Abstract
Using a relatively simple enrichment technique, geosmin and 2-methylisoborneol (MIB)-biodegrading bacteria were isolated from a digestion basin in an aquaculture unit. Comparison of 16S rRNA gene sequences affiliated one of the three isolates with the Gram-positive genus Rhodococcus, while the other two isolates were found to be closely related to the Gram-negative family Comamonadaceae (Variovorax and Comamonas). Growth rates and geosmin and MIB removal rates by the isolates were determined under aerated and nonaerated conditions in mineral medium containing either of the two compounds as the sole carbon and energy source. All isolates exhibited their fastest growth under aerobic conditions, with generation times ranging from 3.1 to 5.7 h, compared to generation times of up to 19.1 h in the nonaerated flasks. Incubation of the isolates with additional carbon sources caused a significant increase in their growth rates, while removal rates of geosmin and MIB were significantly lower than those for incubation with only geosmin or MIB. By fluorescence in situ hybridization, members of the genera Rhodococcus and Comamonas were detected in geosmin- and MIB-enriched sludge from the digestion basin.
INTRODUCTION
Geosmin and 2-methylisoborneol (MIB) are known to cause an earthy, musty taste in water. While primarily known from their undesirable effect in the drinking water industry (25), these compounds also cause significant harm in the fish farming industry. When released into the culture water of aquaculture facilities, geosmin and MIB are absorbed through the gills, skin, or gastrointestinal tract by lipid-rich fish tissues and often render the fish unmarketable (22). In aquaculture systems, preventive measures to combat the accumulation of these compounds are hardly implemented. Instead, posttreatment before marketing, involving purging of off-flavored fish with clean water for several days, is the most common abatement strategy (14, 22). Given the high costs of the latter method and considering the magnitude of the problem, off flavor inflicts considerable financial losses on the aquaculture industry (49).
Much more research has been done to control the accumulation of geosmin and MIB in drinking water than in the aquaculture industry. Most treatment procedures are based on adsorption of geosmin and 2-methylisoborneol (MIB) by surface-rich resins. Powdered activated carbon (6) and granular activated carbon (10) are most commonly used for this purpose. In addition to chemical/physical adsorption, biological degradation of these compounds may take place, as demonstrated in carbon filters (13), clay-based filters (39), and sand filters (19, 20, 32).
In recent studies on recirculating aquaculture systems (16, 17), we concluded that, in addition to chemical/physical adsorption, a substantial quantity of geosmin and MIB is biologically degraded in the anaerobic treatment compartment of these systems. Specifically, it was found that in sludge collected from the digestion basin of the recirculating system, removal of geosmin and MIB was higher in untreated samples than in sterilized samples. Although the latter results pointed to geosmin and MIB biodegradation, direct proof for this process was not provided, and several questions remained unanswered. First, which organisms are capable of geosmin and MIB metabolism? Second, when such microorganisms are indeed present, are they capable of using geosmin and MIB as the sole carbon and energy sources, or are these compounds cometabolized? Third, if not cometabolized, are geosmin and MIB removed by the cells if other easily utilized substrates are present?
The aim of the present work was to isolate and identify geosmin- and MIB-degrading bacteria from the digestion basin of the aforementioned recirculating system and determine their growth and geosmin and MIB degradation when grown solely with geosmin and MIB or with a combination of the latter compounds and other readily available carbon sources.
MATERIALS AND METHODS
Growth and enrichment medium.
Mineral medium supplemented with geosmin and 2-methylisoborneol (MIB) was used as the enrichment medium. The medium (24) contained the following ingredients (liter−1): NH4Cl, 0.05 g; MgSO4 · 7H2O, 0.05 g; CaCl2 · 2H2O, 0.02 g; K2HPO4, 0.1 g; FeCl3 · 6H2O, 0.001 g; and trace element solution, 0.5 ml (45). The same medium, as well as Luria-Bertani (LB) medium (Bacto tryptone,10 g liter−1; yeast extract, 5 g liter−1; NaCl, 10 g liter−1) (33), was used for growth of the isolates. During incubation of crude sludge and during incubation of the isolates in LB medium, geosmin and MIB were supplemented from stock solutions containing 100 μg ml−1 geosmin or MIB in methanol (Supelco, Bellefonte, PA). Stock solutions, therefore, contained 24.4 M methanol and 0.55 mM geosmin or 0.59 mM MIB. Similar stock solutions, containing geosmin and MIB in methanol, were used during part of the enrichment procedure (see below). Bacterial incubations with only geosmin or MIB as the carbon source were performed by adding these compounds from separate stock solutions that were prepared by dissolving 20 mg of either pure MIB or pure geosmin (Wako Chemicals GMBH, Neuss, Germany) in 1 ml of sterile, double-distilled water.
MIB and geosmin biodegradation in crude sludge.
Sludge was derived from a sedimentation/digestion basin, which was part of a zero-discharge marine recirculating system situated on campus (15). To simulate oxygen-limited conditions prevailing in the digestion basin, portions of 5 ml of sludge were placed in 150-ml serum bottles containing 140 ml mineral medium. The bottles were incubated at 30°C on a shaker after flushing with argon for 30 min and subsequent addition of geosmin and MIB (2.5 μg liter−1 in methanol). Periodically, samples (7.5 ml) were withdrawn from the bottles for determination of geosmin and MIB. Bottles containing presterilized sludge were used as controls.
Isolation of bacteria.
Sludge (10 ml) derived from the sedimentation/digestion basin was incubated in 500 ml flasks containing 250 ml of mineral medium. Flasks were flushed with argon for 45 min and supplemented with MIB and geosmin (6 μg liter−1 in methanol) prior to incubation at 28°C. Every fourth day, an aliquot (10 ml) was transferred to fresh geosmin- and MIB-supplemented mineral medium. This procedure was repeated until the medium appeared to be free of visible sludge particles. At this stage, 100 μl of the suspension was spread on agar plates with the same composition as the liquid medium. Successive transfers of distinctively different colonies were conducted until they were fully separated. This enrichment strategy resulted in three morphologically different bacteria. These bacteria showed growth for at least 10 successive transfers on geosmin- and MIB-containing agar plates. Incubation of these isolates in mineral medium (both solid and liquid) in the absence of an organic carbon source resulted in no growth. Proof of their ability to grow on geosmin or MIB was provided by culturing the isolates in the presence of only one of these carbon sources without methanol. Under the latter conditions (i.e., in liquid medium with either geosmin or MIB as the sole carbon source), the isolates have been maintained in culture for more than a year.
Identification of bacteria.
DNA was extracted by heating single colonies suspended in 30 μl of distilled water, pretreated with 1% diethylpyrocarbonate (DEPC), for 10 min at 94°C. 16S rRNA gene sequences of the various isolates were amplified by PCR using the bacterial primer sets 341F with a GC clamp and 907R (34) and 1392R and 11F modified from the method of Lane (29) as detailed in Table 1. Each 50-μl suspension contained the following components: 1.5 U Taq DNA polymerase (Red Taq; Sigma, St. Louis, MO), Taq buffer containing a final magnesium concentration of 1.5 mM, deoxynucleoside triphosphates (dNTPs; 20 nmol each), 50 pmol of each primer, and 1 ml DNA template. PCRs were conducted in a gradient thermal cycler (model PTC-140 200; MJ Research, Watertown, MA) programmed with an initial denaturation step of 95°C for 30 s followed by 33 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 30 s. Cycling was completed with a final elongation stage at 72°C for 2 min. The presence and size of the PCR fragments were determined by agarose gel electrophoresis (2%) using ethidium bromide as the staining agent. The 341F and 907R PCR products were analyzed on a 20 to 80% gradient denaturing gradient gel electrophoresis (DGGE) polyacrylamide gel in order to pinpoint the novel isolates. For identification of the isolates, PCR products of both primer pairs were analyzed with an Applied Biosystems 377 DNA sequencer using the Prism dye terminator cycle sequencing ready reaction kit with Ampli Taq DNA polymerase. Each primer described above was used separately for this process. The sequence products were primarily identified by BLAST analysis (1). The 11F- and 1392R-generated fragments (500 bp) were aligned with a prealigned 16S rRNA sequence database using the aligning tool supplied by the MEGA phylogenetic software package (46), and phylogenetic trees were generated by neighbor-joining and maximum likelihood analyses with a correction method that applied a 50% cutoff filter supplied by the MEGA software package. Bootstrap analyses (1,000 replicates) were then applied to generated trees to select optimal topologies (46).
Table 1.
Primers and probes used in the present study
| Primer or probe | Sequence (5′→3′) | Target site (positions)a | Target organism(s) | Formamide (%) | Fluorescent dye modificationb | Reference |
|---|---|---|---|---|---|---|
| Primers | ||||||
| 11F | GTTTGATCMTGGCTCAG | 16S (11–28) | Domain Bacteria | Lane (29) | ||
| 1392R | ACGGGCGGTGTGTRC | 16S (1392–1407) | Domain Bacteria | Lane (29) | ||
| 341Fc | CCTACGGGAGGCAGCAG | 16S (341–358) | Domain Bacteria | Muyzer et al. (34) | ||
| 907Rc | CCGTCAATTCMTTTGAGTTT | 16S (907–927) | Domain Bacteria | Muyzer et al. (34) | ||
| Probes | ||||||
| EUB338 | GCTGCCTCCCGTAGGAGT | 16S (338–355) | Domain Bacteria | 35 | 5′ CALd | Amann et al. (2) |
| SBACT P338 | GCAGCCACCCGTAGGTGT | 16S (338–355) | Planctomycetales | 30 | 6-FAMe | Daimes et al. (8) |
| HGC69A | GCAGCCACCCGTAGGTGT | 23S (1901–1918) | Actinobacteria | 25 | 6-FAM | Roller et al. (40) |
| VAR1 | CTCCATTCGCGCAAGGTCTT | 16S (209–228) | Variovorax spp. | 35 | 6-FAM | Sanguin et al. (43) |
| RHOCOC3 | CATCCTGCACCAGTAAACCT | 16S (196–215) | Rhodococcus spp. | 35 | 6-FAM | Kyselkova et al. (28) |
| COM1424 | ACCTACTTCTGGCGAGA | 16S (1424–1440) | Comamonas spp. | 25 | 6-FAM | Amann et al. (4) |
rRNA position according to Escherichia coli numbering.
Modifications at 5′ prime end.
DGGE primer attached to 40-bp GC clamp (5′-GCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′).
5′ CAL, CAL Fluor Red 590.
6-FAM, 6-carboxyfluorescein.
FISH.
The presence of the bacteria closely related to the ones isolated was examined in sludge derived from the sedimentation/digestion basin by fluorescence in situ hybridization (FISH) of either 16S or 23S bacterial rRNA sequences with general and genus-specific oligonucleotide probes, synthesized and modified by Sigma-Aldrich, (Germany). Probe modifications and their target organisms are presented in Table 1. Sludge (5 g) was incubated in a 100-ml glass flask containing 75 ml mineral salt medium, which was flushed with argon for 40 min prior to addition of geosmin and MIB (5 μg liter−1). Every 3 days, geosmin and MIB levels were adjusted to secure levels of around 5 μg liter−1. After 3 weeks of incubation, 10 ml of the suspension was added to sterile plastic vials and fixed with paraformaldehyde (final concentration, 2%). Fixation of similar samples with ethanol (1:1) was also performed to allow hybridization with probes against Gram-positive bacteria. Vials were stored overnight at 4°C for fixation, washed twice with phosphate-buffered saline (PBS) (1:1), and stored at −20°C. Portions of 5 μl were introduced onto gelatin-coated diagnostic microscope slides containing eight wells (Erie, Portsmouth, NH) and dried at 37°C for 30 min. Wells containing pure cultures of the bacterial isolates obtained in this study were used as positive controls. Paracoccus sp. was used as a negative control. Hybridization was performed according to a protocol described previously (3). Briefly, 8 μl of hybridization buffer was added to the wells. The concentration of formamide in the buffer differed with each probe used (Table 1). One of the probes (specified in Table 1) was added to each sample at a final concentration of 5 ng ml−1. Parallel runs were conducted with the EUB338 general probe and one of the following specific probes: VAR1, RHCOC3, or COM1424. All slides were incubated for 3 h at 46°C in a moisture chamber for hybridization. Following hybridization, slides were washed with a washing buffer (3) and stained with 1 μg ml−1 of 4′,6′-diamidino-2-phenylindole (DAPI) for 10 min. After final washing with sterilized distilled water, samples were overlaid with AF1 mountant solution (Citifluor, United Kingdom) as an antifading reagent and covered with a coverslip. Final samples were analyzed by epifluorescence microscopy on a Nikon Eclipse 80i microscope (Nikon Instruments, Inc., Melville, NY).
In vitro experiments with bacterial isolates.
Bacterial isolates were incubated in mineral medium supplemented with either geosmin or MIB (without methanol) and in LB medium supplemented with both geosmin and MIB (in methanol). Aerobic incubation was initiated by adding 1-ml aliquots of the bacterial stock solutions to 75 ml of medium in 200-ml serum bottles. The bacterial suspensions were supplemented with geosmin and/or MIB and incubated in a temperature-controlled (37°C) shaker at 200 rpm. Periodically, aliquots were withdrawn from the duplicate bottles for determination of MIB, geosmin, and bacterial concentrations. Since all three strains were isolated from an anoxic digestion basin, proliferation of the isolates and their concomitant reduction of geosmin or MIB were also examined in argon-flushed, nonaerated medium. For this purpose, incubation was conducted in 100-ml serum bottles containing 75 ml of mineral medium. The bottles were flushed with argon for 25 min prior to being sealed with butyl rubber stoppers and crimp capped. Incubations were initiated by injecting geosmin and/or MIB through the bottles' caps. Two serum bottles, similarly treated and inoculated, were dedicated for oxygen determinations. As determined by the Winkler method (5), maximum oxygen levels of 0.03 mg liter−1 were measured at the start of the incubation period and decreased to undetectable levels (below 0.01 mg liter−1) during the exponential phase of growth.
Similar aerobic and nonaerated incubations with isolates minus geosmin or MIB, as well as incubations with autoclaved bacterial suspensions, served as controls.
Bacterial enumeration.
Bacterial cell numbers were determined according to the most probable number (MPN) technique. In this test, serial dilutions (n = 5) were made on enzyme-linked immunosorbent assay (ELISA) plates (Nunc, Denmark) containing the same medium used for bacterial growth. Plates were incubated at 37°C for 48 h, and the number of bacteria in the original sample was determined according to Halvorson and Ziegler (18). In addition, 10-μl aliquots (obtained from each of the 10-fold dilutions on the ELISA plates) were spread onto agar plates containing medium with the same composition as the growth medium. After 48 h of incubation at 37°C, colony-forming units (CFU) in the planted drops, containing 30 to 300 cells, were counted and the original number of CFU in the original sample was calculated based on 5 replicates (27). Bacterial cell numbers (mean values) were calculated based on results obtained with both methods. Bacterial growth rate constants were calculated based on cell proliferation during their exponential phase of growth.
MIB and geosmin analysis.
MIB and geosmin concentrations in liquid samples were analyzed using solid-phase microextraction (SPME) as previously described by Lloyd et al. (31). Briefly, the method is based on extraction of these compounds onto a StableFlex fiber (Supelco, Bellefonte, PA). Individual liquid samples (25 ml) were supplemented with 6 g of NaCl and incubated in crimp cap vials (total volume, 40 ml) in a water bath at 65°C. After 30 min of incubation, SPME fibers were injected through the Teflon-faced silicone septa (Supelco, Bellefonte, PA) of the airtight vials for headspace extraction of MIB and geosmin for 20 min. Concentrations of the compounds (1 ng liter−1 detection limit) were determined by injecting the fibers for 1.5 min at 250°C into the splitless operated injector of an HP5890 (Palo Alto, CA) gas chromatograph with a flame-ionized detector (GC-FID). The GC was operated with an MDN-5 fused silica capillary column (length, 30 m; inside diameter, 0.25 mm) of 0.25-μm film thickness (Supelco, Bellefonte, PA). Helium was used as the carrier gas at constant flow rate of 1 ml min−1. The oven temperature was held at 60°C for 0.5 min from injection and then increased to 100°C at 30°C min−1, followed by increases to 185°C at 20°C min−1 and 250°C at 40°C min−1, and held at this maximum temperature for 2.3 min. The FID temperature was 280°C. Identification of geosmin and MIB peaks detected by GC-FID was verified by parallel analysis of selected samples with a gas chromatograph coupled to a mass spectrometer detector (GC-MS model Saturn 2000; Varian, Inc., Palo Alto, CA).
Nucleotide sequence accession number.
The three 16S rRNA gene sequences determined in this study have been deposited in GenBank under accession no. GQ365214, GQ365216, and GQ365217.
RESULTS
MIB and geosmin biodegradation in sludge samples.
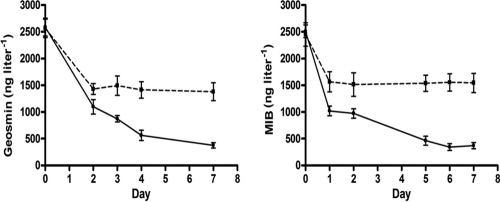
Incubation of sludge from the digestion basin in mineral medium under nonaerated conditions resulted in a larger decrease of MIB and geosmin than those in similar runs with sterilized sludge (Fig. 1). Specifically, during the first 2 days of incubation, the decreases in geosmin and MIB were similar in sterilized and nonsterilized sludges. However, the decrease of these compounds continued beyond this period in nonsterilized sludge at rates of 185.42 and 144.65 ng liter−1 day−1 for geosmin and MIB, respectively. In sterilized sludge, a decrease in these compounds was not observed beyond day 2 of incubation.
Fig 1.
Geosmin (left) and MIB (right) biodegradation under nonaerated conditions in mineral media containing crude (●) and sterilized (■) sludge obtained from a digestion basin. Rates represent mean values (n = 3).
Bacterial identification.
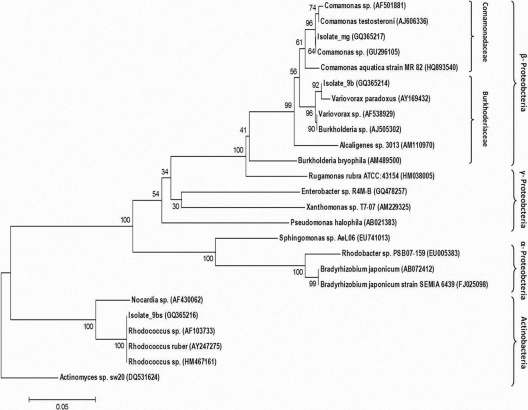
Single bands obtained by denaturizing gel gradient electrophoresis (DGGE) of PCR-amplified DNA extracted from the isolated colonies confirmed that the colonies were comprised of only one bacterial strain (not shown). Phylogenetic tree analysis of the sequenced 16S rRNA gene fragments (Fig. 2), revealed a close relationship of isolate 9b (GQ365214) with Variovorax paradoxus, isolate 9bs (GQ365216) with Rhodococcus sp., and isolate mg (GQ365217) with Comamonas sp. Parallel BLAST analyses of the 16S rRNA gene sequences (not shown) also revealed a close relationship (scores of >98%) of all isolates with these reference bacteria.
Fig 2.
Phylogenetic tree analysis of 16S rRNA gene sequences of three bacterial isolates found capable of geosmin and MIB biodegradation. Bacteria isolated in the current study are noted with their GenBank accession number. Numbers on branches represent bootstrap tests of phylogeny (500 replicates) using the neighbor-joining with maximum likelihood analysis tool.
In vitro MIB and geosmin biodegradation.
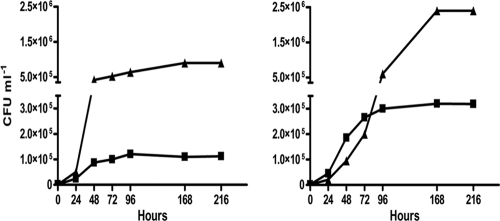
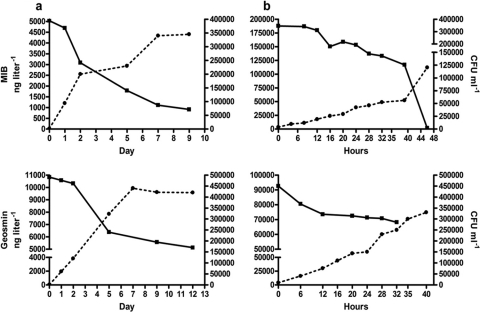
Geosmin and MIB were used by all three isolates as primary carbon sources, as illustrated for the Rhodococcus-like isolate, whose growth was dependent on the initial concentration of either one of these compounds (Fig. 3). Incubation of the three bacterial isolates in aerated culture flasks as well as in argon-flushed, airtight flasks in the presence of either geosmin or MIB as the sole carbon source was accompanied by a significant removal of these compounds as well as growth of the isolates under all conditions tested (Fig. 4, 5, and 6). It should be noted that the geosmin and MIB concentrations in Fig. 4, 5, and 6 represent the net concentrations of geosmin and MIB (obtained by subtracting the decrease in geosmin and MIB concentrations in the sterile control flasks from those in the nonsterilized flasks), as it was found that geosmin and MIB decreased also in the autoclaved control flasks under aerobic conditions. Over the incubation period, this nonbiologically mediated decrease of geosmin and MIB, most likely caused by volatilization of these compounds, accounted for a decrease of 25% of the initial MIB concentrations and 2% of the initial geosmin concentrations in aerated control flasks. No loss of either geosmin or MIB was observed in similar control flasks under airtight conditions (not shown). In control runs (not shown), it was demonstrated that all three isolates did not grow in medium devoid of geosmin and MIB. Generation times during exponential growth of the isolates with either geosmin or MIB under aerobic conditions ranged from 3.1 to 5.8 h, while under nonaerated conditions, longer generation times of 5.8 to 19 h were measured (see the supplemental material). Geosmin removal rates by the various isolates during exponential growth ranged from 3.7 × 10−3 to 1.9 × 10−2 ng h−1 cell−1 under aerobic conditions and from 4.0 × 10−5 to 4.0 × 10−3 ng h−1 cell−1 under nonaerated conditions. MIB removal rates ranged from 1.0 × 10−6 to 1.1 ×10−3 ng h−1 cell−1 under aerobic conditions and 2.5 × 10−5 to 2.2 × 10−3 ng h−1 cell−1 under nonaerated conditions (see the supplemental material).
Fig 3.
Growth of the Rhodococcus sp.-like isolate (expressed as CFU ml−1) during incubation in mineral medium under aerobic conditions with geosmin (left) and MIB (right). Initial geosmin and MIB concentrations were 2.5 μg liter−1 (■) and 2.5 mg liter−1 (▲).
Fig 4.
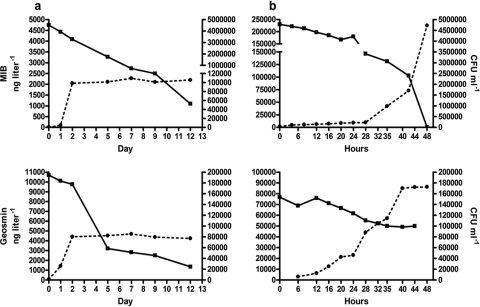
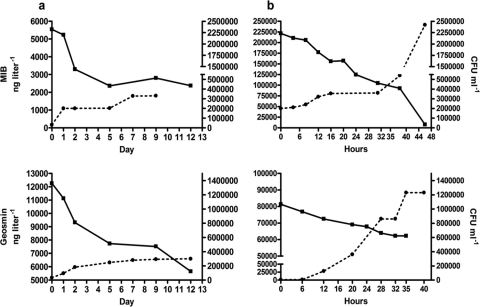
MIB (top) and geosmin (bottom) degradation (■) during growth (●) of a Variovorax paradoxus-like isolate in mineral media containing MIB or geosmin as the sole carbon source under nonaerated (a) and aerated (b) conditions. Rates are mean values of duplicate analyses. Net loss is depicted relative to abiotic control treatments. Differences between the duplicate determinations did not exceed 10%. Note the use of different time scales for the aerated and nonaerated treatments.
Fig 5.
MIB (top) and geosmin (bottom) degradation (■) during growth (●) of Rhodococcus sp.-like isolate in mineral media containing MIB or geosmin as the sole carbon source under nonaerated (a) and aerated (b) conditions. Rates are mean values of duplicate analyses. Net loss is depicted relative to abiotic control treatments. Differences between the duplicate determinations did not exceed 10%. Note the use of different time scales for the aerated and nonaerated treatments.
Fig 6.
MIB (top) and geosmin (bottom) degradation (■) during growth (●) of the Comamonas sp.-like isolate in mineral media containing MIB or geosmin as the sole carbon source under nonaerated (a) and aerated (b) conditions. Rates are mean values of duplicate analyses. Net loss is depicted relative to abiotic control treatments. Differences between the duplicate determinations did not exceed 10%. Note the use of different time scales for the aerated and nonaerated treatments.
Compared to incubation in mineral medium, incubation of the isolates in LB medium under both aerated and nonaerated conditions (Table 2) resulted in higher growth rates of all isolates. Geosmin and MIB removal rates under these conditions were considerably lower (ranging from 1.5 × 10−6 to 4.8 × 10−10) than during incubation of the isolates with only geosmin or MIB as the carbon source. It should be noted that incubation of the isolates in LB medium was conducted with amendments of geosmin and MIB dissolved in methanol. Hence, in addition to tryptone and yeast extract (present in LB medium), methanol was a possible additional carbon source for bacterial growth. Indeed, subsequent runs revealed that, although significantly slower than in LB medium, growth of the isolates was also obtained in medium with methanol as the sole carbon source (not shown). Furthermore, it is important to note that both geosmin and MIB were added to the LB medium—as opposed to the runs with mineral medium, where the isolates were cultured with either geosmin or MIB. Despite the apparent low geosmin and MIB removal rates by the different isolates in LB medium, geosmin as well as MIB removal by each of the isolates was higher than 60% of the initial concentrations under both oxygen-sufficient and -deficient conditions (not shown).
Table 2.
Generation times and geosmin and MIB removal rates during exponential growth of bacterial isolates in LB media supplemented with MIB and geosmin under aerobic and nonaerated conditionsa
| Isolate | Aerobic |
Nonaerated |
||||
|---|---|---|---|---|---|---|
| Generation time (h) | Removal rate (ng h−1 cell−1) |
Generation time (h) | Removal rate (ng h−1 cell−1) |
|||
| Geosmin | MIB | Geosmin | MIB | |||
| Variovorax paradoxus | 1.02 | 1.46 × 10−6 | 7.73 × 10−7 | 2.60 | 2.30 × 10−9 | 1.35 × 10−9 |
| Rhodococcus sp. | 1.15 | NDb | ND | 2.19 | 4.81 × 10−10 | 3.54 × 10−10 |
| Comamonas sp. | 1.11 | ND | ND | 3.46 | 2.94 × 10−9 | 1.36 × 10−9 |
The initial concentration of either geosmin or MIB in the various media was 5 μg liter−1.
ND, not determined.
FISH.
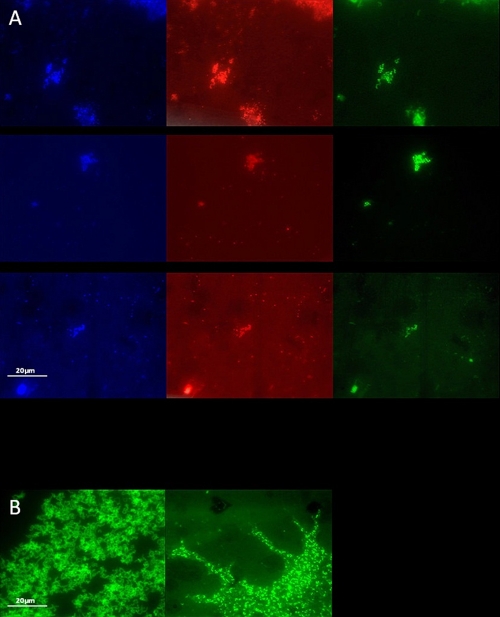
The presence of bacteria closely related to both Rhodococcus sp. and Comamonas sp. in sludge derived from the digestion basin was verified by fluorescence in situ hybridization (FISH) using general and specific rRNA-targeted probes (Fig. 7). The presence of bacteria belonging to the genus Rhodococcus was demonstrated by using both a general 23S rRNA-targeting probe, HGC69A, for Actinobacteria and a specific 16S rRNA-targeting probe, RHCOC3, for the genus Rhodococcus. Bacteria of the genus Comamonas were identified in the sludge by using a specific 16S rRNA-targeting probe, COM1424. The hybridized bacteria showed a high morphological similarity to pure cultures of the Rhodococcus and Comamonas-like isolates, which also hybridized with the aforementioned probes (Fig. 7). Identification of bacteria belonging to the genus Variovorax was more problematic. A general 16S rRNA-targeting probe (SBACT P338) against Planctomycetales (comprising the genus Variovorax) did hybridize with the Variovorax paradoxus-like isolate as well as with bacteria present in the sludge (not shown). However, a specific 16S rRNA-targeting probe, VAR1, failed to hybridize with the isolate.
Fig 7.
FISH analysis of sludge bacteria with DAPI (blue), EUB338 (red) and the targeting probes (green) HGCr69A (top panels), RHCOC3 (middle panels), and COM1424 (bottom panels) (A) and the Rhodococcus sp.-like isolate with the 16S rRNA targeting probe RHCOC3 (left) and the Comamonas sp.-like isolate with the 16S rRNA targeting probe COM1424 probe (right) (B).
DISCUSSION
In the present study, pure cultures of bacteria were obtained that were capable of using geosmin and MIB as the sole organic carbon and energy sources under both aerobic and nonaerated conditions. Closely related bacteria were found to degrade aromatic hydrocarbons, as was found for (i) Variovorax paradoxus, which was shown to metabolize 3-nitrotyrosine (37); (ii) bacteria belonging to the genus Comamonas, found capable of nitrobenzene (36) and quinoline degradation (7); and (iii) bacteria within the genus Rhodococcus, degrading a wide array of aromatic hydrocarbons such as benzene, toluene, and phenol (26). Among the aforementioned genera, only Rhodococcus has been associated with degradation of geosmin and MIB (11, 12, 41) when incubated with additional carbon sources.
Degradation and growth of bacteria with either one of these compounds have been reported in a few studies. It was found that MIB, when supplied as the sole carbon source, was degraded by a consortium of three or more Pseudomonas species isolated from a freshwater lake (24) and by a Bacillus sphaericus isolate from a drinking water reservoir (30). Recently, geosmin, when supplied as the sole carbon source, was found to support growth of a Sphingopyxis sp. isolate from a sand filter in a wastewater treatment plant (21). It seems unlikely that the ability to grow on geosmin and/or MIB as the sole carbon source provides a competitive advantage for these organisms since the sole presence of only those and no other organic carbon compounds in natural environments is not expected. The presence of off-flavor compounds together with other organic carbon sources in natural water bodies might explain why most reported isolates are only capable of geosmin or MIB degradation while metabolizing additional carbon sources (11, 12, 23, 35, 41, 44, 50). From the latter studies, no clear picture emerges as to the bioenergetic value of geosmin and MIB when (co)metabolized with other organic carbon compounds, and as such, the benefits for organisms like those isolated in this study, capable of using these compounds as the sole, hence primary, carbon sources have yet to be determined. Due to the unavailability of suitable bacterial isolates, only a few studies on bacterial geosmin and MIB degradation pathways have been conducted (42, 47). Based on these studies, the degradation pathways of both compounds seem to differ substantially. However, a common degradation pathway for geosmin and MIB using the Bayer-Villiger reaction (48) has been suggested (19). From the above findings, it may be concluded that a better understanding of the bacterial metabolism of geosmin and MIB is hampered by a lack of available suitable bacterial strains. It is anticipated, therefore, that the bacteria reported here, capable of degrading both compounds in the absence of additional carbon sources, may serve as an important tool for a better understanding of the bacterial geosmin and MIB metabolism.
In this study, the bacterial isolates were found to be capable of geosmin and MIB removal also when incubated in argon-flushed and airtight flasks. Given the fact that the bacterial strains were isolated from an anaerobic digestion basin, this finding is not surprising. However, the mineral medium used in this study did not contain any external electron acceptors, and as such, the question remains whether geosmin and MIB were degraded by fermentation processes or whether despite its low ambient concentrations, oxygen was used by the cells for degradation of these compounds.
Limited information is available on geosmin and MIB degradation rates by other bacteria. The geosmin degradation rates obtained in this study were in the same order of magnitude as those of a recently isolated bacterium similar to Sphingopyxix alaskensis (21), while the MIB degradation rates were higher than those observed for a consortium of Pseudomonas species (24). As all three isolates examined in this study failed to grow in autotrophic growth medium but did grow when incubated with either geosmin or MIB in a concentration-dependent mode, it is apparent that these compounds are used by the bacterial isolates as primary carbon sources. By using cell concentrations, cell sizes as estimated from FISH analyses (Fig. 7), and values of bacterial carbon content provided by Norland et al. (38), we estimated that the increase in cell carbon accounted for 20 to 90% of the total carbon which was removed as either geosmin or MIB during the various incubations. This range of carbon incorporation efficiencies (i.e., the amount of new carbon biomass produced per unit of carbon assimilated) is high as respiration accounts for 70% of the assimilated carbon in heterotrophic bacteria (9). Most probably, due to the low initial geosmin and MIB concentrations and the resulting low bacterial biomass during the various incubations, estimates like these are inaccurate. Similar mass balances derived from incubations with higher initial geosmin and MIB concentrations (Fig. 3) resulted in more feasible carbon assimilation rates, which accounted for 6.5 to 25% of the total carbon removed from the flasks.
Threshold levels for both geosmin and MIB, known to affect the taste and odor of fish, are as low as 5 ng liter−1. As such, in order for biological degradation to be of practical importance, geosmin- and MIB-degrading bacteria should exhibit a relatively high affinity for these substrates. We found that when incubated at low ambient geosmin and MIB concentrations (not shown), these compounds were either depleted or at very low levels (lower than 10 ng liter−1) during the late stationary growth phase, and we conclude, therefore, that biological removal of these compounds is also effective in the low concentration range often found in off-flavor-tainted water bodies. Moreover, physical/chemical adsorption of these off-flavor compounds takes place at very low ambient concentrations (e.g., see reference 17). Therefore, at elevated geosmin and MIB levels within the adsorbent matrix, it might be assumed that biological removal of these compounds is not affected by substrate limitation.
Recently, we found that geosmin and MIB could be efficiently removed in a lab-scale up-flow anaerobic sludge blanket (UASB) reactor, containing sludge from which the bacterial strains reported here were isolated (L. Guttman and J. van Rijn, unpublished data). Ongoing research deals with investigating the use of a similar, but larger, reactor in a closed-aquaculture recirculating system.
Supplementary Material
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann R, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amann R, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amann R, et al. 1996. rRNA-targeted oligonucleotide probes for the identification of genuine and former pseudomonads. Syst. Appl. Microbiol. 19:501–509 [Google Scholar]
- 5. American Public Health Association 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, DC [Google Scholar]
- 6. Cook D, Newcombe G, Sztajnbok P. 2001. The application of powdered activated carbon for MIB and geosmin removal: predicting PAC doses in four raw waters. Water Res. 35:1325–1333 [DOI] [PubMed] [Google Scholar]
- 7. Cui MC, Chen FZ, Fu JM, Sheng GY, Sun GP. 2004. Microbial metabolism of quinoline by Comamonas sp. W. J. Microbiol. Biotechnol. 20:539–543 [Google Scholar]
- 8. Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 9. Del Giorgio PA, Cole JJ. 1998. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29:503–541 [Google Scholar]
- 10. Drikas M, Dixon M, Morran J. 2009. Removal of MIB and geosmin using granular activated carbon with and without MIEX pre-treatment. Water Res. 43:5151–5159 [DOI] [PubMed] [Google Scholar]
- 11. Eaton RW, Sandusky P. 2009. Biotransformations of 2-methylisoborneol by camphor-degrading bacteria. Appl. Environ. Microbiol. 75:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eaton RW, Sandusky P. 2010. Biotransformations of (+/−)-geosmin by terpene-degrading bacteria. Biodegradation 21:71–79 [DOI] [PubMed] [Google Scholar]
- 13. Elhadi SLN, Huck PM, Slawson RM. 2006. Factors affecting the removal of geosmin and MIB in drinking water biofilters. J. Am. Water Works Assoc. 98:108–119 [Google Scholar]
- 14. Engle CR, Pounds GL, Van der Ploeg M. 1995. The cost of off-flavor. J. World Aquacult. Soc. 26:297–306 [Google Scholar]
- 15. Gelfand I, et al. 2003. A novel zero-discharge intensive seawater recirculating system for culture of marine fish. J. World Aquacult. Soc. 34:344–358 [Google Scholar]
- 16. Guttman L, van Rijn J. 2008. Identification of conditions underlying production of geosmin and 2-methylisoborneol in a recirculating system. Aquaculture 279:85–91 [Google Scholar]
- 17. Guttman L, van Rijn J. 2009. 2-Methylisoborneol and geosmin uptake by organic sludge derived from a recirculating aquaculture system. Water Res. 43:474–480 [DOI] [PubMed] [Google Scholar]
- 18. Halvorson HO, Ziegler NR. 1933. Application of statistics to problems in bacteriology. I. A means of determining bacteriological populations by the dilution method. J. Bacteriol. 25:101–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho L, Hoefel D, Bock F, Saint CP, Newcombe G. 2007. Biodegradation rates of 2-methylisoborneol (MIB) and geosmin through sand filters and in bioreactors. Chemosphere 66:2210–2218 [DOI] [PubMed] [Google Scholar]
- 20. Hoefel D, et al. 2006. Cooperative biodegradation of geosmin by a consortium comprising three gram-negative bacteria isolated from the biofilm of a sand filter column. Lett. Appl. Microbiol. 43:417–423 [DOI] [PubMed] [Google Scholar]
- 21. Hoefel D, Ho L, Monis PT, Newcombe G, Saint CP. 2009. Biodegradation of geosmin by a novel Gram-negative bacterium; isolation, phylogenetic characterization and degradation rate determination. Water Res. 43:2927–2935 [DOI] [PubMed] [Google Scholar]
- 22. Howgate P. 2004. Tainting of farmed fish by geosmin and 2-methyl-iso-borneol: a review of sensory aspects and uptake/depuration. Aquaculture 234:155–181 [Google Scholar]
- 23. Ishida H, Miyaji Y. 1992. Biodegradation of 2-methylisoborneol by oligotrophic bacterium isolated from a eutrophied lake. Water Sci. Technol. 25:269–276 [Google Scholar]
- 24. Izaguirre G, Wolfe RL, Means EG. 1988. Degradation of 2-methylisoborneol by aquatic bacteria. Appl. Environ. Microbiol. 54:2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jüttner F, Watson SB. 2007. Biochemical and ecological control of geosmin and 2-methylisoborneol in source waters. Appl. Environ. Microbiol. 73:4395–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim D, et al. 2002. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 68:3270–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koch AL. 1981. Growth measurement, p 179–207 In Gerhardt P, et al. (ed), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 28. Kyselkova M, et al. 2008. Development of a 16S rRNA gene-based prototype microarray for the detection of selected actinomycetes genera. Antonie Van Leeuwenhoek 94:439–453 [DOI] [PubMed] [Google Scholar]
- 29. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 30. Lauderdale CV, Aldrich HC, Lindner AS. 2004. Isolation and characterization of a bacterium capable of removing taste- and odor-causing 2-methylisoborneol from water. Water Res. 38:4135–4142 [DOI] [PubMed] [Google Scholar]
- 31. Lloyd SW, Lea JM, Zimba PV, Grimm CC. 1998. Rapid analysis of geosmin and 2-methylisoborneol in water using solid phase micro extraction procedures. Water Res. 32:2140–2146 [Google Scholar]
- 32. McDowall B, Ho L, Saint CP, Newcombe G. 2007. Removal of geosmin and 2-methylisoborneol through biologically active sand filters. Int. J. Environ. Water Management 1:311–320 [Google Scholar]
- 33. Miller JH. 1971. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Narayan IV, Nunez WJ. 1974. Biological control: isolation and bacterial oxidation of the taste and odor compound geosmin. J. Am. Water Works Assoc. 66:32–536 [Google Scholar]
- 36. Nishino SF, Spain JC. 1995. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl. Environ. Microbiol. 61:2308–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishino SF, Spain JC. 2006. Biodegradation of 3-nitrotyrosine by Burkholderia sp. strain JS165 and Variovorax paradoxus JS171. Appl. Environ. Microbiol. 72:1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Norland S, Heldal M, Tumyr O. 1987. On the relation between dry matter and volume of bacteria. Microb. Ecol. 13:95–101 [DOI] [PubMed] [Google Scholar]
- 39. Persson F, Heinecke G, Hedberg T, Hermansson M, Uhl W. 2007. Removal of geosmin and MIB by biofiltration—an investigation discriminating between adsorption and biodegradation. Environ. Technol. 28:95–104 [DOI] [PubMed] [Google Scholar]
- 40. Roller C, Wagner M, Amann R, Ludwig W, Schleifer KH. 1994. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849–2858 [DOI] [PubMed] [Google Scholar]
- 41. Saadoun I, El-Migdadi F. 1998. Degradation of geosmin-like compounds by selected species of Gram-positive bacteria. Lett. Appl. Microbiol. 29:98–100 [DOI] [PubMed] [Google Scholar]
- 42. Saito A, Tokuyama T, Tanaka A, Oritani T, Fuchigami K. 1999. Microbiological degradation of (−)geosmin. Water Res. 33:3033–3036 [Google Scholar]
- 43. Sanguin H, et al. 2006. Development and validation of a prototype 16S rRNA-based taxonomic microarray for Alphaproteobacteria. Environ. Microbiol. 8:289–307 [DOI] [PubMed] [Google Scholar]
- 44. Silvey JKG, Henley AW, Nunez WJ, Cohen RC. 1970. Biological control: control of naturally occurring taste and odors by microorganisms. Proceedings of the National Biological Congress, Detroit, MI. [Google Scholar]
- 45. Stanier JY, Kunisawa R, Mandel M, Cohen-Bazire G. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35:171–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 47. Tanaka A, et al. 1996. Biodegradation of musty odour component, 2-methylisoborneol. Water Res. 30:759–761 [Google Scholar]
- 48. Trudgill PW. 1984. Microbial degradation of the alicyclic ring: structural relationships and metabolic pathways, p 131–180 In Gibson DT. (ed), Microbial degradation of organic compounds. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 49. Tucker CS. 2000. Off-flavor problems in aquaculture. Rev. Fish. Sci. 8:45–88 [Google Scholar]
- 50. Yagi M, Nakashima S, Muramoto S. 1988. Biological degradation of musty odour compounds, 2-methylisoborneol and geosmin, in a bio-activated carbon filter. Water Sci. Technol. 20:255–260 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.