Abstract
Anaplasma species are obligate intracellular rickettsial pathogens that impact the health of humans and animals. Few studies have been carried out on Anaplasma infections in central and southern China. This study was conducted to determine the coinfection rates of Anaplasma ovis, A. bovis, and A. phagocytophilum from 262 field blood samples of goats in these regions. The average prevalences of single infection of A. ovis, A. bovis, and A. phagocytophilum were 15.3, 16.0, and 6.1%, respectively. Coinfection of A. ovis and A. bovis was dominant, with an infection rate of 27.1%. Coinfection of A. ovis and A. phagocytophilum was 1.9% and that of A. bovis and A. phagocytophilum was 4.2%. Three-pathogen coinfection was found in three of four investigated provinces with a prevalence between 0 and 5.3%. The accuracy of the PCR results was corroborated by sequencing. Analysis of the 16S rRNA gene sequences of A. bovis and A. phagocytophilum confirmed the presence of these pathogens at the investigated sites and indicated the possible genetic diversity of A. phagocytophilum. Field blood inoculation of experimental animals led to successful identification and observation of the morphological shapes of A. bovis in the infected monocytes of sheep. Phylogenetic study with msp4 sequences of A. ovis indicated that the A. ovis genotypes from sheep in the north differed from the genotypes of goats in the investigated sites.
INTRODUCTION
Bacteria of the genus Anaplasma are obligate intracellular etiological agents of tick-borne diseases of mammalian hosts (9). The major species that impact animal and human health are Anaplasma marginale, A. ovis, A. centrale, A. bovis, A. phagocytophilum, and A. platys. Among these, A. marginale and A. ovis are the main inter-erythrocytic pathogens of bovine and ovine animals. They are responsible for bovine anaplasmosis and ovine anaplasmosis in tropical and subtropical areas (1, 18). A. centrale is less pathogenic than the closely related A. marginale. A. centrale has been used as a live vaccine for cattle in Israel, South Africa, South America, and Australia (7). A. phagocytophilum, also known as Ehrlichia equi and Ehrlichia phagocytophila, is the causative agent of human granulocytic anaplasmosis, tick-borne fever of ruminants, which infects a wide range of hosts, including humans and wild and domestic animals (8, 9). A. bovis is another leukocyte pathogen of ruminants that is usually found in professional phagocytes, such as monocytes (21). A. platys (formerly E. platys) is a platelet pathogen that infects dogs (24).
A. marginale and A. ovis are the main ruminants pathogens found in northern China (2, 15), and few studies have been carried out in central and southern China. A. phagocytophilum infections have been detected not only in ticks, rodents, and ruminants but also in the blood of human patients (4, 27). Domestic ruminants infected with A. bovis have mainly been reported in African countries, but the pathogen has also been found in Japan and Korea in recent years (12, 17, 20). The 16S rRNA gene sequences of A. bovis have been identified in both goats and cattle in China (29).
Although the presence of Anaplasma spp. in China is certain, to our knowledge, coinfection of these pathogens in domestic ruminants has never been investigated before. In the present study, through PCR-based molecular investigation, Anaplasma spp. had a high prevalence in goats in central and southern China. The PCR results were corroborated by 16S rRNA gene and MSP4 gene sequence analysis, and the genetic diversity of Anaplasma spp. was analyzed. We also provide the first report of a morphological observation of A. bovis from blood samples from small ruminants in China.
MATERIALS AND METHODS
Blood and DNA specimens.
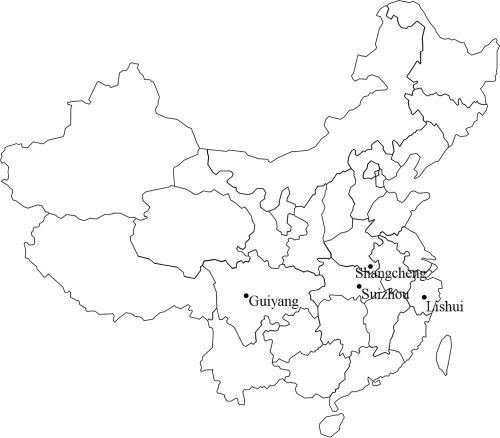
Blood samples were collected from 69 goats in Suizhou in Hubei Province, 46 goats in Shangcheng in Henan Province, 90 goats in Guiyang in Guizhou Province, and 57 goats in Lishui in Zhejiang Province between May and September in 2010. The sampling sites are indicated in Fig. 1. The samples were taken from the jugular vein of each animal and collected in a sterile tube containing an anticoagulant (EDTA). DNA was extracted from the field-blood samples and from experimental sheep blood using a genomic DNA extraction kit (Qiagen, Germany) according to the manufacturer's instructions.
Fig 1.
Distribution of the sampling sites and of goat infections with A. ovis, A. bovis, and A. phagocytophilum.
Inoculation of experimental animals.
Three-month-old sheep purchased from Jingtai in Gansu Province were used as experimental animals. The sheep were born in winter and fed in indoor conditions, which guarantees the animals had almost no chance for contact with hard ticks. Before experiment, the experimental animals were examined weekly for a month by microscopic examination of thin blood smears and by PCR detection (described below) of Anaplasma infection. Only Anaplasma-free animals were used. The blood samples from Suizhou were pooled, and 5 ml was injected into an experimental sheep (animal 1019, female, 6 months old) via the jugular vein. Blood samples from Shangcheng were also pooled and injected into a splenectomized sheep (animal 1090, female, 6 months old) using the same method. The animal experiments were performed according to the approved Institutional Animal Care and Use Committee guidelines. The experimental animals were examined daily for body temperature and clinical signs, and thin blood smears were prepared daily for microscopic examination. Blood was collected from the experimental animals weekly and subjected to DNA extraction as described above.
PCR.
Nested PCRs were carried out to detect A. bovis and A. phagocytophilum infection. During the first round, genomic DNA from field blood samples was amplified using the primers EE1 and EE2 (3). The PCR products were used as templates for the second round using the A. bovis-specific primers AB1f and AB1r, which generate a product of 551 bp, and the A. phagocytophilum-specific primers SSAP2f and SSAP2r, which generate a product of 641 bp (11). A. ovis infection in the field samples was detected with the MSP45 and MSP43 primers (5). These amplify entire major surface protein 4 (MSP4) gene. The reactions were performed in a final volume of 50 μl, containing 1.0 mM concentrations of each primer, 5 μl of PCR buffer, 4 μl of deoxynucleoside triphosphates, 0.25 μl of TaKaRa Taq (5 U/ml) (TaKaRa, China), and 1 μl of DNA sample. Reactions were conducted in an automated DNA C1000 thermal cycler (Bio-Rad, Beijing, China). For the EE1 and EE2 primers, the cycling conditions were denaturation for 4 min at 94°C, followed by 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s. The annealing temperature was stepped down four times by 2°C every two cycles. The final annealing temperature used was 54°C for 28 cycles, followed by a final extension for 5 min at 72°C. For the nested PCR, 1 μl of the product from the first amplification was used for amplification with specific primers; the amplification consisted of 40 cycles, each of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. For MSP4 amplification, after an initial denaturation step of 30 s at 94°C, each cycle consisted of a denaturing step of 30 s at 94°C, an annealing for 30 s at 60°C, and an extension step of 1 min at 68°C. Sheep genomic DNA and distilled water were used as negative controls. The PCR products were subjected to electrophoresis on 1% agarose gels containing 0.5 μg of ethidium bromide/ml and visualized under UV light.
DNA sequencing and data analysis.
Positive PCR products from primers SSAP2f/2r and AB1f/AB1r were cloned into pGEM-T vector (Promega, Madison, WI) and then sequenced by Sangon Biotech Company (Shanghai, China). The obtained sequences were analyzed by a BLASTn search in GenBank or by using the ClustalW method in the MegAlign software (DNAStar, Madison, WI) for determining the accuracy of the PCR method. For genotyping A. phagocytophilum and A. bovis, positive PCR products from the EE1 and EE2 primers from the field samples at different sites and experimental animals were cloned and sequenced. The sizes of the obtained sequences were ∼1,433 bp, and these sequences were used alongside known 16S rRNA sequences longer than 1,300 bp from the other Anaplasma species in GenBank for phylogenetic analysis using MEGA5 program (23). A phylogenetic tree was inferred using the neighbor-joining method (19). For genotyping A. ovis, MSP4 gene sequences obtained from different sampling sites were translated into amino acid sequences and then used for phylogenetic analysis using MEGA5 software.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of A. phagocytophilum from the YDH6, HB322, and HB231 isolates from Hubei, the XT1 isolate from Zhejiang, the GZ691 isolate from Guizhou, and the HN301 isolate from Henan have been deposited in GenBank under accession numbers JN55811 to JN55816. The 16S rRNA gene sequences of A. bovis from the G41 isolate from Henan, from the YX4, B7, R7, and Yu12 isolates from Zhejiang, from the G1, G21, G49, and G55 isolates from Guizhou, and from experimentally infected animals were deposited in GenBank under accession numbers HQ913646, JN558817, JN558819, JN558822 to JN558825, JN558828, JN558829, HQ913644, and HQ913645. The 16S rRNA gene sequences of other Anaplasma species were deposited under accession numbers JN558818, JN558820, JN558821, JN558826, and JN558827. The MSP4 gene sequences obtained from goat blood from the field samples were deposited in GenBank under accession numbers JN572927 to JN572939.
RESULTS
Incidence of Anaplasma coinfection.
As shown in Table 1, of 262 studied samples, 71 (27.1%) were free of Anaplasma infection. The infection rates at different sites varied from 60.9 to 77.8%. The single-pathogen infection rates with A. bovis and A. ovis were 16.0 and 15.3%, respectively, which were higher than the rate for A. phagocytophilum infection (6.1%). Single infection with A. phagocytophilum was not found at the studied sites in Guizhou or Zhejiang. The most common type of infection (27.1%) was A. ovis and A. bovis coinfection, which reached 40% in the samples from Guizhou Province. At other sites, single infection was dominant. The coinfection rates (1.9%) of A. phagocytophilum and A. ovis were lower than the coinfection rates (4.2%) of A. phagocytophilum and A. bovis. Three-pathogen coinfection occurred in only 2.3% of the animals studied. Generally, A. bovis was the most common of the three pathogens, with an infection rate of 49.6%. It was followed by A. ovis with a rate of 46.6% and A. phagocytophilum with 14.5%.
Table 1.
Numbers of goats multiply infected with Anaplasma pathogens at various geographic sites
| Site |
No. (%) of goats infected with: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No pathogens | One pathogen |
Two pathogens |
Three pathogens |
Total | ||||||
| Province | County | A. ovis | A. phagocytophilum | A. bovis | A. ovis + A. phagocytophilum | A. ovis + A. bovis | A. phagocytophilum + A. bovis | A. ovis + A. phagocytophilum + A. bovis | ||
| Henan | Shangcheng | 18 (39.1) | 4 (8.7) | 6 (13.0) | 6 (13.0) | 2 (4.3) | 4 (8.7) | 4 (8.7) | 2 (4.3) | 46 (100) |
| Hubei | Suizhou | 19 (27.5) | 5 (7.2) | 10 (14.5) | 14 (20.3) | 3 (4.3) | 18 (26.1) | 0 (0) | 0 (0) | 69 (100) |
| Guizhou | Guiyang | 20 (22.2) | 16 (17.8) | 0 (0) | 15 (16.7) | 0 (0) | 36 (40.0) | 2 (2.2) | 1 (1.1) | 90 (100) |
| Zhejiang | Lishui | 14 (24.6) | 15 (26.3) | 0 (0) | 7 (12.3) | 0 (0) | 13 (22.8) | 5 (8.8) | 3 (5.3) | 57 (100) |
| Total | 71 (27.1) | 40 (15.3) | 16 (6.1) | 42 (16.0) | 5 (1.9) | 71 (27.1) | 11 (4.2) | 6 (2.3) | 262 (100) | |
Identification of A. bovis infection.
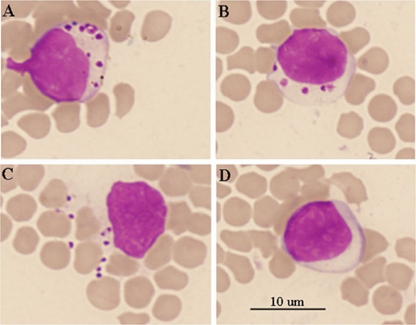
Sheep 1019 was inoculated with blood from the field samples collected in Suizhou, and daily examination was carried out for a month. Physical examination revealed no signs of debility or enlarged peripheral lymph nodes, and body temperatures ranged from 39.2 to 39.9°C, which is within the normal range. Morulae of A. bovis were observed by light microscopy in host monocytes in blood smears stained with Giemsa at 10 days postinoculation (dpi) (Fig. 2). The number of morulae in the monocytes varied from several to dozens (Fig. 2A and B). Morulae were also occasionally observed outside the monocytes (Fig. 2C). The morulae disappeared rapidly, and none were observed by the following day. PCR successfully amplified the 16S rRNA gene segment from the blood DNA samples at 10 dpi, indicating successful infection. Sheep 1090 was a splenectomized sheep and was inoculated with the field-blood samples from Shangcheng. Daily examination revealed no clinical signs except for pyrexia that developed by 9 dpi, with a body temperature of 41.3°C, increasing to 41.5°C by 10 dpi. The fever subsequently declined, and the body temperature returned to normal. No morulae were observed in host cells in blood smears at any point during the examination period. However, the 16S rRNA gene segment was successfully amplified by 9 dpi, and the target gene was detectable by PCR 4 months after inoculation (data not shown), indicating the presence of persistent infection.
Fig 2.
Microscopic examination of blood smears from the experimental sheep (animal 1019) stained with Giemsa. (A to C) Inclusions of A. bovis in ovine monocytes are visible. (D) Uninfected control monocyte.
Phylogenetic analysis of Anaplasma species on the basis of the 16S rRNA gene sequences.
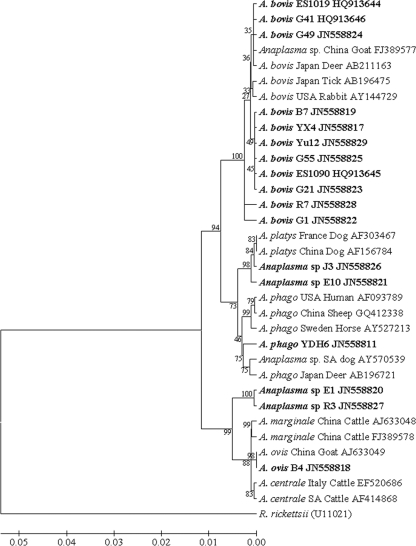
Nearly the full length (∼1,432 bp) of the 16S rRNA gene sequences from selected positive samples from different sampling sites was used to corroborate the results of the PCR and to determine Anaplasma species identity. Phylogenetic analysis revealed that not only were A. ovis, A. bovis, and A. phagocytophilum present in the investigated goats in different studied sites but A. platys-like (GenBank accession numbers JN558826 and JN558821) and A. marginale-like (GenBank accession numbers JN558820 and 558827) pathogens were also present (Fig. 3). ClustalW analysis showed that most of the Chinese isolates of A. bovis had similarities between 99.0 and 99.9%, but the R7 isolate (maximum of 99.3%) from Zhejiang and the G1 isolate (maximum of 98.9%) from Guizhou showed relatively little similarity (data not shown). Most of the Chinese isolates formed two main clusters in the phylogenetic tree (Fig. 3). One showed a close relationship with the sequence from Japanese deer (GenBank accession number AB211163). Another presented a independent cluster with six isolates, B7, YX4, and Yu12 isolates from Zhejiang, G55 and G21 from Guizhou, and ES1090 from experimental sheep inoculated with goat blood from Henan. No geographic segregation of A. bovis isolates was observed.
Fig 3.
Phylogram of Anaplasma inferred with long (∼1,432 bp) sequences of the 16S rRNA gene using the neighbor-joining method. The optimal tree with a branch length sum of 0.16815385 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed using the maximum-composite-likelihood method. Sequences newly identified in the present study are indicated in boldface.
A. phagocytophilum infections in goats from different sites were confirmed by 16S rRNA gene sequencing. The sequence of the YDH6 isolates was the longest (1,332 bp). It was used for phylogenetic analysis (Fig. 3), which was classified into a cluster together with isolates from South African dogs (GenBank accession number AY570539) and Japanese deer (GenBank accession number AB196721). This cluster was separated from the cluster containing the U.S. human isolate, a Swedish horse isolate, and northeastern Chinese isolates (GenBank accession number GQ412338). Two Chinese isolates distributed into two clusters indicated the presence of different A. phagocytophilum strains in China. ClustalW analysis of the partial 16S rRNA gene sequence (641 bp) of different isolates (YDH6, HB322, and HB231 isolates from Hubei, XT1 isolate from Zhejiang, GZ691 isolate from Guizhou, and HN301 isolate from Henan in GenBank under accession numbers JN55811 to JN55816) showed that the similarities of these isolates were between 96.3 and 99.7% (data not shown), further indicating the genetic diversity of A. phagocytophilum in China.
Genetic identification of A. ovis using MSP4 gene sequences.
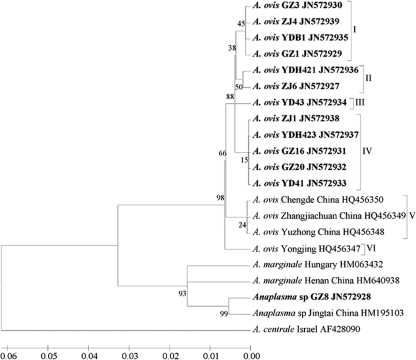
A. ovis infections were confirmed by sequencing msp4 from randomly selected positive samples from each sampling site. The A. ovis msp4 sequences identified in the present study were used with sequences reported previously for the characterization of the genetic diversity of A. ovis strains relative to other Anaplasma spp. (Fig. 4). As a result, the sequences from goats in southern of China constituted genotypes I, II, III, and IV, while previously obtained sequences (GenBank accession numbers HQ456347 to HQ456350) from sheep in the north included genotypes V and VI. No geographic segregation of A. ovis genotype was observed. In addition, one sequence in a GZ8 isolate from Guizhou and a previously obtained sequence from a Jingtai isolate from Gansu showed similarity to A. marginale, indicating that goats and sheep are the likely alternative hosts of A. marginale in China.
Fig 4.
Phylogenetic analysis of A. ovis strains based on the protein sequences of MSP4 gene was inferred using the neighbor-joining method. The optimal tree with a branch length sum of 0.22235680 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed using the Poisson correction method. All positions containing gaps and missing data were eliminated. Sequences newly obtained in the present study are indicated in boldface.
DISCUSSION
Northwestern China is the country's main grazing area for sheep and goats. Investigation of ovine anaplasmosis using 2,813 field samples from five northwestern provinces had been carried out using serological methods from 1986 through 1991. The results showed that the seroprevalence varied from 10.0 to 60.8% (14). Since then, no similarly large-scale investigation has been conducted. However, reports of A. ovis can occasionally be found in the literature (16, 29). The results of the present study showed that the prevalences of A. ovis in the central and south provinces Henan, Hubei, Guizhou, and Zhejiang were 26.1, 37.7, 58.9, and 54.4%, respectively. The average prevalence was 46.6%. These figures are similar to those found decades ago, indicating wide distribution and enzootic stability. For confirmation of the PCR results, selected positive PCR products from different sites were used to sequence the MSP4 gene, which has proved useful for phylogenetic studies of A. ovis and for the genetic characterization of Anaplasma spp. (5, 7). Phylogenetic analysis of msp4 sequences from goats in Hubei, Guizhou, and Zhejiang and previously obtained msp4 sequences from sheep in northern China revealed six A. ovis msp4 genotypes in goats and sheep (Fig. 4). Genotypes I, II, III, and IV consisted exclusively of A. ovis msp4 from goats in the south, whereas genotypes V and VI included A. ovis msp4 from sheep in the north. The results suggest that A. ovis msp4 genotypes may vary between sheep and goats. Similar variations have been observed in between sheep and deer and in the hosts of other Anaplasma spp. (5, 8, 25). However, no phylogeographic information was obtained from msp4 sequence analysis in the present study. Interestingly, one msp4 sequence (GenBank accession number JN572928) from an isolate of Anaplasma sp. in Guizhou showed great similarity to A. marginale, and similar results were observed in sheep in Gansu (16), indicating that sheep and goats are very likely natural reservoir hosts of A. marginale. This is not surprising because A. marginale infection of ruminants other than cattle has been documented (1, 26). Experimental infection of splenectomized sheep with A. marginale has been shown to cause subclinical symptoms (13).
Investigation of A. bovis infections showed that the prevalences of the investigated goats in Henan, Hubei, Guizhou, and Zhejiang were 34.8, 46.4, 60.0, and 49.1%, respectively, with an average of 49.6%, which was higher than the prevalences of A. bovis in deer (9.0%) and Haemaphysalis longicornis (12.0%) in Japan but similar to the prevalence in Japanese cattle (53.3%) (11, 17). A. bovis infection rates (average, 49.6%) in goats were higher than A. ovis infection rates (average, 46.6%) in central and southern China. Infections with A. bovis were confirmed by sequencing the 16S rRNA gene sequence and inoculation of experimental animals with the blood from field samples in the Henan and Hubei provinces. The results demonstrated that A. bovis caused subclinical symptoms in sheep. Both splenectomized and intact sheep experimentally infected with A. bovis developed either a short fever or no fever at all and recovered rapidly. However, studies with larger numbers of animals must be conducted to properly evaluate the pathogenicity of this organism in sheep and goats. In intact experimental sheep, inclusions were only found in the monocytes for a very short period (1 day). They later disappeared. This may lead to the underdiagnosis of A. bovis infection when light microscopic exanimation of blood smears is used for diagnosis. Regarding morphology, in 1996 Sreekumar et al. identified A. bovis cells of different shapes and sizes in bovine monocytes, including small (1-μm), medium (1- to 3-μm), and large (3- to 6-μm) forms (21). The forms found in ovine monocytes in the current study were exclusively small and roughly spherical (Fig. 2A, B, and C), similar to those found in bovine monocytes (21). Some organisms were also observed outside the cells (Fig. 2C). However, it is likely that the intracellular organisms increase the fragility of the cells, and we cannot exclude the possibility that these extracellular organisms may have been released during blood smear preparation when their hosts were crushed. These forms resemble the cellular debris phagocytosed by the monocytes, which might also lead to confusion when identifying the organism by microscopic examination.
The 16S rRNA gene sequences of A. bovis from both field isolates and experimental animals were used to infer a phylogenetic tree (Fig. 3), and most of the sequences were composed of two major clusters. One presented an independent cluster, and another was placed together with the sequences from a Japanese deer (GenBank accession number AB211163) and from an unidentified Anaplasma species in goat in the Chongqing region in southwestern China (29). Regarding the unidentified Anaplasma species found in southwestern China, in 2010 Zhou et al. obtained 16S rRNA gene sequences from seven isolates from sheep and goats (29). A phylogenetic study placed all seven isolates in two clusters in an inferred phylogenetic tree, separate from the clusters comprising A. centrale, A. marginale, A. ovis, and A. phagocytophilum (29). However, Zhou et al. did not include sequences from A. bovis in their study. They therefore missed a chance to infer A. bovis infection in that region. PCR detection found a high prevalence of A. bovis in the sites studied here, but none of the animals investigated showed any clinical signs. This is in agreement with the findings of Zhou et al. indicating the limited pathogenicity of A. bovis in sheep and goats. This could explain why A. bovis has gone unnoticed in these regions. We also demonstrated that A. bovis infection could persist in experimentally infected sheep for over 4 months, indicating that sheep and goats may be an important natural reservoir for this organism. The possible presence of A. bovis should therefore be taken into account as a potential cause of illness in other species.
A. phagocytophilum has been identified in rodents, sheep, goats, dogs, ticks, and human patients in northern China (24, 27, 28). A large-scale investigation of ticks (n = 2,429) showed that the prevalence of A. phagocytophilum was between 1.1 and 6.0% at different sites near the China-Russia border (10). Our results showed that the prevalence in goats ranged from 3.3 to 30.4% in the four investigated provinces, higher than in ticks in the north. Although only three positive samples were found in Guizhou Province, the 16S rRNA gene sequence obtained from GZ691 isolate confirmed the accuracy of the PCR. The 16S rRNA gene sequence analysis indicated that A. phagocytophilum is probably genetically diverse within China, but the limited data in this study do not provide phylogeographic information. Further genetic identification must be performed using encoding genes, since high conservation of the rRNA gene means they provide poor information to distinguish closely related species and strains (8).
To our knowledge, this is the first study on the coinfection of Anaplasma species in ruminants in China. A. ovis, A. bovis, and A. phagocytophilum were shown to be present at the investigated sites. Surprisingly, A. bovis rather than A. ovis was found to be the dominant Anaplasma species in goats. Coinfection of A. ovis and A. bovis is much more common than other combinations. Whether this is due to competition among Anaplasma species remains to be elucidated, although it has been shown that some Anaplasma genotypes can prevent the multiplication of other genotypes. This has been observed in A. marginale, A. ovis, and A. phagocytophilum (5, 6, 22). In addition, sequence analysis revealed that goats are the most likely potential hosts of A. marginale and Anaplasma species closely related to A. platys.
In conclusion, we demonstrated here the presence of A. ovis, A. bovis, and A. phagocytophilum in goats in central and southern China. Coinfection of these three species was found in all of the investigated provinces. A. bovis infection in goats was found here for the first time, and its prevalence was higher than A. ovis infection. After identification of A. bovis through inoculation of the experimental animals, A. bovis pathogens were observed in the monocytes of infected sheep. Phylogenetic studies showed that A. phagocytophilum is probably highly diverse within China. Genetic identification of the msp4 gene of A. ovis indicated that its genotypes are likely host specific. These results may promote our understanding of the etiological situation of Anaplasma spp. in southern China and the construction of risk assessment models in the investigated regions.
ACKNOWLEDGMENTS
We thank Shijie Jiang, Qiong Liu, and Chunping Liu for their assistance.
This study was financially supported by the 973 Program (2010CB530206), the Key Project of Gansu Province (1002NKDA035 and 0801NKDA033), the NSFC (30800820, 30972182, 31072130, and 31001061), 948 (2010-S04), the Beef and Yak Production System Programme, the Specific Fund for Sino-Europe Cooperation, MOST (China), and the State Key Laboratory of Veterinary Etiological Biology Project (SKLVEB2008ZZKT019). The research was also facilitated by EPIZONE (FOOD-CT-2006-016236), ASFRISK (211691), ARBOZOONET (211757), and PIROVAC (KBBE-3-245145) of the European Commission, Brussels, Belgium.
Footnotes
Published ahead of print 4 November 2011
REFERENCES
- 1. Aubry P, Geale DW. 2010. A review of bovine anaplasmosis. Transbound. Emerg. Dis. 58:1–30 [DOI] [PubMed] [Google Scholar]
- 2. Bai Q, Chen Z, Ying S, Liu G, Zhou J. 1987. Study on isolation and preservation of single species of haematocytozoon in bovine: separation of bovine A. marginale single isolate. Chin. J. Vet. Sci. Tech. 3:12–15 (In Chinese.) [Google Scholar]
- 3. Barlough JE, Madigan JE, DeRock E, Bigornia L. 1996. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet. Parasitol. 63:319–329 [DOI] [PubMed] [Google Scholar]
- 4. Cao WC, et al. 2006. Natural Anaplasma phagocytophilum infection of ticks and rodents from a forest area of Jilin Province, China. Am. J. Trop. Med. Hyg. 75:664–668 [PubMed] [Google Scholar]
- 5. de la Fuente J, et al. 2007. Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet. Microbiol. 119:375–381 [DOI] [PubMed] [Google Scholar]
- 6. de la Fuente J, Garcia-Garcia JC, Blouin EF, Saliki JT, Kocan KM. 2002. Infection of tick cells and bovine erythrocytes with one genotype of the intracellular ehrlichia Anaplasma marginale excludes infection with other genotypes. Clin. Diagn. Lab. Immunol. 9:658–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de la Fuente J, et al. 2005. Genetic diversity of Anaplasma species major surface proteins and implications for anaplasmosis serodiagnosis and vaccine development. Anim. Health Res. Rev. 6:75–89 [DOI] [PubMed] [Google Scholar]
- 8. de la Fuente J, et al. 2005. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 43:1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dumler JS, et al. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145–2165 [DOI] [PubMed] [Google Scholar]
- 10. Jiang JF, et al. 2011. Anaplasma phagocytophilum infection in ticks, China-Russia border. Emerg. Infect. Dis. 17:932–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawahara M, et al. 2006. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 72:1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim CM, Kim MS, Park MS, Park JH, Chae JS. 2003. Identification of Ehrlichia chaffeensis, Anaplasma phagocytophilum, and A. bovis in Haemaphysalis longicornis and Ixodes persulcatus ticks from Korea. Vector-Borne Zoonotic Dis. 3:17–26 [DOI] [PubMed] [Google Scholar]
- 13. Kuttler KL. 1981. Infection of splenectomized calves with Anaplasma ovis. Am. J. Vet. Res. 42:2094–2096 [PubMed] [Google Scholar]
- 14. Lu W, Lu WX, Zhang Q, Yu F, Dou H, Yin H. 1997. Ovine anaplasmosis in northwest China. Trop. Anim. Health Prod. 29:16S–18S [Google Scholar]
- 15. Ma L, Hua N, Chen W. 1982. Investigation of ovine anaplasmosis. J. Xinjiang Anim. Husbandry Sci. Tech. 2:2–13 (In Chinese.) [Google Scholar]
- 16. Ma M, et al. 2011. Development and evaluation of a loop-mediated isothermal amplification method for rapid detection of Anaplasma ovis. J. Clin. Microbiol. 49:2143–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ooshiro M, Zakimi S, Matsukawa Y, Katagiri Y, Inokuma H. 2008. Detection of Anaplasma bovis and Anaplasma phagocytophilum from cattle on Yonaguni Island, Okinawa, Japan. Vet. Parasitol. 154:360–364 [DOI] [PubMed] [Google Scholar]
- 18. Palmer GH, Abbott JR, French DM, McElwain TF. 1998. Persistence of Anaplasma ovis infection and conservation of the msp-2 and msp-3 multigene families within the genus Anaplasma. Infect. Immun. 66:6035–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 20. Sakamoto L, Ichikawa Y, Sakata Y, Matsumoto K, Inokuma H. 2010. Detection of Anaplasma bovis DNA in the peripheral blood of domestic dogs in Japan. Jpn. J. Infect. Dis. 63:349–352 [PubMed] [Google Scholar]
- 21. Sreekumar C, Anandan R, Balasundaram S, Rajavelu G. 1996. Morphology and staining characteristics of Ehrlichia bovis. Comp. Immunol. Microbiol. Infect. Dis. 19:79–83 [DOI] [PubMed] [Google Scholar]
- 22. Stuen S, Dahl H, Bergstrom K, Moum T. 2005. Unidirectional suppression of Anaplasma phagocytophilum genotypes in infected lambs. Clin. Diagn. Lab. Immunol. 12:1448–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 24. Wen B, Cao W, Pan H. 2003. Ehrlichiae and ehrlichial diseases in china. Ann. N. Y. Acad. Sci. 990:45–53 [DOI] [PubMed] [Google Scholar]
- 25. Yabsley MJ, et al. 2005. Evidence of tick-borne organisms in mule deer (Odocoileus hemionus) from the western United States. Vector-Borne Zoonotic Dis. 5:351–362 [DOI] [PubMed] [Google Scholar]
- 26. Zaugg JL, Goff WL, Foreyt W, Hunter DL. 1996. Susceptibility of elk (Cervus elaphus) to experimental infection with Anaplasma marginale and A. ovis. J. Wildl. Dis. 32:62–66 [DOI] [PubMed] [Google Scholar]
- 27. Zhan L, et al. 2010. Anaplasma phagocytophilum from rodents and sheep, China. Emerg. Infect. Dis. 16:764–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang L, et al. 2011. Investigation of anaplasmosis in Yiyuan County, Shandong Province, China. Asian Pac. J. Trop. Med. 4:568–572 [DOI] [PubMed] [Google Scholar]
- 29. Zhou Z, et al. 2010. Phylogenetic analysis of the genus Anaplasma in southwestern China based on 16S rRNA sequence. Res. Vet. Sci. 89:262–265 [DOI] [PubMed] [Google Scholar]