the supplemental material. Based on lorica attributes, hosts belonged to the genus Eutintinnus (two species), Favella (three species), or Tintinnopsis (four species) (27) (Fig. 1). The Eutintinnus and Favella species were identified to the species level. Two of the four Tintinnopsis morphospecies were from the Rhode River, MD; one was tentatively identified as T. cf. subacuta (a Tintinnopsis species that looks like T. subacuta), and a second (referred to here as Tintinnopsis sp. from the Rhode River) was identified only to the genus level. A third Tintinnopsis species from Annapolis harbor, MD, was identified as T. major, and the fourth Tintinnopsis morphotype based on a specimen from Villefranche-sur-Mer, France, was identified only to the genus level.
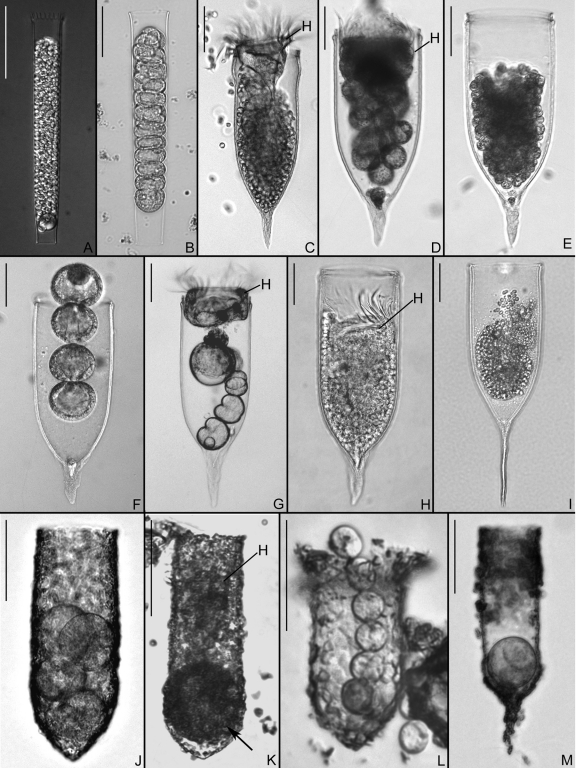
Fig 1.
Representative images of host species and parasites for which rRNA sequences were obtained. Bars, 50 μm. (A) Eutintinnus pectinis lorica filled with sporocytes of Euduboscquella cachoni. (B) Lorica of Eutintinnus tenuis containing Euduboscquella cachoni in early sporogenesis. (C) Favella arcuata lorica, with sporocytes of Euduboscquella sp. located below host cell (H). (D) Favella panamensis from Assawoman Bay, MD, with host cell (H) positioned above sporocytes of Euduboscquella crenulata. (E) Lorica of F. panamensis from Indian River Lagoon, FL, housing sporocytes of Euduboscquella cf. crenulata. (F) Favella panamensis lorica from Solomons Island, MD, with Euduboscquella cf. crenulata in a second sporogenic division. (G) Host cell (H) and sporocytes of Euduboscquella cf. crenulata in F. panamensis lorica from York River, MD. (H) F. panamensis from Masan Bay, Republic of Korea, with numerous sporocytes of Euduboscquella cf. crenulata posterior to host cell (H). (I) Lorica of Favella markusovszkyi, with a mass of sporocytes from Euduboscquella sp. (J) Tintinnopsis sp. lorica from Rhode River, MD, containing four sporocytes of Euduboscquella sp. (K) Lorica of Tintinnopsis cf. subacuta from Rhode River, MD, with a tomont of Euduboscquella sp. (arrow) located below host cell (H). (L) Tintinnopsis major lorica from Annapolis Harbor, MD, containing Euduboscquella sp. sporocytes. (M) Lorica of Tintinnopsis sp. from the Bay of Villefranche-sur-Mer, France, harboring a tomont of Euduboscquella sp.
Ciliate phylogeny.
Amplification and sequencing of ribosomal DNA from single-host ciliate cells yielded unambiguous sequences with good matches to ciliates in BLASTn searches, although apparent prey sequences were amplified in a few cases (data not shown). In this study, all SSU rRNA sequences from replicate individuals of the same ciliate morphotype were identical. For example, Favella panamensis yielded a single specific SSU rRNA sequence when different individuals were sampled from the same location and when individuals from different locations such as Masan Bay, Republic of Korea (Fig. 1H), or the U.S. Atlantic coast (Fig. 1D to G) were compared; overall, the SSU rRNA sequences for nine individual F. panamensis isolates from 5 sites were identical. In addition, two Tintinnopsis morphotypes from a single sample, one tentatively identified as T. cf. subacuta (Fig. 1K) and the second as Tintinnopsis sp. (Fig. 1J), yielded identical SSU rRNA sequences from eight individual cells. Only one ciliate host, identified as Tintinnopsis major, was not subjected to molecular analysis due to an insufficient sample.
Comparison of the ciliate host sequences to GenBank sequences by the use of BLASTn often yielded very close matches, although an exact identity between novel and GenBank sequences was found only in the case of Favella ehrenbergii and F. panamensis (AY143572), which may be synonymous (26). In other cases, such as that of E. pectinis, very similar sequences (4 base differences across 1,685 sites) attributed to the same species were found in GenBank. In one case, for the host identified here as Favella arcuata, collected from Assawoman Bay, MD, the most similar sequences in GenBank were associated with three genera, Rhabdonella (AY143566), Metacylis, (AY143567 and AY143568), and Favella (AF399162 and FJ196073) (30, 43, 46).
For host phylogenetic analysis, the 61 SSU rRNA sequences from tintinnida in GenBank were reduced to 39 nonredundant sequences by eliminating redundant identical or nearly identical sequences from the same species and by including the six novel sequences from this study. The ciliate host phylogeny clearly distinguishes between four well-supported clades: Eutintinnus (100% bootstrap), Tintinnidium (and others; 100%), Favella (pro parte; 100%), and a large, less-well-supported “Tintinnopsis” clade (87%) that includes sequences attributed to 7 or 8 genera (Fig. 2). There was poor support within the broadly defined Tintinnopsis clade, with multiple polyphyletic genera, and at least one sequence attributed to the genus Tintinnopsis (DQ487200) was outside this clade (30, 43, 45, 46).
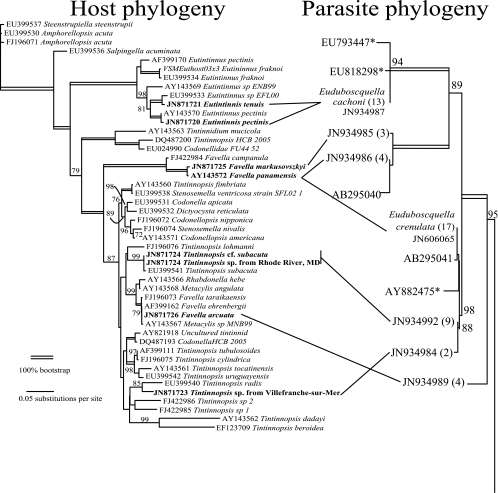
Fig 2.
Comparison of host and parasite SSU rRNA phylogenies. On the left, the maximum-likelihood phylogeny for the tintinnid hosts is shown, with the novel sequences in boldface type. On the right is a portion of the corresponding phylogeny for the parasite, showing only the Euduboscquella clade (see Fig. 3 for the complete phylogeny), with lines indicating the hosts. Next to the parasite accession numbers are the numbers of cells for which identical SSU sequences were determined. Bootstrap values are shown above the branches when greater than 70%.
Basic morphology of parasites.
The parasites exhibited development typical of the genus Euduboscquella, with parasites often found after emergence from the host or in late stages of sporogenesis, sometimes with the host remaining (Fig. 1). The characteristic chain of dividing cells (Fig. 1D, F, G, J, and L) or the mass of spores (Fig. 1A to C, E, H, and I) was readily identified as representing Euduboscquella parasites. The species E. crenulata was identified based on both trophont morphology and sporogenic products. Similarly, the complete host engulfment as well as sporogenic division patterns of E. cachoni were features diagnostic of the species. The remaining parasites were identified only to the genus level as Euduboscquella.
Syndinean group I SSU phylogeny.
Sequences attributed to syndineans have been classified into seven major groups (I to VII), and Euduboscquella was placed within syndinean group I, which was in turn divided into clades 1 to 8 in a previous study (17) (Fig. 3). Within group I, the only clades with sequences attributed to described species or genera were of clade 3 with Ichthyodinium and clade 4 with Euduboscquella.
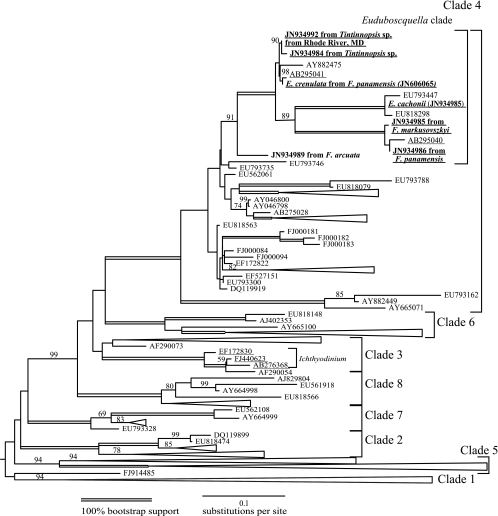
Fig 3.
Maximum-likelihood phylogeny of syndinean group I based on SSU rRNA. This tree contains 694 SSU sequences, of which 593 were from environmental clone libraries. These sequences were selected from a total of 1,258 based on a length of >690 bases, and redundant sequences (within two bases) were removed. Clade labels were based on data from reference 17. The two clades containing sequences attributed to Euduboscquella and Ichthyodinium were the only sequences from identified taxa, and those GenBank numbers are underlined. Bootstrap values greater than 60% are shown above the branches. See the supplemental material for an uncompressed version of the tree in Newick format.
Here, a total of nine different group I data sets were analyzed, subsampling the total available environmental clone library data. In six of the nine data sets, only duplicate sequences were used to avoid artifacts (Table 1, Fig. 3). Generally, as more sequences were included, most of the clades were well supported and consistent across the different analyses. Clades 2 and 4 consistently had 100% bootstrap support in all 9 different data sets. Clades 1 and 5 were generally well supported (94 to 100% support), and within both clades, two well-supported clades were found. Similarly, when multiple sequences were present, clades 6, 7, and 8 had high support (100%). Clade 3 was consistently poorly supported (maximum of 63% bootstrap) and was not monophyletic in some analyses, as was previously reported (17). More broadly, clades 3, 4, 6, and 8 together consistently formed a supported monophyletic group (79 to 100% bootstrap), although the relationships between clades 1, 2, 5, and 7 and the clade combining 3, 4, 6, and 8 were not consistent. In the largest, most inclusive data set, with 694 sequences of at least 690 bases, clades 1 and 5, each containing over 200 sequences, were the most common, followed by clade 4 with 85 sequences and clade 3 with 41. Clades 2, 6, and 7 were much smaller, containing at most 8 to 11 sequences (Fig. 3; see also Table S1 in the supplemental material).
Table 1.
Bootstrap support for different syndinean group I SSU rRNA datasets with maximum likelihood analysis
| Minimum sequence size (bp)a | Redundancyb | Total no. of sequences | Clade 1 |
Clade 2 |
Clade 3 |
Clade 4 |
Clade 5 |
Clade 6 |
Clade 7 |
Clade 8 |
Unassigned |
Euduboscquella (% support) | % bootstrap support (clades 3, 4, 6, and 8) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % support | No. of sequences | % support | No. of sequences | % support | No. of sequences | % support | No. of sequences | % support | No. of sequences | % support | No. of sequences | % support | No. of sequences | % support | No. of sequences | No. of sequences | |||||
| 700 | 2 | 113 | 100 | 25 | 100 | 3 | 10 | 5 | 100 | 13 | 100 | 10 | NA | 1 | NA | 0 | NA | 0 | 0 | 78 | 100 |
| 100 | 2 | 73 | 100 | 31 | 100 | 4 | NA | 6 | 100 | 19 | 100 | 12 | NA | 1 | NA | 0 | NA | 0 | 0 | 55 | 100 |
| 1,400 | 2 | 57 | 100 | 49 | 100 | 5 | 42 | 6 | 100 | 22 | 100 | 30 | NA | 1 | NA | 1 | NA | 0 | 0 | 56 | 100 |
| 700 | 5 | 170 | 100 | 65 | 100 | 9 | 31 | 7 | 100 | 30 | 100 | 52 | 100 | 3 | 100 | 2 | NA | 1 | 1 | 81 | 95 |
| 1,000 | 5 | 110 | 100 | 39 | 100 | 7 | 16 | 7 | 100 | 16 | 100 | 23 | 100 | 3 | NA | 1 | NA | 1 | 0 | 65 | 79 |
| 1,400 | 5 | 87 | 100 | 29 | 100 | 7 | 22 | 6 | 100 | 21 | 100 | 20 | 100 | 2 | NA | 1 | NA | 1 | 0 | 74 | 92 |
| 1,400 | NAc | 200 | 100 | 88 | 100 | 10 | NA | 19 | 100 | 24 | 100 | 49 | 100 | 7 | NA | 1 | 100 | 2 | 1 | 100d | 99 |
| 690 | NA | 696 | 81 | 282 | 100 | 24 | 63 | 41 | 100 | 85 | 95 | 224 | 100 | 10 | 100 | 8 | 100 | 11 | 11 | 73 | 100 |
| 690 | NA | 692 | 94 | 279 | 100 | 24 | 13 | 41 | 100 | 85 | 94 | 223 | 100 | 10 | 100 | 8 | 100 | 11 | 11 | 91 | 99 |
Sequences shorter than the cutoff were excluded.
Data represent the number of allowable nucleotide differences allowed between redundant sequences.
NA, only sequences differing by 2 or more bases were retained.
Data include 20 environmental clone sequences.
Clade Euduboscquella.
The Euduboscquella clade was consistent but often not well supported in the different analyses (55 to 91% bootstrap) and was always embedded within clade 4 (Fig. 2 and 3). Within the Euduboscquella clade, there were four different groups. First, the Euduboscquella parasite of F. arcuata from Assawoman Bay (Fig. 3) was most distant, even branching with 20 environmental sequences in one analysis but most often forming an outgroup to the Euduboscquella clade. The second Euduboscquella clade comprised a cluster of five very similar sequences, three from this study (SERC39, VSM21, and E. crenulata from F. panamensis), one attributed to Duboscquella in a previous study, and a single environmental sequence. The third and fourth clades each had three very similar members; one included E. cachoni (Fig. 1A and B) and two environmental sequences and the second two novel sequences and a sequence attributed to Duboscquella in a previous study. The relationships between these four Euduboscquella clades were well supported in most SSU analyses (Table 1).
Sequence variability between and within individual parasite cells.
Sequence variability within and between individuals was present. Replicate rRNA sequences between parasites from the same host species and from same location often contained ambiguous sites in the ITS1 (0 to 15 sites), ITS2 (1 to 10), and LSU (0 to 5) regions (Table 2; see also the supplemental material). As an example of a parasite with little variation, two parasitized Tintinnopsis spp. were found in a single sample from Villefranche-sur-Mer, France (Fig. 1M), and there was variation between the sequences from these two individuals. In the pairwise comparison, there were a total of 7 differences: none in the SSU, four in ITS1, three in ITS2, and none in the partial LSU. However, two of the ITS1 differences were ambiguous bases (double peaks) from one individual, suggesting a degree of sequence polymorphism within an individual.
Table 2.
Nucleotide changes in different regions based on comparisons of individual sequences and cloning
| Cell category and site | Host | No. of cells | No. of sequences | No. of nucleotide changesb |
|||
|---|---|---|---|---|---|---|---|
| ITS1 | ITS2 | LSU | Total | ||||
| Variability between individual parasite cells | |||||||
| U.S. Atlantic coast | Favella panamensis | 17 | 17 | 3 | 2 | 1 | 6 |
| Villefranche-sur-Mer, France | Favella markusovszkyi | 3 | 3 | 0 | 2 | 3 | 5 |
| Masan Bay, Republic of Korea | Favella panamensis | 4 | 4 | 1 | 1 | 1 | 3 |
| Villefranche-sur-Mer, France | Tintinnopsis sp. | 2 | 2 | 4 | 3 | 0 | 7 |
| Rhode River, MD | T. cf. subacuta | 4 | 4 | 6 | 9 | 4 | 19 |
| Rhode River, MD | Tintinnopsis sp. | 4 | 4 | 5 | 6 | 5 | 16 |
| Assawoman Bay, MD | Favellaarcuata | 4 | 14 | 1 | 6 | 3 | 10 |
| Rhode River, MD | Eutintinnus tenuis | 5 | 35 | 15 (5) | 10 (5) | 3 | 28 (10) |
| Annapolis harbor, MD | Eutintinnus pectinis | 2 | 27 | 8 (4) | 9 (3) | 0 | 17 (7) |
| Variability within individual parasite cells | |||||||
| Assawoman Bay | Favella arcuata | 1 | 6a | 1 | 6 | 2 | 9 |
| Assawoman Bay | Favella arcuata | 1 | 6a | 0 | 1 | 2 | 3 |
| Rhode River | T. cf. subacuta | 1 | 24a | 1 | 0 | 3 | 4 |
| Rhode River | Eutintinnus tenuis | 1 | 24a | 12 (4) | 8 (4) | 3 | 20 (4) |
| Annapolis Harbor | Eutintinnus pectinis | 1 | 22a | 3 (2) | 9 (2) | 0 | 12 (4) |
Number of clones sequenced.
Numbers in parentheses represent inferred insertions and deletions in the alignment.
Overall, cloning of rRNA amplicons was used to estimate polymorphism or sequence variation within an individual for 8 different individual infected hosts from five different ciliate hosts. Polymorphic sites were considered only when sequences from two or more clones or direct sequences shared a nucleotide change; variation found in only a single cloned sequence was not considered. Using rRNA from the Euduboscquella sp. found in Favella arcuata from Assawoman Bay (JN934989 in Fig. 2 and 3), a 2,955-base amplicon from the SSU to the LSU was generated and cloned from two individual loricae from the same site collected 1 week apart. Six clones were sequenced from each individual. The sequences differed by zero bases or one base in ITS1, one to six bases in ITS2, and two bases in the LSU, with a total of nine and six polymorphic sites in ITS-to-LSU sequences from these two individuals. Compared to results determined for two directly sequenced individual cells (Table 2), across all sequences from four individuals (two by direct sequencing and 12 cloned sequences), a total of 10 polymorphic sites were observed, eight of which were polymorphic according to comparisons of the two directly sequenced cells, and only two additional sites were found in the sequences from clones (Table 2). The sequence variation was restricted to single nucleotide polymorphism, and no insertions or deletions were detected. None of these base changes were compensatory across base-paired “stems” in the ITS2 or LSU regions.
E. cachoni in Eutintinnus tenuis and E. pectinis had a larger degree of sequence variation in the ITS region within an individual cell. Direct sequencing of SSU, ITS, and LSU PCR products was attempted using parasites from 10 individual parasitized loricae, and only two from E. tenuis produced interpretable contiguous sequences across the ITS region. However, for all 10 individuals, SSU and partial LSU sequences were obtained, and the sequencing chromatograms implied length variation in the ITS regions. In E. cachoni, there was a single ambiguous base in the SSU region, with 6 sequences having a G and four having an ambiguous G or T at the site, although there was not a correlation with host species.
Sequences for cloned PCR products for E. cachoni revealed differences across the problematic ITS region from the end of the SSU rRNA to the LSU. Multiple sequences were obtained by cloning for five different host loricae containing E. cachoni: three from E. tenuis (Fig. 1A) and two from E. pectinis (Fig. 1B). For the five different loricae, 3, 4, 6, 24, and 22 clones were sequenced and compared to the two directly sequenced ITS PCR products from E. tenuis. Comparisons of only the 22 and 24 more deeply sequenced cloned sequences representing the two different hosts are presented in Table 2. Although the two host species, E. tenuis and E. pectinis, were clearly distinct, as determined on the basis of SSU comparisons and morphology (Fig. 1A and B, 2, and 3), for the parasite, the sequence variation within an individual cell was as broad as the variation between individuals isolated from the two host species. In phylogenies determined using ITS regions as a whole or in part, sequences from parasites from the two different host species intermingled and did not form distinct clades (see Fig. S1 and S2 in the supplemental material). Variation occurred both between and within individual loricae, with up to 20 variable sites found in comparisons of clones from one individual, including two to four insertion deletion sites. Preliminary rarefaction (data not shown) suggested incomplete sampling of overall diversity. Across all 62 ITS-to-LSU regions for E. cachoni from 7 infected hosts found at two sites in two host species, the ITS1 region contained 5 instances of one to four base insertion deletions, as well as 16 polymorphic sites, and the ITS2 region contained 5 distinct insertion deletions and 16 polymorphic sites, while the LSU region contained 3 polymorphic sites. When mapped onto a provisional ITS2 structure, the sequence variation in the ITS2 region was mostly centered on the two “loops” at the end of stems III and IV; none of the sequence variations caused compensatory changes on both sides of a base-paired stem in ITS2.
In contrast to E. cachoni, the parasites from Tintinnopsis sp. (Fig. 1J) and T. cf. subacuta (Fig. 1K) from the Rhode River, MD, had little apparent sequence variation within an individual cell but relatively large variation between individuals (Table 2 and Table 3). These two Tintinnopsis host morphotypes yielded identical parasite SSU rRNA sequences in all 8 cases (3 from Tintinnopsis sp. and 5 from T. cf. subacuta), as well as identical host SSU sequences (6 from T. cf. subacuta and 2 from Tintinnopsis sp.). The parasite SSU sequences were also identical to the sequence amplified from a single T. major isolated from a nearby site about a month later, for which only a single parasitized host was found. Within the four parasite sequences from T. cf. subacuta, there were a total of 19 variable sites, while the four sequences from Tintinnopsis sp. from the Rhode River contained 16 variable sites (Table 2). Across all 9 individuals from the three different host morphotypes, there were a total of 27 variable sites in the parasite ITS-to-LSU regions over 1,415 bases. One individual from T. cf subacuta was selected for cloning, and the 24 cloned ITS sequences showed little within-cell variation, with one variant site found in the ITS1, none in the ITS2, and three in the LSU region. While the variation within an individual cell was small, the variable sites discovered in cloning were novel compared with variable sites from single-cell sequencing. Overall, the variable ITS sequences from this parasite did not sort with host morphotype.
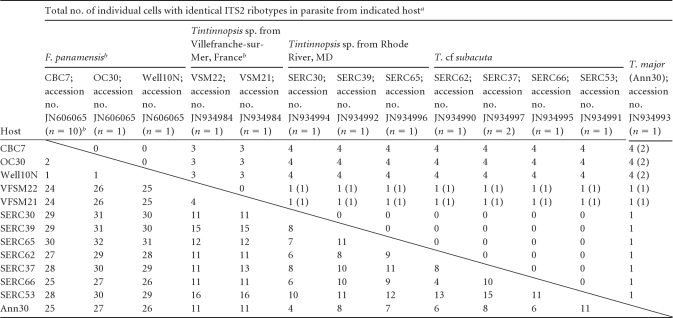
Table 3.
Compensatory and total nucleotide changes (above and below diagonal line, respectively) in the ITS2 region for a selected Euduboscquella clade
Compensatory and total nucleotide changes are shown above and below the diagonal line, respectively. Numbers in parentheses represent compensatory changes in the “stem” between 5.8S and LSU.
Single nonredundant sequences with ambiguous bases as needed were deposited in GenBank.
Compensatory base changes in the ITS2 region.
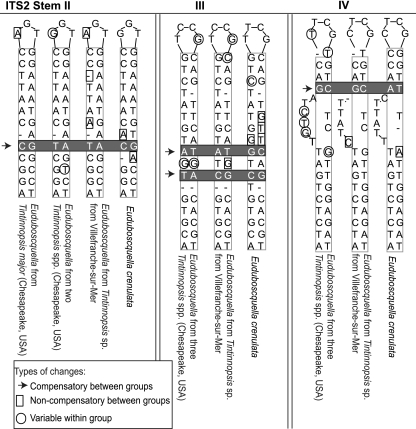
The parasites from all four Tintinnopsis hosts formed a tight cluster with E. crenulata from F. panamensis in the SSU tree, making comparisons of provisional ITS2 alignments and structures within the group possible (Fig. 4). The typical base-paired stem between the end of the 5.8S and beginning of the LSU rRNA was clearly distinguishable, and the provisional ITS2 structures followed the general four-stem pattern (23). The ITS2 differences were sorted into compensatory and noncompensatory changes. Compensatory changes were seen when both nucleotides in base-paired stem regions changed compared to noncompensatory changes occurring in single-stranded loops or on only one side of a stem (Table 3 and Fig. 4). Within this clade, there were 13 unique ITS2 ribotypes, 3 associated with E. crenulata in F. panamensis, 2 with the Tintinnopsis sp. host from Villefranche-sur-Mer, 7 from Tintinnopsis sp. and T. cf. subacuta hosts from the Rhode River, and 1 from T. major from Annapolis harbor. Overall, there were up to 30 ITS2 changes across all ribotypes, but most of the changes were noncompensatory. Between one and four compensatory base changes distinguished four groups of sequences, with each group having some noncompensatory variation: (i) E. crenulata with 2 noncompensatory changes across 3 ribotypes; (ii) the parasite of Tintinnopsis sp. from Villefranche-sur-Mer with 4 noncompensatory changes in the two ribotypes; (iii) the parasites from T. cf. subacuta and Tintinnopsis sp. from Rhode River, MD, with 4 to 15 noncompensatory changes among the 8 ribotypes; and (iv) the single T. major parasite that differed from the other Tintinnopsis species from the Rhode River by a single compensatory change in stem II. Overall, E. crenulata was more distantly related to the other parasites. Among these 13 ITS2 ribotypes, the unpaired loops at the end of the stem regions were most variable, with many noncompensatory changes.
Fig 4.
Alignments of homologous ITS2 regions of different Euduboscquella sequences. Compensatory changes in three different base-paired stems from the ITS2 of selected Euduboscquella species define four different parasite ITS2 ribotypes. Only stem II from the parasite from Tintinnopsis major isolated from Annapolis Harbor, MD, is shown. The sole ITS2 compensatory change between the sequence from the parasite from T. major isolated from Annapolis Harbor, MD, and the sequences from the other two Tintinnopsis host morphotypes isolates from the Rhode River, MD, was found in stem II. No compensatory changes between parasites from these three host morphotypes in stems III or IV were found. There was also a single compensatory change in stem III found for the parasites from the three Tintinnopsis hosts from the Chesapeake Bay, MD (T. major, T. cf. subacuta, and Tintinnopsis sp. from the Rhode River), and the parasite isolated from a fourth Tintinnopsis sp. from Villefranche-sur-Mer.
There were a number of differences between the Masan Bay parasite from F. ehrenbergii and the parasite from F. markusovszkyi found in Villefranche-sur-Mer with respect to the ITS2 regions. Between these two parasites, there were only seven SSU differences (JN934985 versus JN934986) (Fig. 2 and 3), but there was a large 34-base extension of stem IV in the ITS2 from F. ehrenbergii in the Masan Bay parasite, and there were four compensatory changes in stems I to III among the Euduboscquella isolates from these two sites and hosts.
DISCUSSION
Syndinean group I phylogeny based on SSU rRNA.
The reported diversity of syndinean dinoflagellates is largely based on data from environmental clone libraries, and here we compare results from directed sequencing of a specific syndinean genus, Euduboscquella. Sequences from environmental clone libraries may suffer from a number of potential artifacts, including amplification or sequencing errors leading to single nucleotide changes and chimeric amplicons combining partial sequences from two or more distinct organisms (3). Within the dinokaryotes, the divergence of rRNA genes has a broad range. In some dinoflagellates, multiple SSU rRNA sequences are present in a single individual such that multiple environmental clone sequences could be attributed to a single species (38). At the other extreme, there is an example of identical rRNA sequences (SSU through LSU) for two dinokaryote species (31). Determining whether a particular environmental clone represents the sequence from an organism in the environment can be difficult, particularly when reference sequences from described species are not present, as was often the case for syndineans.
Here we employed a graduated strategy using only duplicate sequences to estimate the phylogeny of syndinean group I and took advantage of a simple heuristic. Since a chimeric breakpoint would probably not happen at the same place twice, duplicated or redundant sequences would be less likely to be chimeric (28). Similarly, using differently sized sequences in the analysis might also alter the tree topology or support. However, the results from analysis performed with increasing numbers of sequences indicated that inclusion of unique and shorter sequences did not decrease bootstrap support or alter the tree topology. The phylogenies were quite stable across the range of sequence parameters. Overall, these results suggest that sequence artifacts have not created misleading clades in this group and, instead, that the environmental clone libraries represent genuine undiscovered species diversity in the ocean.
The syndinean group I SSU rRNA tree contains a number of diverse clades, often with well-supported branches, although the larger-scale relationships between clades 1, 2, 5, and 7 and the monophyletic group that includes clades 3, 4, 6, and 8 remain ambiguous. The SSU tree for syndinean group I strongly contrasts with the poorly resolved and unsupported relationships among dinokaryotic dinoflagellates based on SSU rRNA sequences (21, 36, 39). Within the Euduboscquella clade, there were very few environmental clone sequences and few overlaps between the two sampling strategies, and more individual cell sequencing and environmental clone libraries would be needed to exhaustively sample overall diversity of the group.
The single-cell PCR techniques used here were quite laborious, particularly compared to the bulk high-throughput sequencing of environmental clone data sets, and required infection diagnosis of individual cells. However, the single-cell PCR technique allowed amplification of a large contiguous section of rRNA from the spore products of a single infected host. The variation within and between individuals could be estimated by comparing cloned sequences from an individual to sequences from multiple individuals parasitizing the same host. Simultaneously, diagnostic features could be used for genus or species level identification of both host and parasite.
Syndinean dinoflagellates have been distinguished from the other major dinoflagellate lineage, namely, the dinokaryotes, based primarily on nuclear features, with the dinokaryotes having high DNA content and chromosomes condensed during the interphase (15). Recently, the type for the genus Duboscquella was found to have dinokaryotic features, and so the remaining, truly syndinean tintinnid parasites were moved into the new genus Euduboscquella (12a). Although Euduboscquella has been most often described in studies of isolates from tintinnids, other dinoflagellate parasites of tintinnids include species of the genera Duboscquodinium, Duboscquella, and Tintinnophagus. In those three genera, extracellular growth, cell division, motile spores, and available rRNA sequence data indicated that these genera were dinokaryotic dinoflagellates and not syndineans (11, 12a).
All the parasite sequences described here can be assigned to the genus Euduboscquella. The genus Euduboscquella includes intracellular parasitic dinoflagellates that divide only after exiting the host cell and show characteristic features during the trophic growth stage (4, 8, 12a). The division products from a single infection produce either motile dinospores or nonmotile spores, although the full morphological variation within species remains poorly characterized (4, 9, 12a). In comparison, the syndinean genus Amoebophrya, found in syndinean clade II, produces a single type of spore and undergoes nuclear division inside the host cell (4, 5). Discrimination between different Euduboscquella species has often been based on specific trophont features found only during the intracellular life stage—features not accessible in many of the specimens seen here. Based on trophont and sporogenic stages, two species, E. cachoni and E. crenulata, were identified to the species level, although we hesitate to attribute species epithets to the remaining parasites. We chose to instead focus on describing the molecular diversity of the genus based on rRNA sequences.
Comparison of ITS2 changes.
The very short branch lengths between individual parasite sequences based on SSU phylogenies required additional sequence data from the more rapidly evolving ITS regions to clearly resolve relationships. Although multiple different SSU rRNA gene copies are found in some dinoflagellate genera (38), in the genus Euduboscquella, SSU rRNA variation within an individual was not found in this study. However, it was first necessary to quantify and estimate the ITS diversity of different copies within individuals and then compare the differences between individuals.
In three of the SSU rRNA clades, including (i) E. cachoni infecting Eutintinnus tenuis and E. pectinis, (ii) the parasites of Tintinnopsis spp. and E. crenulata from F. panamensis, and, finally, (iii) the parasites of F. panamensis and F. markusovszkyi, there were parasite sequences attributed to two or more host species. The fourth, most distant SSU clade contained only the parasite from F. arcuata. Within these clades, ITS2 sequence variation could be mapped onto the secondary structure to apply the compensatory base change model to these data. This model views complementary changes to both sides of a base-paired stem as an indication of a species barrier, based on the sexual incompatibility of individuals with different ITS2 structures (13) In the genus Euduboscquella, tests of the biological species concept based on sexual incompatibility and comparison to the compensatory model would be premature. However, the ITS2 compensatory change model is useful for describing molecular diversity within the genus and agrees well with the SSU-based tree.
In the case of E. cachoni, there was extensive sequence diversity (up to 20 sites) within individuals but no compensatory base changes between or within individuals were observed, suggesting that each individual contains multiple distinct rRNA gene copies. The sequence diversity within an individual was also greater than the diversity between individuals and between different host species, suggesting the presence of a single parasite species infecting two hosts.
Within the parasites of Tintinnopsis spp. and E. crenulata from F. panamensis, there were one to four compensatory base changes between (i) E. crenulata from F. panamensis, (ii) parasites of Tintinnopsis sp. from Villefranche-sur-Mer, (iii) parasites from Tintinnopsis sp. and T. cf. subacuta from the Rhode River, and (iv) T. major (a singleton sequence). The ITS2 compensatory base changes support the SSU phylogeny that defined three different subclades of Euduboscquella with very similar SSU sequences (E. crenulata, JN934984, and JN934992 on Fig. 2). The sequences attributed to E. crenulata were generated from multiple individuals isolated over 3 years from four distinct sites on the U.S. Atlantic coast, and yet, based on the rRNA sequence data, there was little variation in these parasite populations. In contrast, the parasite sequences from Tintinnopsis sp. and T. cf. subacuta were all generated from a single sample from the Rhode River, MD, and there was substantial ribosomal sequence diversity between individual cells, rather than variation within an individual, based on comparison to the cloned sequences. Since only two identical ITS2 sequences were found within this group, additional undescribed diversity in ITS ribotypes was likely, suggesting that the overall diversity was only partially sampled. Despite this variability, none of the changes were compensatory, again agreeing well with the compensatory base change model. Interestingly, there were no SSU sequence differences between the different host morphotypes. These two host morphotypes were here considered to represent two different species based on morphology, but the identical SSU sequences suggested that more molecular data would be required to distinguish the host species. Alternatively, the two host morphotypes may indeed belong to the same host species.
Similarly, according to the results of comparisons of the parasites of F. panamensis from Masan Bay, Republic of Korea, with the parasites from F. markusovszkyi, there were a number of distinct compensatory changes. The compensatory model seems to fit the data well; whether applied to variation within or between individuals, compensatory changes agreed well with subtle SSU sequence differences.
Comparing host and parasite phylogenies.
Considering host and parasite phylogenies, the species and generic level taxonomy of both host and parasite may continue to change in the future (1, 2). However, comparing the existing host and parasite molecular phylogenies suggests three discordant patterns in the data. First, some parasites, such as E. cachoni, appear to infect multiple host species, although host preferences and prevalences were not explored. Second, a single host species, F. panamensis, was infected by two distinct parasites, E. crenulata from multiple sites in the U.S. Atlantic coast and a different parasite found in the same host from Masan Bay, Republic of Korea. Finally, the similarity between F. markusovszkyi and F. panamensis hosts was mirrored by their parasites and might represent an example of host parasite coevolution. Model-based tests of coevolution would be premature, given the sparse sampling of diversity. Overall, these data suggest that discriminating Euduboscquella parasite species solely on the basis of host identification should be discouraged.
Comparison of syndinean clades I and II.
The results for Euduboscquella from syndinean group I can be compared with the results for the genus Amoebophrya from syndinean group II. While some Euduboscquella seem to infect more than one host and more than one Euduboscquella may infect the same host species, very little work has been done on the genus Euduboscquella in comparison to Amoebophrya. Within the genus Amoebophrya, environmental and culture studies have suggested host parasite specificity in some cases (7, 12, 19, 22). However, there also seem to be cases where Amoebophrya may not be host specific (24, 37). In addition, while Euduboscquella species formed a specific clade within clade 4 of group I, sequences attributed to Amoebophrya were found in a large (in terms of both diversity and number) fraction of syndinean clade II. However, it remains to be seen whether these preliminary results truly capture the diversity of these different parasite genera or, more broadly, what features distinguish the organisms in syndinean clades I and II. Future work should help to reveal whether some of the more abundantly represented clades from environmental clone libraries include many unique parasite species.
However, estimating the overall diversity of syndineans, or of Euduboscquella and Amoebophrya, can only be described as a work in progress. The results presented here agree well with the idea that even subtle differences in SSU sequence may be correlated with distinct species at the ITS2 and ultimately at the morphological level, suggesting that the diversity of environmental clone sequences represents a diverse array of potential parasite species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NSF Assembling the Tree of Life grant (EF-06299624) to C.F.D., D.W.C., and colleagues.
We thank Sabine Agatha of Fachbereich Organismische Biologie, Universität Salzburg, for expert ciliate host identification. Cindy Gilmour from the Smithsonian Environmental Research Center, the captain and crew of the R.V. Sharpe, W. Lee and H. Reichert of the Smithsonian Marine Station, and Myung-Gil Park of Chonnam National University provided gracious hospitality and assistance with acquiring samples. John Dolan of the Observatoire Oceanologique at Villefranche-sur-Mer also provided facilities, assisted with sampling, and was supported by the French ANR-Biodiversité project AQUAPARADOX.
This publication is contribution 868 from the Smithsonian Marine Station.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Agatha S, Riedel-Lorjé JC. 2006. Redescription of Tintinnopsis cylindrica Daday, 1887 (Ciliophora, Spirotricha) and unification of tintinnid terminology. Acta Protozool. 45:137–151 [PMC free article] [PubMed] [Google Scholar]
- 2. Agatha S, Strüder-Kypke M. 2007. Phylogeny of the order Choreotrichida (Ciliophora, Spirotricha, Oligotrichea) as inferred from morphology, ultrastructure, ontogenesis, and SSrRNA gene sequences. Eur. J. Protistol. 43:37–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berney C, Fahrni J, Pawlowski J. 2004. How many novel eukaryotic ‘kingdoms’?Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cachon J. 1964. Contribution à ľétude des péridiniens parasites. Cytologie, cycles évolutifs. Ann. Sci. Nat. Zool. 6:1–158 [Google Scholar]
- 5. Cachon J, Cachon M. 1987. Parasitic dinoflagellates, p 571–610 In Taylor FJR. (ed), The biology of dinoflagellates, vol 21 Blackwell Scientific Publications, Oxford, United Kingdom [Google Scholar]
- 6. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 7. Chambouvet A, Morin P, Marie D, Guillou L. 2008. Control of toxic marine dinoflagellate blooms by serial parasitic killers. Science 322:1254–1257 [DOI] [PubMed] [Google Scholar]
- 8. Chatton E. 1920. Les peridiniens parasites. Morphologie, reproduction, ethologie. Arch. Zool. Exp. Gen. 59:1–475 [Google Scholar]
- 9. Coats DW. 1988. Duboscquella cachoni n. sp., a parasitic dinoflagellate lethal to its tintinine host Eutintinnus pectinis. J. Protozool. 35:607–617 [Google Scholar]
- 10. Coats DW. 1999. Parasitic life styles of marine dinoflagellates. J. Eukaryot. Microbiol. 46:402–409 [Google Scholar]
- 11. Coats DW, Kim S, Bachvaroff TR, Handy SM, Delwiche CF. 2010. Tintinnophagus acutus n. g., n. sp. (Phylum Dinoflagellata), an ectoparasite of the ciliate Tintinnopsis cylindrica Daday 1887, and its relationship to Duboscquodinium collini Grassse 1952. J. Eukaryot. Microbiol. 57:468–482 [DOI] [PubMed] [Google Scholar]
- 12. Coats DW, Park MG. 2002. Parasitism of photosynthetic dinoflagellates by three strains of Amoebophyra (Dinophyta): parasite survival, infectivity, generation time, and host specificity. J.Phycol. 38:520–528 [Google Scholar]
- 12a. Coats DW, Bachvaroff TR, Delwiche CF. Revision of the family Duboscquellidae with description of Euduboscquella crenulata n. gen., n. sp. (Dinoflagellata, Syndinea), an intracellular parasite of the ciliate Favella panamensis Kofoid and Campbell, 1929. J. Eukaryot. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 13. Coleman AW, Vacquier VD. 2002. Exploring the phylogenetic utility of ITS sequences for animals: a test case for abalone (Haliotis). J. Mol. Evol. 54:246–257 [DOI] [PubMed] [Google Scholar]
- 14. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fensome RA, et al. 1993. A classification of living and fossil dinoflagellates. Micropaleontology special publication no. 7. American Museum of Natural History, New York, NY [Google Scholar]
- 16. Gottschling M, Plötner J. 2004. Secondary structure models of the nuclear internal transcribed spacer regions and 5.8S rRNA in Calciodinelloideae (Peridiniaceae) and other dinoflagellates. Nucleic Acids Res. 32:307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guillou L, et al. 2008. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata). Environ. Microbiol. 10:3349–3365 [DOI] [PubMed] [Google Scholar]
- 18. Gunderson JH, Goss SH, Coats DW. 1999. The phylogenetic position of Amoebophrya sp. infecting Gymnodinium sanguineum. J. Eukaryot. Microbiol. 46:194–197 [DOI] [PubMed] [Google Scholar]
- 19. Gunderson JH, John SA, Boman WC, Coats DW. 2002. Multiple strains of the parasitic dinoflagellate Amoebophrya exist in Chesapeake Bay. J. Eukaryot. Microbiol. 49:469–474 [DOI] [PubMed] [Google Scholar]
- 20. Harada A, Ohtsuka S, Horiguchi T. 2007. Species of the parasitic genus Duboscquella are members of the enigmatic marine alveolate group I. Protist 158:337–347 [DOI] [PubMed] [Google Scholar]
- 21. Hoppenrath M, Leander BS. 2010. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS One 5:e13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Janson S, Gisselson L-Å, Salomon PS, Granéli E. 2000. Evidence for multiple species within the endoparasitic dinoflagellate Amoebophrya ceratii as based on 18S rRNA gene-sequence analysis. Parasitol. Res. 86:929–933 [DOI] [PubMed] [Google Scholar]
- 23. Keller A, et al. 2009. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430:50–57 [DOI] [PubMed] [Google Scholar]
- 24. Kim S. 2006. Patterns in host range for two strains of Amoebophrya (Dinophyta) infecting thecate dinoflagellates: Amoebphrya spp. ex Alexandrium affine and ex Gonyaulax polygramma. J. Phycol. 42:1170–1173 [Google Scholar]
- 25. Kim S, et al. 2008. Genetic diversity of parasitic dinoflagellates in the genus Amoebophrya and its relationship to parasite biology and biogeography. J. Eukaryot. Microbiol. 55:1–8 [DOI] [PubMed] [Google Scholar]
- 26. Kim SY, Yang EJ, Gong J, Joong KC. 2010. Redescription of Favella ehrenbergii (Claparéde and Lachmann, 1858) Jörgensen, 1924 (Ciliophora: Choreotrichia), with phylogenetic analyses based on small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 57:460–467 [DOI] [PubMed] [Google Scholar]
- 27. Kofoid CA, Campbell AS. 1929. A conspectus on the marine and freshwater Ciliata belonging to the suborder Tintinnoinea, with descriptions of new species principally from the Agassiz expedition to the eastern tropical Pacific 1904–1905. Univ. Cal. Publ. Zool. 34:1–403 [Google Scholar]
- 28. Lahr DJ, Nguyen TB, Barbero E, Katz LA. 2010. Evolution of the actin gene family in testate lobose amoebae (Arcellinida) is characterized by two distinct clades of paralogs and recent independent expansions. Mol. Biol. Evol. 28:223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larkin M, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 30. Li Z, et al. 2009. Phylogenetic investigation on five genera of tintinnid ciliates (Ciliophora, Choreotrichia), based on the small subunit ribosomal RNA gene sequences. Prog. Nat. Sci. 19:1097–1101 [Google Scholar]
- 31. Logares R, et al. 2007. Phenotypically different microalgal morphospecies with identical ribosomal DNA: a case of rapid adaptive evolution? Microb. Ecol. 53:549–561 [DOI] [PubMed] [Google Scholar]
- 32. Maddison WP, Maddison PR. 2002. MacClade version 4: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland, MA [Google Scholar]
- 33. Moon-van der Staay S, De Wachter R, Vaulot D. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607–610 [DOI] [PubMed] [Google Scholar]
- 34. Morgan DR, Soltis DE. 1995. Phylogenetic relationships among members of Saxifragaceae sensu lato based on rbcL sequence data. Ann. Mo. Bot. Gard. 80:631–660 [Google Scholar]
- 35. Ryazanova TV, Eliselkina MG, Kukhlevsky AD, Kharlamenko VI. 2010. Hematodinium sp. infection of red Paralithodes camtschaticus and blue Paralithodes platypus king crabs from the Sea of Okhotsk, Russia. J. Invertebr. Pathol. 105:329–334 [DOI] [PubMed] [Google Scholar]
- 36. Saldarriaga JF, Taylor FJ, Keeling PJ, Cavalier-Smith T. 2001. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J. Mol. Evol. 53:204–213 [DOI] [PubMed] [Google Scholar]
- 37. Salomon PS, Janson S, Granéli E. 2003. Multiple species of the dinophagous dinoflagellate genus Amoebophrya infect the same host species. Environ. Microbiol. 5:1046–1052 [DOI] [PubMed] [Google Scholar]
- 38. Scholin CA, Anderson DM. 1993. The existence of two distinct small-subunit rRNA genes in the toxic dinoflagellate Alexandrium fundyense. J. Phycol. 29:209–216 [Google Scholar]
- 39. Shalchian-Tabrizi K, et al. 2006. Heterotachy processes in rhodophyte-derived second hand plastid genes: implications for addressing the origin and evolution of dinoflagellate plastids. Mol. Biol. Evol. 23:1504–1515 [DOI] [PubMed] [Google Scholar]
- 40. Skovgaard A, Massana R, Balague V, Saiz E. 2005. Phylogenetic position of the copepod-infesting parasite Syndinium turbo (Dinoflagellata, Syndinea). Protist 156:413–423 [DOI] [PubMed] [Google Scholar]
- 41. Skovgaard A, Meneses I, Angélico MM. 2009. Identifying the lethal fish egg parasite Ichthyodinium chabelardi as a member of marine alveolate group I. Environ. Microbiol. 11:2030–2041 [DOI] [PubMed] [Google Scholar]
- 42. Small HJ, Neil DM, Taylor AC, Atkinson RJ, Coombs GH. 2006. Molecular detection of Hematodinium spp. in Norway lobster Nephrops norvegicus and other crustaceans. Dis. Aquat. Organ. 69:185–195 [DOI] [PubMed] [Google Scholar]
- 43. Snoeyenbos-West O, Salcedo T, McManus GB, Katz LA. 2002. Insights into the diversity of choreotrich and oligotrich ciliates (class: Spirotrichea) based on genealogical analyses of multiple loci. IJSEM 52:1901–1913 [DOI] [PubMed] [Google Scholar]
- 44. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 45. Strüder-Kypke M, Lynn DH. 2008. Morphological versus molecular data—phylogeny of tintinnid ciliates (Ciliophora, Choreotrichia) inferred from small subunit rRNA gene sequences. Denisia 23:417–424 [Google Scholar]
- 46. Strüder-Kypke M, Lynn DH. 2003. Sequence analyses of the small subunit rRNA gene confirm the paraphyly of oligotrich ciliates sensu lato and support the monophyly of the subclasses Oligotrichia and Choreotrichia (Ciliophora, Spirotrichea). J. Zool. (London) 260:87–97 [Google Scholar]
- 47. Suzuki N, Kurihara T, Matsuoka A. 2009. Sporogenesis of an extracellular cell chain from the spheroidal radiolarian host Haliommilla capillaceum (Haeckel), Polycystina, Protista. Mar. Micropaleontol. 72:157–164 [Google Scholar]
- 48. Yuasa K, et al. 2007. Infection by a protozoan endoparasite of the genus Ichthyodinium in embryos and yolk-sac larvae of yellowfin tuna Thunnus albacares. Fish Pathol. 42:59–66 [Google Scholar]
- 49. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.