Abstract
Degradation of the quorum-sensing (QS) signals known as N-acylhomoserine lactones (AHL) by soil bacteria may be useful as a beneficial trait for protecting crops, such as potato plants, against the worldwide pathogen Pectobacterium. In this work, analytical chemistry and microbial and molecular approaches were combined to explore and compare biostimulation of native and introduced AHL-degrading Rhodococcus erythropolis populations in the rhizosphere of potato plants cultivated in farm greenhouses under hydroponic conditions. We first identified gamma-heptalactone (GHL) as a novel biostimulating agent that efficiently promotes plant root colonization by AHL-degrading R. erythropolis population. We also characterized an AHL-degrading biocontrol R. erythropolis isolate, R138, which was introduced in the potato rhizosphere. Moreover, root colonization by AHL-degrading bacteria receiving different combinations of GHL and R138 treatments was compared by using a cultivation-based approach (percentage of AHL-degrading bacteria), pyrosequencing of PCR-amplified rrs loci (total bacterial community), and quantitative PCR (qPCR) of the qsdA gene, which encodes an AHL lactonase in R. erythropolis. Higher densities of the AHL-degrading R. erythropolis population in the rhizosphere were observed when GHL treatment was associated with biocontrol strain R138. Under this condition, the introduced R. erythropolis population displaced the native R. erythropolis population. Finally, chemical analyses revealed that GHL, gamma-caprolactone (GCL), and their by-products, gamma-hydroxyheptanoic acid and gamma-hydroxycaproic acid, rapidly disappeared from the rhizosphere and did not accumulate in plant tissues. This integrative study highlights biostimulation as a potential innovative approach for improving root colonization by beneficial bacteria.
INTRODUCTION
N-Acylhomoserine lactones are intercellular signals used by numerous alpha-, beta-, and gammaproteobacteria to regulate gene expression at the population and community levels (18, 38). The mechanism connecting cell population to gene expression via AHLs is termed quorum sensing (QS) (17). Pectobacterium carotovorum is a causative agent of blackleg and soft rot diseases in several crops, including potato plants and tubers. Over the 2004-2009 period, quality refusals oscillated between 2 and 5% and 3 and 8% of the total seed tuber production in France and Netherlands, respectively; blackleg disease, which is caused by Pectobacterium and Dickeya, represented 10 to 30% and 60 to 80% of the causes of refusal in the same countries. In Pectobacterium, production of virulence factors such as pectinolytic and cellulolytic enzymes and harpins is positively controlled by AHLs (23, 29). In Pectobacterium carotovorum, inhibition of AHL synthesis or degradation of the AHLs produced results in the absence of the expression of the QS-regulated genes and consequently in a decrease of the virulence symptoms on potato plants (23, 31).
During the past decade, several approaches have been proposed to disrupt QS regulation and hence limit the virulence of P. carotovorum and other bacterial pathogens in which pathogenicity is controlled by QS (11, 14, 40). The antivirulence approaches targeting QS are collectively called quorum quenching (40). Among quorum-quenching (QQ) strategies targeting Pectobacterium, introduction of AHL-degrading bacteria remains the most fully explored. In the potential QQ agents Bacillus thuringiensis and Rhodococcus erythropolis (13, 32), several AHL-degrading enzymes were discovered. While B. thuringiensis expresses a unique lactonase, AiiA (13), R. erythropolis expresses three QQ activities, a lactonase (QsdA), an acylase, and a reductase (28, 33, 34). Aside from the use of biocontrol agents (13, 32), QQ molecules (24, 27), and QQ transgenic plants (12, 16), a biostimulation approach was recently proposed (6, 7). It consists of the application of a biodegradable agent, gamma-caprolactone (GCL), to stimulate in the rhizosphere of Solanum tuberosum the growth of endogenous AHL-degrading bacteria (hence the name “biostimulation” to describe this technique). GCL exhibits some similarity with the conserved core of the AHL (gamma-butyrolactone) ring. Notably, over 95% of the GCL-stimulated QQ bacteria belonged to R. erythropolis, revealing that GCL treatment stimulated the growth of one the most efficient QQ bacteria.
In this study, analytical chemistry, microbiology, and molecular approaches were combined (i) to investigate the structural characteristics of biostimulating molecules, discover a novel biostimulation agent, gamma-heptalactone (GHL), and evaluate its growth-stimulating effect on QQ bacteria in the rhizosphere of potato plants, its impact on total bacterial community by rrs pyrosequencing, and its fate, as determined by fine high-pressure liquid chromatography–tandem mass spectrometry (HPLC-MS/MS) tools, and (ii) to characterize a QQ bacterium, Rhodococcus erythropolis R138, which was used alone and in combination with the biostimulating agent GHL for enhancing the level of QQ bacteria in the rhizosphere of potato plants in farm greenhouses. In the combined treatment with GHL and R138, the fates of the introduced biological and biochemical agents were analyzed, as well as the dynamics of the bacterial community, by rrs pyrosequencing. This integrative approach revealed that a combination of biostimulating and biological treatments is a potential innovative strategy for efficiently stimulating the colonization of a crop rhizosphere by AHL-degrading bacteria.
MATERIALS AND METHODS
Chemicals and bacterial cultures.
All chemicals were purchased from Sigma-Aldrich-Fluka, and their structures are shown in Fig. 1A. 4-Hydroxy-caproic acid (HCA) and 4-hydroxyheptanoic acid (HHA) were obtained by incubating GCL and GHL, respectively, in the presence of NH4OH (0.5 M; pH 9) for 24 h at 25°C for lactone ring opening. Bacterial cultures were grown in rich media: tryptic soy agar (TSA), purchased from AES (France), TY (tryptone 5 g liter−1, yeast extract 3 g liter−1), and synthetic medium AB (5), in which ammonium chloride (1 g liter−1) was used as a sole nitrogen source and mannitol (2 g liter−1) as a sole carbon source, except where another carbon source is specified. Pectobacterium atrosepticum CFBP 6276 was cultivated in PGA medium (31). R. erythropolis strain R138R was cultivated in the presence of rifampin at 100 mg liter−1 (Rif100). Agar was added at 15 g liter−1.
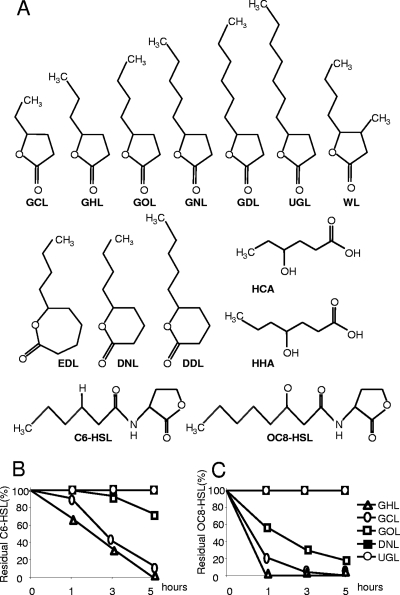
Fig 1.
Lactones and derivatives used in this study and kinetics of AHL inactivation by bacterial consortia stimulated by lactones. (A) Structural formulae of γ-caprolactone (GCL), γ-heptalactone (GHL), γ-octalactone (GOL), γ-nonalactone (GNL), γ-decalactone (GDL), undecanoic γ-lactone (UGL), Whiskey lactone (WL), ε-decalactone (EDL), δ-nonalactone (DNL), δ-decalactone (DDL), 4-hydroxyhexanoic acid (HCA), 4-hydroxyheptanoic acid (HHA) (the latter two compounds result from lactonolysis of GCL and GHL, respectively), N-hexanoyl-homoserine lactone (C6-HSL), and N-3-oxo-octanoyl-homoserine lactone (OC8-HSL). The bacterial consortia, obtained after enrichment on the indicated compounds or mannitol as a sole carbon source, were tested for their capacity to inactivate C6-HSL (B) and OC8-HSL (C). At each time point, residual AHL was measured and expressed as a percentage of residual AHL in the reference assay with the mannitol-only consortium. Kinetics obtained with DNL and UGL are superimposed.
Assimilation test and colorimetric quantification of GCL and GHL.
The capacity of bacterial strains to assimilate GCL and GHL in vitro was determined by inoculating bacterial strains in medium AB supplemented with mannitol (2 g liter−1) and/or GCL or GHL (2 g liter−1). Bacterial growth was monitored by spectrophotometry at 600 nm, while a colorimetric assay (39) allowed rapid quantification of the introduced lactones.
Identification and quantification of GCL, HCA, GHL, and HHA by HPLC-MS.
Plant tissues were crushed under liquid nitrogen and extracted with 25 ml of phosphate-buffered saline (PBS; NaCl, 8 g liter−1; KCl, 0.20 g liter−1; Na2HPO4, 1.44 g liter−1; KH2PO4, 0.24 g liter−1; adjusted to pH 7.2) and centrifuged at 10,000 × g for 20 min (4). Plant extracts and samples from nutrient solution were filtered through polyethersulfone columns (10 kDa; Vivaspin 500). Chromatographic separation of samples (10 μl) was performed by HPLC (Waters Allians 2690) combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Waters ZQ mass spectrometer with a single-quadrupole system and electrospray ionization). For each of the analyzed compounds, a calibration curve is defined with pure compound. Mobile phase A was water–0.1% formic acid, and mobile phase B was acetonitrile–0.1% formic acid. Four experimental procedures were defined for detecting GCL alone, GCL plus its by-product HCA, GHL alone, and GHL plus its by-product HHA.
To detect GCL molecule alone, a Waters Sunfire C18 (150 mm by 3 mm by 5 μm) column was used. The mobile phase flow rate was 0.7 ml min−1. An isocratic mixture of 70% mobile phase A and 30% B was applied for 10 min to elute GCL and then 100% B for 5 min to wash the column. An equilibration time of 10 min was allowed before the next injection. The retention time of GCL was 4.6 min.
GCL and its hydrolyzed form HCA in the same run were detected with a Waters Gemini C18 (150 mm by 2 mm by 5 μm) column was used. The mobile-phase flow rate was 0.2 ml min−1, and the retention times of GCL and HCA were 11.8 and 8.6 min, respectively. A gradient was used to optimize molecule separation. The gradient started at 5% B, was kept at 5% for 1 min, was increased linearly to 60% for 14 min, and then was increased until to 95% for 3 min. The mobile phase composition was kept at 95% for 2 min and returned to 5% for 2 min. An equilibration time of 8 min was applied before the next injection.
To detect GHL molecules alone, a Waters Gemini C18 (150 mm by 2 mm by 5 μm) column was used. The mobile-phase flow rate was 0.2 ml min−1 with a retention time of 7.3 min. An isocratic mixture of 70% A and 30% B was applied for 10 min to elute GHL and then increased to 100% B for 5 min to wash the column. Before the next injection, an equilibration time of 10 min was allowed.
GHL and HHA in the same run were detected with the Waters Gemini column and flow rate used for GHL alone. A gradient was applied to optimize molecule separation. The gradient started at 5% B and increased linearly to 30% over 10 min. The mobile-phase composition was kept at 30% B for 10 min, increased to 95% for 5 min, and returned to 5% B over 1 min. An equilibration time of 8 min was allowed before the next injection. The retention times of GHL and HHA were, respectively, 17.1 and 12.6 min.
Bacterial enrichment in the presence of lactones.
For comparing biostimulation of AHL degradation by different lactones, 1 g of a nonsterile soil from the CNRS experimental field (Mérantaise) at Gif-sur-Yvette (France) was resuspended in 10 ml of sterile 0.8% (wt/vol) NaCl. Soil suspensions were diluted (1/50) into AB medium supplemented with actidione (100 mg liter−1) and mannitol (as a control) or one of the tested lactones. After 2 cycles of enrichment as described by Cirou et al. (6), the bacterial consortia were washed twice in 0.8% NaCl, their cell density was adjusted to an optical density at 600 nm (OD600) of 1.0, and their capacities to inactivate hexanoylhomoserine lactone (C6-HSL) at 25 μM and 3-oxo-octanoylhomoserine lactone (OC8-HSL) at 200 nM were compared, as described by Carlier et al. (3). Residual C6-HSL and OC8-HSL were quantified using the biosensor strains Chromobacterium violaceum CV026 (25) and Agrobacterium tumefaciens NT1(pZNLR4) (4).
Biocontrol tests on potato tubers.
Overnight cultures of P. atrosepticum and AHL-degrading strains were washed in 0.8% NaCl. Each tuber of S. tuberosum var. Allians was inoculated with 2 × 107 CFU of P. atrosepticum and/or 2 × 107 CFU of each AHL-degrading strain. Three to nine tubers were inoculated for each condition. Five days postinfection, the tubers were cut in the middle and photographed. Maceration was rated as follows: 1, no maceration; 2, moderate maceration (no more than 5 mm around the infection site); 3, strong maceration (more than 5 mm). The Kruskal-Wallis test (α = 0.05) allowed the statistical analysis of the maceration categories.
Plant culture in hydroponic conditions in a greenhouse.
S. tuberosum var. Allians was cultivated in a greenhouse (Comité Nord, Bretteville-du-Grand-Caux, Normandy, France) under natural light at 10 to 15°C (night) and 25 to 30°C (day) from April to July. The nutritive solution Hydrobloom (Cellmax, United Kingdom), with nitrogen at 0.8 g liter−1 and potassium at 1.48 g liter−1 as major components, was diluted from a commercial stock solution (1:250) with nonsterile water from the public water system. One hundred in vitro plants were placed into holes (3 cm apart) in the batch cover. Each batch (40 by 60 by 8 cm) contained 13 liters of nutritive solution.
Analysis of cultured bacterial populations collected from potato rhizosphere.
One gram of roots (fresh weight) was suspended in 10 ml of sterile 10 mM MgSO4, diluted, and spread on TSA medium for counting total populations and, when appropriate, on AB medium supplemented with Rif100 and GCL as a sole carbon source for counting strain R138R only. At each time, three samples were analyzed from each batch. From each of the samples, 30 TSA isolates were individually cultivated on 96-microwell plates and tested for production and inactivation of AHL on thin-layer chromatography C18 reverse-phase silica plate (Whatman), as well as growth on GCL-Rif minimal medium. AHL production was revealed with the biosensor A. tumefaciens NT1(pZNLR4), while C6-HSL disappearance (C6-HSL was introduced at 25 μM) was measured with the biosensor C. violaceum CV026, as described previously (8).
Genotyping of cultured bacteria by rrs sequencing.
For rrs genotyping, the 5′ end of the rrs gene was amplified with primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and 518r (5′-ATTACCGCGGCTGCTGG-3′) and sequenced with primer pA by GATC (Germany). Blastn analyses were performed with a 400-bp-minimal-length query against the EMBL database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Though some sequence comparisons permitted identification of isolates at the species level, such as R. erythropolis, the genus level was retained for homogeneity.
DNA extraction from potato rhizosphere.
The roots of two potato plants were taken from hydroponic culture batches, crushed under liquid nitrogen, and solubilized with 500 μl of saline buffer (containing 10 mM MgSO4). Total genomic DNA of this homogenate was extracted with the appropriate DNA isolation kit (Mo Bio Power Soil) according to the manufacturer's protocol. DNA quantity and quantity were evaluated with a spectrophotometer (NanoDrop ND1000; Labtech, France).
Relative abundance of qsdA determined by qPCR.
In DNA extracted from rhizosphere samples, the relative abundance of the qsdA gene of R. erythropolis was estimated by quantitative PCR (qPCR) (LightCycler 480 in 96-well plates; Roche). Primers (5′-ACGAGCATGTCTTCGTTCTG and 5′-GGATCGACGATCGTGCTGAT) were designed for a conserved region of qsdA and generated a fragment of 145 bp. A calibration curve was defined with genomic DNA of R. erythropolis R138. The composition of the PCR mix for each sample was as follows: 5 μl of SYBR green I master mix (Roche), reverse primer (0.5 μM), forward primer (0.5 μM), and 1 μl of DNA sample at 8 ng μl−1.
Amplification of rrs target for pyrosequencing.
Extracted total DNA was adjusted to a final DNA concentration of 15 ng/μl in 1/10× TE buffer using a NanoVue spectrophotometer (GE Healthcare) and verified by ethidium bromide fluorescence after electrophoresis through a 1% agarose gel in TAE (2 mM Tris-acetate [pH 8], 5 mM Na-EDTA) buffer. Then, multiple 50-μl PCRs were performed using the universal 16S rRNA gene bacterial primers 8F (BxxxxxxAGAGTTTGATCMTGGCTCAG) and 357R (AxxxxxxCTGCTGCCTYCCGTA), where B and A represent the adaptors B and A for pyrosequencing using the Gold reaction (GS20; Roche/454 Life Sciences) and “xxxxxx” represents 6-nucleotide sequence tags designed for sample identification bar coding (2, 19). PCR amplification conditions were adapted for the use of three different thermostable DNA polymerases. The first PCR used Phusion high-fidelity DNA polymerase (Finnzymes), as follows: 98°C for 2 min, followed by 25 cycles of 98°C for 30 s, 48°C for 20 s, and 72°C for 12 s, and a final elongation step at 72°C for 5 min. The second PCR used Pfu DNA polymerase (Fermentas), as follows: 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 48 s, and a final elongation step at 72°C for 5 min. The third PCR used high-fidelity PCR enzyme mix (Fermentas), as follows: 94°C for 3 min followed by 30 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 24 s, and a final elongation step at 72°C for 5 min. Each 50-μl PCR mixture contained 15 ng DNA, a 0.1 μM concentration of each primer (Sigma), 0.2 mM deoxynucleoside triphosphate mix (Fermentas), and 1.25 U of Phusion high-fidelity and Pfu DNA polymerases or 2 U of high-fidelity PCR enzyme mix (Fermentas)., and the buffers supplied with each polymerase were used. Each DNA sample was subjected to 5 to 10 PCRs per thermostable DNA polymerase, and two different polymerases were used per sample. The resultant PCR products were pooled and loaded on a 1% agarose gel in TAE buffer (2 mM Tris-acetate [pH 8], 5 mM Na-EDTA). After DNA visualization by ethidium bromide staining and long-wave UV illumination, the 16S amplified DNA fragments were cut from the gel and purified using a NucleoSpin Extract II kit (Macherey-Nagel) according to the manufacturer's instructions. For pyrosequencing, 50 ng of PCR products from each sample was mixed.
bTEFAP FLX pyrosequencing.
Pyrosequencing was performed using a Roche/454 FLX pyrosequencer (GATC Biotech, Konstanz, Germany). The sequences obtained for each sample were grouped according to the tag used, and after removal of the tags, the average sequence was 227 nt. The sequences were selected by their length (>150 nt) and their quality score (90% of nucleotides with a quality score of >25) using the Greengenes website (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi). Then sequences with more than two errors in the primer or more than one ambiguous base were removed using the pyrosequencing pipeline of the Ribosomal Database Project (RDP) (http://pyro.cme.msu.edu/index.jsp).
Analysis of rrs sequences recovered from pyrosequencing.
The 16S sequences obtained from pyrosequencing were analyzed using the Ribosomal Database Project website (http://rdp.cme.msu.edu/). The 16S sequences were assigned to classes or genera using the Classifier tool (http://rdp.cme.msu.edu/classifier/classifier.jsp). The Shannon indices, cluster values, and CHAO1 values were obtained using the RDP's pyrosequencing pipeline (http://pyro.cme.msu.edu/). Statistical calculations were performed using R software, version 2.10.1 (http://www.r-project.org/). The package ade4TkGUI was used for component analysis, between-group analysis, and permutation test (10,000 bootstraps). The hierarchical clustering of genera and treatment data was performed using Cluster software 2.11 and displayed with Treeview software 1.6 (http://rana.lbl.gov/EisenSoftware.htm). Prior to this analysis, the total number of sequences per sample was arbitrarily fixed at 100,000, and the number of sequences per genus was adjusted. These values were log10 transformed and used in the Cluster software. Uncentered correlation was used as a parameter for the clustering.
RESULTS
Lactone specificity for biostimulation of microbial quorum-quenching activity.
We investigated whether various lactones (with gamma, delta, and epsilon rings and acyl chains of different lengths) might stimulate the capability of a native bacterial community to inactivate the QS signals hexanoylhomoserine lactone (C6-HSL) and 3-oxo-octanoylhomoserine lactone (OC8-HSL), the latter being a key signal involved in the regulation of virulence in Pectobacterium. Synthetic AB medium supplemented with one of the tested lactones (Fig. 1A), GCL (positive control), or mannitol (negative control) as a sole carbon source was inoculated into planted soil (1 g liter−1). After two enrichment cycles at 24°C, the bacterial consortia were collected and compared for their ability to inactivate C6-HSL and OC8-HSL (Fig. 1B and C). The efficient stimulation of the AHL degradation activity was observed only with GCL (the positive control) and GHL, and to a lesser extent with γ-octalactone (GOL). Remarkably, the application of whiskey lactone (WL), γ-nonalactone (GNL), γ-decalactone (GDL), ε-decalactone (EDL), and δ-decalactone (DDL) inhibited growth of soil bacteria under the assay conditions. Further biostimulation assays on the cultures of S. tuberosum were therefore performed with GCL and GHL.
GHL treatment stimulates native quorum-quenching population of R. erythropolis.
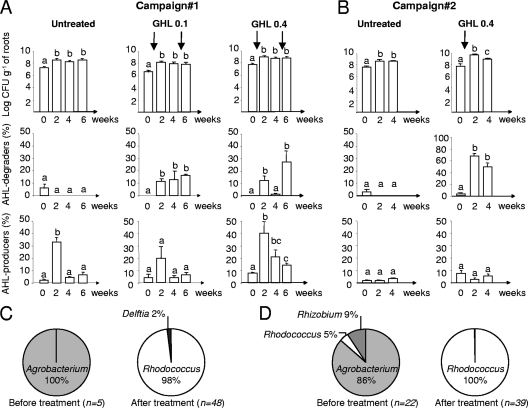
In two independent campaigns (one per year), we investigated GHL biostimulation of AHL-degrading bacteria in hydroponic cultures of S. tuberosum var. Allians. GHL was introduced at two concentrations (0.1 and 0.4 g liter−1) at the beginning of the experiment, i.e., 1 h after sampling (time zero) (Fig. 2A and B). In one experiment only, a second GHL addition (0.1 or 0.4 g liter−1) was performed 1 h after sampling at 4 weeks to evaluate at 6 weeks the impact of two successive biostimulations on the abundance of QQ bacteria. The total number of cultured bacteria increased slightly after GHL treatments. The highest density of AHL degraders, which represented up to 60% of cultured bacteria, was observed following treatment with GHL at 0.4 g liter−1. GHL treatments did not repeatedly affect the percentage of AHL producers. An increase of AHL producers in campaign 1 at 2 weeks appeared likely incidental. Noticeably, a strong bias in the structure of the cultured QQ community was observed after GHL treatment. While most of the isolated QQ bacteria belonged to the genus Agrobacterium before treatment, the vast majority (98 to 100%) belonged to the genus Rhodococcus, more specifically to R. erythropolis species, after treatment (Fig. 2C and D). The R. erythropolis population therefore appears to be the main culturable QQ population whose growth is stimulated after GHL treatment.
Fig 2.
Abundance and diversity of cultured AHL-degrading populations under GHL treatment. (A and B) Total cultured bacteria on TSA medium (CFU g [fresh weight] of roots−1), the percentage of AHL degraders, and the percentage of AHL producers were measured in untreated and GHL-treated (at 0.1 or 0.4 g liter−1) rhizospheres in the course of two independent campaigns; sampling was performed just before (time 0) and 2, 4, and 6 weeks after GHL treatment. In each histogram, all mean values were compared, and statistically significantly different values (Mann-Whitney, 0.05) are indicated by different letters. (C and D) Diversity of AHL degraders recovered from experiments 1 (C) and 2 (D) before and after GHL treatment. n, total number of rrs-identified isolates.
Dynamics of the bacterial community receiving GHL treatment.
Total DNA extracted from rhizospheres of campaign 2, collected before (week 0 [W0]) and after GHL (W2 and W4) treatment, were used as templates for 16S rrs amplification. The amplicons were pyrosequenced using the 454-Roche system. Two replicates were performed and analyzed separately and after merging (see Table S1 in the supplemental material). Among a total of 108,411 sequences, 75,485 were retained for further analyses, of which the first step was removal of chloroplast sequences. The remaining 55,055 bacterial rrs sequences were first analyzed by hierarchical clustering to define the number of clusters at 95% sequence identity. These clusters served as operational taxonomy units (OTUs) for calculations of CHAO1 and Shannon indexes (see Table S1 in the supplemental material). Next, to define the relationship between the bacterial class or genus and the treatment conditions, sequences were individually identified at the genus and class levels (confidence threshold, 80%) and again compared (Fig. 3B; also, see Fig. S1 in the supplemental material).
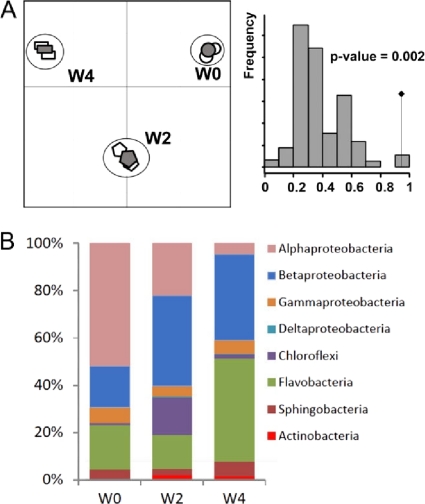
Fig 3.
Dynamics of bacterial diversity under GHL treatment revealed by rrs pyrosequencing. (A) (Left) Correspondence analysis of the two replicates (empty symbols) and merge data (filled symbols) in samples collected before treatment (W0) and 2 (W2) and 4 (W4) weeks after GHL treatment. (Right) Permutation test (n = 10,000) supporting correspondence analysis. The observed value is indicated by a black diamond. (B) Distribution of bacterial classes in merged W0, W2, and W4 data.
Correspondence analysis of genera assigned sequences revealed that the two replicates and merged samples clustered together at W0, W2, and W4 (Fig. 3A). We therefore used only merged data for comparison of the W0, W2, and W4 samples. Cluster numbers and CHAO1 values decreased from W0 to W4. Remarkably, the Shannon index was higher in the W2 samples than in those at W0 and W4, suggesting a strong and temporary effect of GHL treatment on the bacterial biodiversity 2 weeks after its addition to the plant culture system (see Table S1 in the supplemental material).
While Alphaproteobacteria (51%) dominated in W0 samples, most of the sequences belonged to Betaproteobacteria (39%) at W2 and Flavobacteria (44%) and Betaproteobacteria (36%) at W4 in GHL-treated samples. Actinobacteria (2.3 and 1.8% in W2 and W4 samples), which are represented mainly by the genus Rhodococcus (2.2 and 1.7% in the W2 and W4 samples), emerged after GHL treatment (Fig. 3B). In GHL-treated samples at W2 and W4, Rhodococcus reached the 8th and 6th positions among the most abundant genera (Table 1), while it was positioned 62nd in the untreated sample (W0). The other dominant genera were Flavobacterium before treatment and Acidovorax, Herpetosiphon, Flavobacterium, and Hydrogenophaga after GHL treatment (Table 1).
Table 1.
rrs pyrosequencing data showing the major genera obtained after GHL treatment
| Time and genusa | Abundance (%) |
|---|---|
| W0 | |
| Flavobacterium | 14 |
| Sphingobium | 5.9 |
| Devosia | 5.1 |
| Acidovorax | 4.9 |
| Rhizobium | 4.2 |
| Shinella | 3.8 |
| Emticicia | 2.5 |
| Bosea | 2.4 |
| Pseudomonas | 2.1 |
| Acinetobacter | 1.8 |
| Brevundimonas | 1.6 |
| Novosphingobium | 1.5 |
| Methylophilus | 1.3 |
| Delftia | 0.8 |
| Massilia | 0.8 |
| Rhodococcus | 0.06 |
| W2 | |
| Acidovorax | 20.5 |
| Herpetosiphon | 15.7 |
| Flavobacterium | 13.1 |
| Hydrogenophaga | 7.4 |
| Shinella | 6.6 |
| Rhizobium | 4.3 |
| Devosia | 3.5 |
| Rhodococcus | 2.2 |
| Pseudomonas | 2.1 |
| Hyphomicrobium | 1.7 |
| Pseudoxanthomonas | 1.4 |
| Bosea | 1.3 |
| Novosphingobium | 1.1 |
| Sphingobium | 0.8 |
| Azospirillum | 0.7 |
| W4 | |
| Flavobacterium | 41.5 |
| Hydrogenophaga | 26.3 |
| Acidovorax | 5.6 |
| Pseudoxanthomonas | 3.2 |
| Emticicia | 1.8 |
| Rhodococcus | 1.7 |
| Herpetosiphon | 1.5 |
| Simplicispira | 1.3 |
| Terrimonas | 1.2 |
| Pseudomonas | 0.9 |
| Thermomonas | 0.9 |
| Sphingobium | 0.64 |
| Devosia | 0.5 |
| Rhizobium | 0.4 |
| Shinella | 0.4 |
Bold type is used to highlight the change in position for Rhodococcus at the three time points.
The dynamics of the identified genera were compared using a hierarchical clustering approach (see Fig. S1 in the supplemental material). In group I are genera whose frequency increased after treatment, such as Rhodococcus (frequency at W2/frequency at W0 = 37, and W4/W0 = 28) and Hydrogenophaga (W2/W0 = 25 and W4/W0 = 90.7). In group II are genera whose frequency was weakly affected by GHL treatment, such as Flavobacterium (W2/W0 = 0.9 and W4/W0 = 3) and Acidovorax (W2/W0 = 4 and W4/W0 = 1.1). In group III are those whose relative abundance decreased after GHL treatment, such as Sphingobium (W2/W0 = 0.14 and W4/W0 = 0.11). All genera belonging to the Actinobacteria (Rhodococcus), Chloroflexi (Herpetosiphon), and Deltaproteobacteria, including the bacterial parasite Bdellovibrio, fell into group I.
Rapid degradation of GHL and its by-product 4-hydroxy-heptanoic acid.
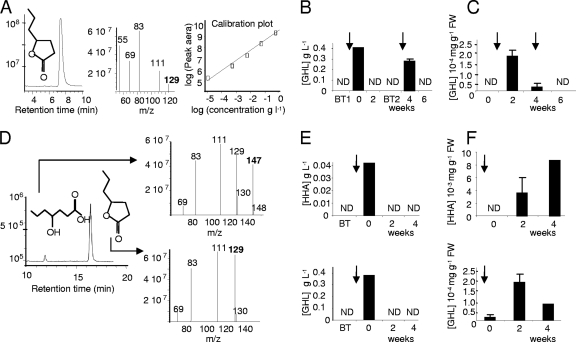
Appropriate HPLC-MS/MS conditions were defined for detection of GHL in the range from 1 g liter−1 to 10−5 g liter−1 (Fig. 4A). In samples of campaign 1 corresponding to GHL treatments at 0.4 g liter−1, a rapid decrease of the introduced GHL was observed in the nutritive solution (Fig. 4B). In plant tissues (Fig. 4C), GHL was detected at a very low level (2 × 10−4 mg g−1 [fresh weight {FW}]) 2 weeks after treatment, and was undetectable (below the detection limit of 1 × 10−5 mg g−1 [FW]) at the end of experiment (after 6 weeks). In samples collected from campaign 2, analytical procedures were upgraded (Fig. 4D) to simultaneously detect GHL and its by-product 4-hydroxyheptanoic acid (HHA). In nutritive solution, 1 h after GHL treatment, HHA reached 0.04 g liter−1, representing approximately 10% of the introduced GHL (Fig. 4E), while it represented only 1% of pure GHL in stock solution. This feature reveals a rapid opening of the lactone ring of GHL. Two weeks after treatment, GHL and HHA were not detected in the nutritive solution (Fig. 4E). In plant tissues, the concentration of GHL (up to 2 × 10−4 mg g−1 [FW]) and HHA (up to 9 × 10−3 mg g−1 [FW]) were very low after treatment and below the detection limit in untreated samples (Fig. 4F). The introduced GHL, and its by-product HHA, did not accumulate in plant tissues.
Fig 4.
Fate of GHL and its by-product HHA in GHL-treated plant cultures. (A to C) Quantification of GHL in experiment 1; (D to F) quantification GHL and HHA in experiment 2. (A) Pure GHL (M+H m/z of 129) was released after 7.3 min and used for preparing a calibration curve (GHL ranging from 1 g liter−1 to 0.01 mg liter−1). This curve was used to estimate the GHL concentration in nutritive solutions (B) and plant tissues (C). (D) Pure HHA (M+H m/z of 147) and GHL (M+H m/z of 129) showed retention times of 12.2 and 17.1 min, respectively, and were used for measuring two calibration curves. The concentrations of GHL and HHA were evaluated in nutritive solutions (E) and in plant tissues (F). In panels A and D, the signal intensity is expressed in arbitrary units; in panels B, C, E, and F, vertical arrows indicate GHL treatments at 0.4 g liter−1. All values are the mean of two replicates. ND, not detected.
Characteristics of R. erythropolis strain R138.
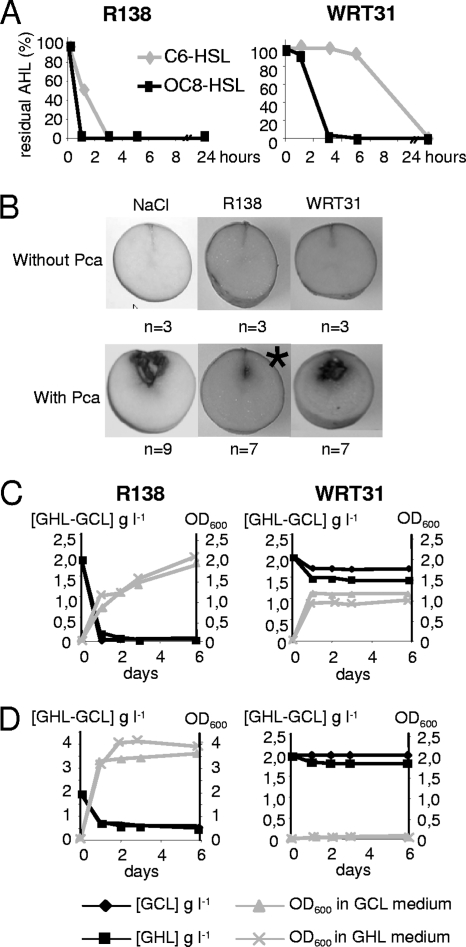
An appropriate bacterial strain was chosen by comparing properties of several nonpathogenic AHL degraders collected from the plant rhizosphere, namely, Agrobacterium sp. strain WRT31 (9), R. erythropolis strain W2 (32), and R. erythropolis isolates R78, R127, and R138, all recovered from GCL-treated cultures of S. tuberosum (6). All strains were compared for their AHL-degrading capacity (Fig. 5A), biocontrol activity against the potato pathogen P. carotovorum 6276 (Fig. 5B), degradation of GCL and GHL in the presence of mannitol as additional energy and carbon sources (Fig. 5C), and assimilation of GCL and GHL as sole carbon sources (Fig. 5D). All Rhodococcus isolates exhibited characteristics similar to those of isolate R138. The Agrobacterium isolate WRT31, which less efficiently inactivated QS signals (compared to Rhodococcus isolate R138), was not able to protect potato tubers against Pectobacterium or to degrade or assimilate GCL and GHL. The biocontrol strain R. erythropolis R138 was therefore chosen for further experiments.
Fig 5.
Properties of the AHL-degrading isolate Rhodococcus sp. strain R138. The isolates Rhodococcus sp. strain R138 and Agrobacterium sp. strain WRT31 were compared for the kinetics of C6-HSL and OC8-HSL inactivation (A), biocontrol activity against P. atrosepticum (Pca) using NaCl (0.8%) for suspending bacterial cells (B), and degradation of GCL and GHL (2 g liter−1) in the presence (C) or absence (D) of mannitol (2 g liter−1) as an additional carbon source. (C and D) Bacterial growth (OD600; gray symbols) and concentration of residual GCL and GHL (g liter−1; black symbols). The asterisk indicates a statistically significant decrease of the maceration symptoms using the Kruskal-Wallis test (α = 0.05), where n is the total number of tubers tested per condition.
Combination of biostimulation and application of biological agent.
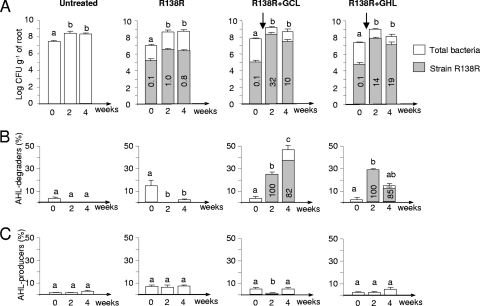
To monitor the introduced strain R138 among the native bacterial community, including native R. erythropolis population, a Rif-resistant mutant was selected and named R138R. The growth in minimal AB and rich TY media, AHL degradation, GCL and GHL assimilation, and biocontrol activities of R138 and R138R were similar. A selective medium based on minimal AB medium with GCL (2 g liter−1) as a sole carbon source and supplemented with Rif100 was used to reisolate the introduced strain R138R from hydroponic rhizospheres. In control experiments, no bacteria were recovered from the rhizospheres of S. tuberosum using this defined GCL-Rif AB medium, confirming the selectivity of this medium for R138R.
A novel campaign in the farm greenhouses allowed comparison of four plant growth conditions: an untreated control, the inoculation of strain R138R alone, and a combined treatment that included strain R138R and GCL or strain R138R and GHL at 0.4 g liter−1. Strain R138R was inoculated at 2 × 105 CFU g of roots−1, therefore representing 0.1% of total culturable bacteria. In the absence of GCL and GHL supplementation, R138R was still present 4 weeks postinoculation in the rhizosphere, but at a low abundance, representing 0.1 to 1% of the total culturable bacteria (Fig. 6A). Consequently, the percentage of AHL degraders did not significantly increase after a unique inoculation of the biological agent (Fig. 6B). In contrast, the combination of biocontrol strain R138R and biochemical agent GHL (or GCL) stimulated growth of the introduced Rhodococcus strain; cell density reached 32% of total culturable bacteria (Fig. 6A). Concomitantly, with this dual treatment, a significant increase in AHL degraders was observed (Fig. 6B). The introduced strain represented 80 to 100% of the isolated AHL degraders, suggesting that R138R displaced native AHL degraders, including the native R. erythropolis population. However, in the course of this campaign, the percentage of AHL producers remained low and varied from 3 to 7% of total cultivated bacteria (Fig. 6C).
Fig 6.
Biostimulation of the introduced R. erythropolis strain by GCL and GHL. Four conditions were compared: no treatment (control), inoculation of strain R138R only, and inoculation of strain R138R with GCL and GHL treatment (0.4 g liter−1). For each condition, the number of total bacteria (CFU g [fresh weight] of roots−1) (A) as well as the percentage of AHL degraders (B) and AHL producers (C) was estimated. Gray columns indicate the percentage of strain R138R among total cultivable bacteria. Different letters indicate a significant difference (Mann-Whitney; α = 0.05) over time under the same condition. Vertical arrows indicate the introduction of strain R138R and GCL or GHL.
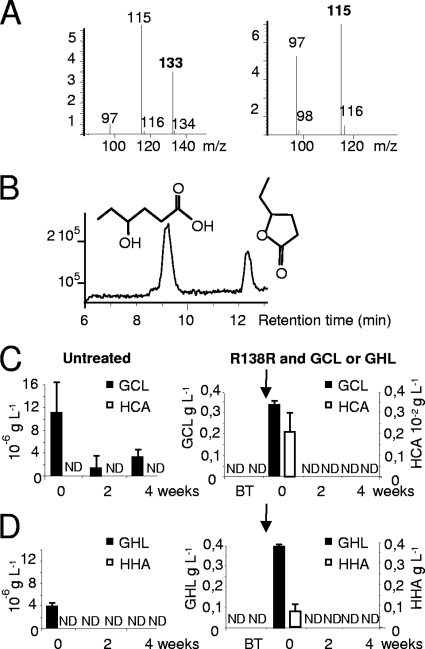
Novel HPLC-MS conditions were defined to identify, quantify, and monitor concentrations of GCL and of its by-product, 4-hydroxy-caproic acid (HCA) (Fig. 7A and B). In the control samples (nutritive solution and plant tissues), the concentrations of GCL and GHL were very low, and levels of their respective by-products, HCA and HHA, remained below the detection limits (Fig. 7C and D). In samples treated with both R138R and GCL or GHL, a rapid decrease in levels of the two lactones was observed in nutrient solution and plant tissues, and the concentrations of their lactone-opened derivatives, HCA and HHA, were very low or below the detection limit (Fig. 7C and D).
Fig 7.
Quantification of GCL, HCA, GHL, and HHA after combined treatments with R. erythropolis. (A and B) Pure GCL (M+H m/z of 115) and HCA (M+H m/z of 133) were released after retention times of 11.8 and 8.6 min (B), respectively, and used for measuring a calibration curve; the intensity of the signal is expressed in arbitrary units. (C and D) Concentrations of GCL, HCA, GHL, and HHA were measured in nutritive solution from untreated batches and those inoculated with strain R138R and treated with GCL (C) and GHL (D) at 0.4 g liter−1. Vertical arrows indicate introduction of strain R138R and GCL or GHL. ND, not detected.
Structure of the bacterial community upon biological treatment with R. erythropolis.
Pyrosequencing of amplified rrs was used for comparing bacterial community recovered after biological treatment with R. erythropolis, alone and combined with GHL treatment, with respect to that detected under untreated condition. Sampling was performed 2 weeks after treatment. Procedures identical to those described above were applied for the analysis of pyrosequencing data. Two replicates were performed and analyzed separately and after merging (see Table S2 in the supplemental material). Chloroplast sequences were removed from the initial stock of 89,143 sequences, and the CHAO1 and Shannon index values of the remaining 48,934 bacterial rrs sequences were determined (see Table S2 in the supplemental material) and compared (Fig. 8).
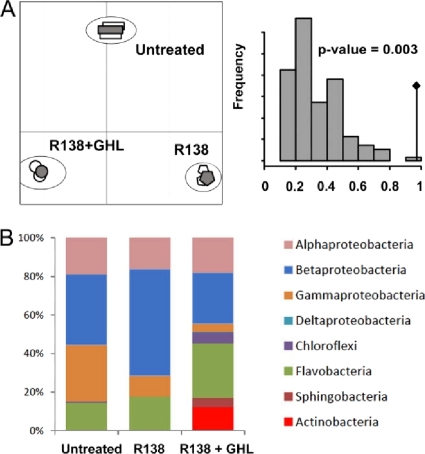
Fig 8.
Bacterial diversity revealed by rrs pyrosequencing under bioaugmentation and its combination with GHL. (A) (Left) Correspondence analysis of the two replicates (empty symbols) and merged data (filled symbols) collected 2 weeks after the bioaugmentation with strain R138R alone, treatment with R138R and GHL, and no treatment. (Right) Permutation test (n = 10,000) supporting correspondence analysis. The observed value is indicated by a black diamond. (B) Distribution of the main bacterial classes, comparing the three sets of conditions.
Correspondence analysis on genus-associated sequences showed that the two replicates and merged data clustered for each of the analyzed conditions (Fig. 8A). Consequently, only merged data were used thereafter for a deeper comparison of the three plant growth conditions. The CHAO1 values were identical for untreated samples and R138R-treated samples and were higher for samples from the combined treatment (see Table S2 in the supplemental material). Notably, the Shannon index was similar under all conditions (3.10 to 3.22).
Alphaproteobacteria, Betaproteobacteria, Flavobacteria, and to a lesser extent Gammaproteobacteria were the major bacterial classes in all samples (Fig. 8B). Actinobacteria (12.3%) clearly emerged under combined conditions. Remarkably, Rhodococcus represented 9.8% of the identified rrs sequences after a combined treatment and reached the third position among the most abundant genera (Table 2). The ratio of the abundance of Rhodococcus with biological treatment (strain R138R) to that without treatment (B/U = 2.7) and the ratio of the abundance with combined biological and biochemical treatments (R138R and GHL) to that under untreated conditions (C/U = 140) were calculated. These ratios revealed that growth of Rhodococcus members was strongly stimulated in the potato rhizosphere by the combined treatment.
Table 2.
rrs pyrosequencing data collected 2 weeks after treatments showing the major genera obtained with bioaugmentation (R138R) with or without GHL
| Treatment and genusa | Abundance (%) |
|---|---|
| None | |
| Acidovorax | 25.7 |
| Pseudomonas | 21.6 |
| Flavobacterium | 13.2 |
| Rhizobium | 6.1 |
| Aeromonas | 4.7 |
| Shinella | 3.9 |
| Rhodoferax | 2.5 |
| Devosia | 1.4 |
| Pseudoxanthomonas | 0.9 |
| Sphingobium | 0.8 |
| Roseateles | 0.5 |
| Bosea | 0.5 |
| Methylophilus | 0.4 |
| Herpetosiphon | 0.4 |
| Methyloversatilis | 0.4 |
| Rhodococcus | 0.07 |
| Strain R138R | |
| Hydrogenophaga | 27.5 |
| Flavobacterium | 15.1 |
| Acidovorax | 11.6 |
| Pseudomonas | 3.8 |
| Pseudoxanthomonas | 3.7 |
| Pelomonas | 3.5 |
| Devosia | 2.6 |
| Sphingobium | 2.5 |
| Methyloversatilis | 2.1 |
| Aeromonas | 1.9 |
| Methylophilus | 1.1 |
| Rhizobium | 1.0 |
| Roseateles | 0.8 |
| Shinella | 0.8 |
| Rhodoferax | 0.7 |
| Rhodococcus | 0.19 |
| Strain R138R and GHL | |
| Flavobacterium | 24.7 |
| Acidovorax | 13.8 |
| Rhodococcus | 9.8 |
| Herpetosiphon | 5.5 |
| Delftia | 3.0 |
| Shinella | 3.0 |
| Sphingobium | 2.8 |
| Novosphingobium | 2.2 |
| Rhizobium | 1.8 |
| Hydrogenophaga | 1.7 |
| Pseudomonas | 1.7 |
| Emticicia | 1.6 |
| Pseudoxanthomonas | 1.3 |
| Devosia | 1.2 |
| Ferruginibacter | 1.0 |
Bold type is used to highlight the change in position for Rhodococcus in the three treatments.
The identified genera were classified into three groups according to variations in their frequency in untreated samples, samples with the biocontrol strain alone, and samples with the combined treatment (see Fig. S2 in the supplemental material): group I includes genera such as Rhodococcus and Herpetosiphon (C/U = 13.7), whose frequency increased with the combined treatment; in group II were those such as Flavobacterium (B/U = 1.1 and C/U = 1.9) and Acidovorax (B/U = C/U = 0.5), among which only weak variations were observed when the treatments were compared; and group III consisted of a single genus, Lacibacter, which exhibited a strong decrease under treatment conditions compared to no treatment. Among all identified genera, Rhodococcus was the genus whose abundance increased the most with the combined treatment.
Relative abundance of the qsdA gene of R. erythropolis.
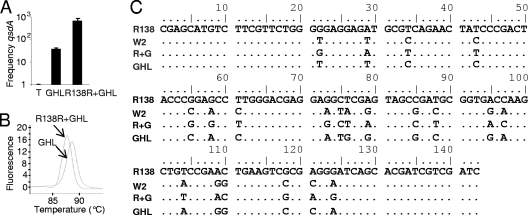
The relative abundance of the qsdA gene, which encodes a QQ lactonase in R. erythropolis, was measured by qPCR. Three contrasting conditions, the same as those analyzed by rrs pyrosequencing, were compared: (i) untreated rhizosphere, in which qsdA levels were assigned a reference level of 1; (ii) rhizosphere receiving GHL biostimulation, in which qsdA reached a relative level of 35; and (iii) rhizosphere receiving treatment with GHL plus R138, in which qsdA reached a relative level of 670 (Fig. 9A). In the same samples, rrs pyrosequencing revealed that the relative abundance of the R. erythropolis population reached a relative level of 31 with GHL treatment and 146 with GHL-R138R treatment, where the values are normalized to the no-treatment value (set at 1).
Fig 9.
Relative level of the qsdA gene of R. erythropolis. (A) The relative abundance of the qsdA gene after biostimulation with GHL and its combination with strain R138R was estimated by qPCR using the no-treatment control as a reference (arbitrarily set at 1). (B) Melting curve of the amplified qsdA fragments obtained from GHL-treated and GHL-R138R-treated samples. (C) Sequence alignment of the amplified fragments with the qsdA gene of R. erythropolis strains R138 and W2.
The melting peaks of qPCR products obtained from samples undergoing biostimulation with native and introduced R. erythropolis differed (Fig. 9B), suggesting that the sequences of the PCR fragments may not be strictly identical. The nucleotide sequences of PCR products were determined, found to be homogeneous in each of samples, and then compared with those of qsdA in R. erythropolis strains R138 and W2 (Fig. 9C). The qsdA fragment amplified from samples receiving the R138R-GHL treatment was identical to that of strain R138, while that from samples receiving GHL biostimulation of the native R. erythropolis population differed from all others. Consequently, sequencing of the qsdA gene allowed us to distinguish native and introduced R. erythropolis populations. The data also suggest that, with combined treatment, the introduced R. erythropolis strain displaced the endogenous R. erythropolis population.
DISCUSSION
Over a 4-year period of assays, GCL (6) and GHL (this work) have been found to successfully stimulate growth of the native AHL-degrading bacteria, mainly R. erythropolis, in hydroponic cultures of S. tuberosum in greenhouses, which are used by farmers for early propagation of tubers used for breeding. In this work, structural analysis of biostimulating molecules revealed the importance of the lactone ring (gamma-lactone versus delta- and epsilon-lactone) and that of the length of the lateral chain, which should be shorter than 4 carbons to efficiently stimulate AHL-degrading bacteria. Biostimulation activity of GHL was measured under plant growth conditions using two molecular approaches: quantification of qsdA (encoding the AHL lactonase) and quantification of Rhodococcus rrs sequences by pyrosequencing. The calculated B/U ratios measured the variation of the abundance of the Rhodococcus rrs population and that of the qsdA gene in GHL biostimulated rhizosphere samples versus untreated controls. Using rrs pyrosequencing and qsdA qPCR, the B/U ratios reached 30 and 50, respectively. When B/U ratios were calculated using cultivation-based data (i.e., percentage of AHL-degrading bacteria among total TSA-isolated bacteria in GHL-treated and untreated samples), the values ranged between 10 and 20 in the different experiments. The B/U values measured by different approaches (cultivation-based method, qPCR, and rrs pyrosequencing) are on the same order of magnitude, supporting the effectiveness of GHL treatment for enhancing the AHL-degrading R. erythropolis population.
One of the major results of this study is the proposition of a combined treatment which associates GCL or GHL with a selected R. erythropolis isolate. The chosen bacterial strain, R138, recovered from a GCL-treated potato rhizosphere, assimilates GCL and GHL, inactivates QS signals, and exhibits biocontrol activity in a potato tuber assay. Although the R. erythropolis isolate was introduced at a low level in the reported experiments (105 CFU g [FW] of roots−1, i.e., 0.1% of the total TSA-cultured bacterial community), extensive biostimulation of Rhodococcus populations was observed, with calculated C/U ratios (combined treatment versus no treatment) of >100: 1.4 × 102 and 7 × 102 using rrs pyrosequencing and qsdA qPCR, respectively. The effectiveness ratio reached 3 × 102 when GHL biostimulation and no biostimulation were compared by viable-colony (CFU) counting. The combined-treatment strategy strongly improved root colonization by the chosen R. erythropolis strain, compared to biological (strain R138) and biochemical (GHL) treatments alone. However, both CFU counting and identification of the amplified qsdA gene by DNA sequencing showed that the introduced R. erythropolis isolate displaced the native R. erythropolis population. This observation supports the efficient competitiveness of the introduced biocontrol R. erythropolis strain.
This work also provides deep exploration of the bacterial diversity in the potato rhizosphere by rrs pyrosequencing, as well as its dynamics under bioaugmentation, biostimulation, and a combination of both. All genera which were previously identified, mainly by cultivation-based methods, as major components in the potato rhizosphere community (11), including Flavobacterium, Pseudomonas, and Rhodococcus, were also identified by rrs pyrosequencing. To our knowledge, the bacterial community of the hydroponic potato rhizosphere had not been analyzed previously by rrs pyrosequencing. One interesting part of this work lies in the evaluation of the relative abundance of plant-associated genera under different environmental constraints. When R. erythropolis R138 was introduced alone into the potato rhizosphere, a weak impact on the bacterial community was observed. In contrast, GHL biostimulation, and especially its combination with the introduction of R. erythropolis R138, strongly biased the bacterial community by promoting the growth of Rhodococcus but also that of other genera, such as Herpetosiphon (Chloroflexi) and Delftia (Betaproteobacteria). Remarkably, Delftia isolates recovered from the GHL-treated potato rhizosphere, as well as from soil (20), were identified as AHL-degrading bacteria. Rhodococcus was the main, but not unique, AHL-degrading population whose growth was promoted by GCL or GHL treatment. We are currently investigating the enzymatic pathways involved in assimilation of GCL and GHL in R. erythropolis R138, as it is reasonable to speculate that this trait may be related to its capability to outcompete the other bacteria in rhizosphere colonization under biostimulation.
GCL (CAS 695-06-7) and GHL (CAS 105-21-5) are natural plant molecules (15, 41) whose functions are not known. GCL and GHL are both registered as flavor compounds (FL-10.021 and FL-10.020, respectively) by the European Food Safety Authority. Because this work suggests that these two molecules may be used in agronomy, we evaluated the fate of the introduced GHL and GCL, as well as their by-products HHA and HCA. None of them accumulated in plant tissues. However, a remarkable effect of GHL treatment was that it allowed the density of Rhodococcus cells to remain sustainably high over a 4-week period, although GHL and its by-product HHA were undetectable in the hydroponic fluid at 2 and 4 weeks. These experiments suggest that GHL promotes the installation of populations, including that of R. erythropolis, which are competitive enough to survive in the rhizosphere at high density, even when GHL was completely degraded. R. erythropolis is able to assimilate GCL and GHL. We hypothesize that the AHL lactonase QsdA (34) could also be involved in the opening of the ring of GHL and GCL, but we cannot exclude the possibility that other Rhodococcus enzymes or other microbial populations are involved in this degradation.
R. erythropolis isolates are also able to degrade a large spectrum of compounds, including pyrazines, atrazine, and polychlorinated biphenyl (21, 30, 35), phenol and catechol (37), and the n-alkanes hydrocarbons and oils (10, 22). They are thus proposed for bioremediation of contaminated soils and waters. The GHL biostimulation, and its combination with the addition of appropriate R. erythropolis strains, may therefore also be considered as a way to maintain a high density of a rhodococcal population and hence a high bioremediation activity in the environment. Reciprocally, plant protection strategies could benefit from investigations of the use of R. erythropolis as a bioremediation agent, especially from reports describing safety evaluation and encapsulated formulations of R. erythropolis (1, 35, 36). Finally, we are aware that the introduction of an AHL-degrading agent in the rhizosphere may potentially alter the efficiency of other biocontrol agents, in which beneficial traits are controlled by QS (26). The appropriate use of QQ microbes versus that of QS-dependent beneficial microbes should therefore be carefully evaluated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Dessaux (CNRS) for critical reading of the manuscript.
A. Cirou received a Ph.D. grant from Comité Nord Plants de Pommes de terre (CNPPT) and Association Nationale de la Recherche et de la Technologie (CIFRE 2007/0117); A. Charrier and A. Sarrazin were undergraduate students of Université Diderot-Paris7; D. Faure, M. DuBow, O. Thoison, and S. Mondy were supported by Centre National de la Recherche Scientifique (CNRS); M. DuBow was supported by the AQUAPHAGE program of Agence Nationale de la Recherche (ANR); D. Faure was supported by Région Ile-de-France (R2DS), Programme National d'Ecotoxicologie (PNETOX), and Programme Ecosphère Continentale et Côtière (EC2CO).
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aoshima H, et al. 2007. Safety evaluation of a heavy oil-degrading bacterium, Rhodococcus erythropolis C2. J. Toxicol. Sci. 32:69–78 [DOI] [PubMed] [Google Scholar]
- 2. Binladen J, et al. 2007. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One 2:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carlier A, et al. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant Microbe. Interact. 11:1119–1129 [DOI] [PubMed] [Google Scholar]
- 5. Chilton MD, et al. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. U. S. A. 71:3672–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cirou A, Diallo S, Kurt C, Latour X, Faure D. 2007. Growth promotion of quorum-quenching bacteria in the rhizosphere of Solanum tuberosum. Environ. Microbiol. 9:1511–1522 [DOI] [PubMed] [Google Scholar]
- 7. Cirou A, et al. 2011. Gamma-caprolactone stimulates growth of quorum-quenching Rhodococcus populations in a large-scale hydroponic system for culturing Solanum tuberosum. Res. Microbiol. 162:945–950 doi:10.1016/j.resmic.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 8. d'Angelo-Picard C, et al. 2004. Dynamics of bacterial populations in the rhizosphere of tobacco plants producing—or not—the quorum sensing signals hexanoyl- and 3-oxo-hexanoyl-homoserine lactone. FEMS Microbiol. Ecol. 51:19–29 [DOI] [PubMed] [Google Scholar]
- 9. D'Angelo-Picard C, Faure D, Penot I, Dessaux Y. 2005. Diversity of N-acylhomoserine lactone-producing anddegrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 7:1796–1808 [DOI] [PubMed] [Google Scholar]
- 10. de Carvalho CC, Wick LY, Heipieper HJ. 2009. Cell wall adaptations of planktonic and biofilm Rhodococcus erythropolis cells to growth on C5 to C16 n-alkane hydrocarbons. Appl. Microbiol. Biotechnol. 82:311–320 [DOI] [PubMed] [Google Scholar]
- 11. Diallo S, et al. 2011. Mechanisms and recent advances in biological control mediated through the potato rhizosphere. FEMS Microbiol. Ecol. 75:351–364 [DOI] [PubMed] [Google Scholar]
- 12. Dong YH, et al. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 14:813–817 [DOI] [PubMed] [Google Scholar]
- 13. Dong YH, Zhang XF, Xu JL, Zhang LH. 2004. Insecticidal Bacillus thuringiensis silences Erwinia carotovora virulence by a new form of microbial antagonism, signal interference. Appl. Environ. Microbiol. 70:954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faure D, Dessaux Y. 2007. Quorum sensing as a target for developing biocontrol strategies towards the plant pathogen Pectobacterium. European J. Plant Pathology 119:353–365 [Google Scholar]
- 15. Franco M, Peinado RA, Medina M, Moreno J. 2004. Off-vine grape drying effect on volatile compounds and aromatic series in must from Pedro Ximénez grape variety. J. Agric. Food Chem. 52:3905–3910 [DOI] [PubMed] [Google Scholar]
- 16. Fray RG. 2002. Altering plant-microbe interaction through artificially manipulating bacterial quorum sensing. Ann. Bot. 89:245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuqua WC, Winans SC, Greenber EP. 1994. Quorum sensing in bacteria: the LuxR/LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gray KM, Garey JR. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379–2387 [DOI] [PubMed] [Google Scholar]
- 19. Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jafra S, et al. 2006. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can. J. Microbiol. 52:1006–1015 [DOI] [PubMed] [Google Scholar]
- 21. Leigh MB, et al. 2006. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl. Env. Microbiol. 72:2331–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li GQ, et al. 2008. Improved biodesulfurization of hydrodesulfurized diesel oil using Rhodococcus erythropolis and Gordonia sp. Biotechnol. Lett. 30:1759–1764 [DOI] [PubMed] [Google Scholar]
- 23. Liu H, et al. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manefield M, Welch M, Givskov M, Salmond GP, Kjelleberg S. 2001. Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora. FEMS Microbiol. Lett. 27:131–138 [DOI] [PubMed] [Google Scholar]
- 25. McClean KH, et al. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711 [DOI] [PubMed] [Google Scholar]
- 26. Molina L, et al. 2003. Degradation of pathogen quorum-sensing molecules by soil bacteria: a preventive and curative biological control mechanism. FEMS Microbiol. Ecol. 45:71–81. [DOI] [PubMed] [Google Scholar]
- 27. Palmer AG, Streng E, Blackwell HE. 2011. Attenuation of virulence in pathogenic bacteria using synthetic quorum-sensing modulators under native conditions on plant hosts. ACS Chem. Biol. [Epub ahead of print] doi:10.1021/cb200298g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park SY, et al. 2006. N-acylhomoserine lactonase producing Rhodococcus spp. with different AHL-degrading activities. FEMS Microbiol. Lett. 261:102–108 [DOI] [PubMed] [Google Scholar]
- 29. Pirhonen M, Flego D, Heikinheimo R, Palva ET. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rappert S, et al. 2007. Degradation of 2,5-dimethylpyrazine by Rhodococcus erythropolis strain DP-45 isolated from a waste gas treatment plant of a fishmeal processing company. Biodegradation 18:585–596 [DOI] [PubMed] [Google Scholar]
- 31. Smadja B, et al. 2004. Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum). Mol. Plant Microbe Interact. 17:1269–1278 [DOI] [PubMed] [Google Scholar]
- 32. Uroz S, et al. 2003. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 149:1981–1989 [DOI] [PubMed] [Google Scholar]
- 33. Uroz S, et al. 2005. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology 151:3313–3322 [DOI] [PubMed] [Google Scholar]
- 34. Uroz S, et al. 2008. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 74:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vancov T, Jury K, Van Zwieten L. 2005. Atrazine degradation by encapsulated Rhodococcus erythropolis NI86/21. J. Appl. Microbiol. 99:767–775 [DOI] [PubMed] [Google Scholar]
- 36. Vancov T, Jury K, Rice N, Van Zwieten LL, Morris S. 2007. Enhancing cell survival of atrazine degrading Rhodococcus erythropolis NI86/21 cells encapsulated in alginate beads. J. Appl. Microbiol. 102:212–220 [DOI] [PubMed] [Google Scholar]
- 37. Vesely M, Knoppová M, Nesvera J, Pátek M. 2007. Analysis of catRABC operon for catechol degradation from phenol-degrading Rhodococcus erythropolis. Appl. Microbiol. Biotechnol. 76:159–168 [DOI] [PubMed] [Google Scholar]
- 38. Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GPC. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365–404 [DOI] [PubMed] [Google Scholar]
- 39. Yang YH, et al. 2006. High-throughput detection method of quorum-sensing molecules by colorimetry and its applications. Anal. Biochem. 356:297–299 [DOI] [PubMed] [Google Scholar]
- 40. Zhang LH. 2003. Quorum quenching and proactive host defense. Trends Plant Sci. 8:238–244 [DOI] [PubMed] [Google Scholar]
- 41. Zhang B, et al. 2010. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J. Agric. Food Chem. 58:6157–6165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.