Abstract
Ammonia-oxidizing bacteria (AOB) and archaea (AOA) are important for nitrogen cycling in marine ecosystems. Little is known about the diversity and abundance of these organisms on the surface of marine macroalgae, despite the algae's potential importance to create surfaces and local oxygen-rich environments supporting ammonia oxidation at depths with low dissolved oxygen levels. We determined the abundance and composition of the epiphytic bacterial and archaeal ammonia-oxidizing communities on three species of macroalgae, Osmundaria volubilis, Phyllophora crispa, and Laminaria rodriguezii, from the Balearic Islands (western Mediterranean Sea). Quantitative PCR of bacterial and archaeal 16S rRNA and amoA genes was performed. In contrast to what has been shown for most other marine environments, the macroalgae's surfaces were dominated by bacterial amoA genes rather than those from the archaeal counterpart. On the basis of the sequences retrieved from AOB and AOA amoA gene clone libraries from each algal species, the bacterial ammonia-oxidizing communities were related to Nitrosospira spp. and to Nitrosomonas europaea and only 6 out of 15 operational taxonomic units (OTUs) were specific for the host species. Conversely, the AOA diversity was higher (43 OTUs) and algal species specific, with 17 OTUs specific for L. rodriguezii, 3 for O. volubilis, and 9 for P. crispa. Altogether, the results suggest that marine macroalgae may exert an ecological niche for AOB in marine environments, potentially through specific microbe-host interactions.
INTRODUCTION
Macroalgae and other marine eukaryotes, such as corals and sponges, harbor complex epiphytic microbial communities (39). These communities are distinct from the planktonic ones, indicating that macroalgae are specific and unique habitats for microorganisms (7, 25, 26). The composition and diversity of bacterial communities in macroalga-related biofilms have been surveyed (7, 15, 25, 26, 47, 53) and have in some cases been shown to depend on the specific attraction that algal exudates may exert on certain bacteria but also on the antimicrobial effect of some compounds produced by the algae (27). Macroalgal epiphytic bacterial communities have been reported to be host specific (25, 29), and their composition can even differ between the algal rhizoid, cauloid, meristem, and phyloid, indicating a close host-bacterium interaction (47). However, although the bacterial community composition of the green alga Ulva australis was unique compared to that of the planktonic community, there was a high variability among individuals (7), which suggests that algal surface colonization is driven by selection processes in combination with stochastic recruitment of bacteria from the planktonic community.
Algal surfaces are important microbial habitats in the marine environments, but most experimental studies have focused on bacterial communities and no attention has been paid to archaea as members of the epiphytic communities. Conversely, archaea have been widely studied in marine environments, especially after the discovery of the ammonia-oxidizing archaea (AOA). The AOA are key players in nitrogen cycling in marine systems by performing the first step of nitrification, a process that the ammonia-oxidizing bacteria (AOB) also carry out (13, 56). AOB and AOA belong to the phyla proteobacteria and thaumarchaeota, respectively (6, 24). Phylogenetic studies on ammonia oxidizers are mainly based on the analysis of the amoA gene, coding for the α subunit of the ammonia monooxygenase (20, 44). The ammonia oxidizers are present in marine environments either as planktonic populations (21, 34) or on host-associated marine microbial communities of corals (3) and sponges (35, 48). In all examples of marine environments, the AOA outnumber the AOB. For algal species, the ammonia oxidizers have not been described. However, we hypothesize that macroalgae may offer a selective habitat for ammonia oxidizers and hot spots for nitrification since algae produce oxygen, which is needed for ammonia oxidation. This may be important, especially at depths with low dissolved oxygen levels. However, epiphytic communities in general, let alone the AOA and AOB within these communities, are basically unknown at mesophotic depths (30 to 200 m), since all the studies have focused on macroalgae inhabiting shallower depths (25, 26, 47, 53).
To address the importance of macroalgae as habitats for AOA and AOB in the mesophotic zone, we determined their composition and abundance on the surface of three dominant seascape-building macroalgae in the northwestern Mediterranean Sea. Host specificity was analyzed by sequencing bacterial and archaeal amoA genes from the surface of Osmundaria volubilis, Phyllophora crispa, and Laminaria rodriguezii.
MATERIALS AND METHODS
Site description and sampling.
Samples were collected from the MEDITS_ES05 2008 bottom trawl survey (4), conducted in June 2008 along the shelf and slope (50 to 100 m depth) of Majorca and Minorca Islands, on board the R/V Cornide de Saavedra (Fig. 1). Estimation of the physical and chemical parameters of water masses around the Balearic Islands is regularly performed by the Spanish Institute of Oceanography following the European Framework Directive (5, 31) with the monitoring system RADMED (54). Water around the Balearic Islands has an Atlantic origin, with homoeothermic winter and summer stratification (13 to 26°C). Surface salinity values vary from 37.5 to 38.2 practical salinity units (psu). These waters are oligotrophic, with low nitrate concentrations (0.75 μM), and show maximum fluorescence values of 1.5 mg/m3 in spring and summer at 60 to 80 m depth. Fluorescence is generated by photosynthetic pigments, organic matter, and suspended particles. The maximum dissolved oxygen concentration in the mixed water mass is reached in winter, with about 6.5 ml/liter. During summer, these values are maintained at the thermocline and decrease down to 4.5 ml/liter at 200 m depth. The water masses in the area are slightly alkaline, with the pH ranging from 8.1 to 8.3.
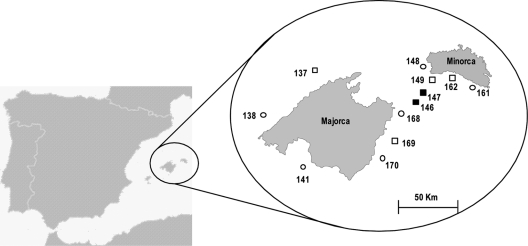
Fig 1.
Geographic situation of sampling locations in the Balearic Islands. Seascapes dominated by Laminaria rodriguezii are shown with closed symbols. Seascapes dominated by Osmundaria-Phyllophora communities are shown with open symbols. Different shapes indicate the depth intervals for each sample: ○, 50 to 64 m; □, 65 to 81 m.
The sampling scheme and gear were the same as those used throughout the northern Mediterranean coast in the international MEDITS surveys (4). This sampling strategy is appropriate for screening of algae at depths that cannot be reached by scuba diving (19). A total of 12 samplings were obtained, resulting in 26 samples of algal thalli from the red algae Osmundaria volubilis (Linnaeus) R. E. Norris and Phyllophora crispa (Hudson) P. S. Dixon and the brown alga Laminaria rodriguezii Bornet, which are important seascape builders in this area (1, 19). When possible, we separated three replicates with intact phyloids of similar size for each species and sampling point. The specimens were gently cleaned in seawater and stored at −20°C until further analysis.
DNA extraction and quantification.
Clean phyloids of several individuals were cut in pieces of approximately 2 cm2 and distributed in three subsamples of 1 g of fresh material. For biofilm detachment from the algal surfaces, the cut algae were immersed in 2 ml of 0.1 M sodium pyrophosphate (Na4P2O7 · 10H2O). Biofilm was dislodged by two rounds of sonication for 20 s at maximum intensity (Selecta, Barcelona, Spain), followed by 30 s on ice (23). Algal suspensions were centrifuged at 10,000 × g for 2 min, and supernatants were discarded. Cell pellets were resuspended in 0.6 ml of sodium phosphate buffer. Nucleic acids were extracted using a FastDNA Spin kit for soil (MP, Biomedicals) and stored at −20°C.
Quantitative PCR.
Selective quantification of AOA and AOB was done by targeting the amoA genes using the primer pairs amoA1F-amoA2R (44) and crenamoA23F-crenamoA616R (51), while the primers 341F (37) and 534R (30) and crenar771F and crenar957R (38) were used for quantification of the 16S rRNA genes of the domain bacteria and the archaeal phyla crenarchaeota and thaumarchaeota, respectively. Quantitative PCR (qPCR) was performed as described previously with minor modifications (17, 38). Quantifications were carried out in a Bio-Rad IQ5 thermal cycler (Bio-Rad, Richmond, CA), using a Dynamo Flash SYBR green qPCR kit (Finnzymes, Oy, Espoo, Finland). Twenty-microliter reaction mixtures contained 1× SYBR green master mix, 1 μg/μl bovine serum albumin (BSA), and 10 ng of DNA. Primer concentrations used were 1 μM for 16S rRNA and 0.5 μM for amoA. The standard curves were obtained from serial dilutions of linearized plasmids (pGEM-T Easy; Promega, Madison, WI) containing standard sequences. The PCR efficiency ranged between 80 and 100%. Negative controls resulted in undetectable values in all cases. To detect possible inhibitory effects, 105 copies of the pGEM-T Easy plasmid (Promega) were mixed with sample DNA and quantified with plasmid-specific primers T7 and SP6. The obtained cycle thresholds (CTs) were not significantly different from those obtained when quantifying the plasmid alone.
Statistical analysis of gene abundances.
Sampling locations were grouped into two different seascape categories according to the dominant algal species, in agreement with previous results that statistically tested the presence and dominance of different algal species around the islands (19). These categories corresponded to Laminaria-dominated assemblages in samplings 146 and 147 and Osmundaria-Phyllophora-dominated assemblages at the rest of the locations. Additionally, samples were grouped into two depth categories (above 65 m and below 65 m) (Fig. 1). Differences in gene abundances in relation to the depth, algal species, and algal seascape categories were calculated using the Kruskal-Wallis test for nonnormal distributions. Pairwise comparisons of the means of the 4 genes quantified were analyzed using a Mann-Whitney U test; and linear regressions, significance, and Spearman correlation coefficients among gene abundances were calculated. All the statistical analyses were performed with SPSS for Windows (version 15.0; SPSS, Inc.).
PCR, cloning, and sequencing of amoA genes.
All the DNA extractions obtained from the same algal species were pooled to minimize individual diversity and to determine the host-specific AOA and AOB amoA gene diversity. Clone libraries of archaeal and bacterial amoA genes were constructed for the three algal species. PCR amplifications were performed in a GenAmp PCR system 9700 (Applied Biosystems, Foster City, CA) using the same primers used for qPCR. In both cases, reaction mixtures contained 2.5 U of DreamTaq DNA polymerase (Fermentas, Hanover, MD), 1× PCR buffer, 0.2 μM each primer, 0.2 mM deoxynucleoside triphosphates, 0.1 μg/μl BSA (New England BioLabs, Beverly, MA), and 2.2 mM MgCl2 in a total volume of 50 μl. Thermal conditions used were 5 min at 94°C, 42 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C, and a final extension of 10 min at 72°C. PCR products were excised from an agarose gel, purified using an Ultraclean DNA purification kit (MoBio, Carlsbad, CA), and then cloned using the pGEM-T Easy vector and Escherichia coli JM109 competent cells (Promega). Clones were screened for correct insert size by PCR with primers T7 and SP6 and sequenced using T7 primer (Macrogen, Seoul, South Korea). In total, 411 sequences were recovered.
Sequences analysis and phylogeny.
Sequences were checked for chimeras and manually refined by using the BioEdit (version 7.0) package (16). Alignment of sequences and clustering (cutoff = 0.05) were done using the platform mothur (version 1.11.0) (46). Operational taxonomic units (OTUs) were defined at the same cutoff value and used to construct rarefaction curves, to estimate richness (Chao1), and to calculate diversity indices (Shannon) (9, 33). This procedure was repeated using single clone libraries for each algal species and for all the sequences retrieved from the three algal species to compare for shared OTUs between clone libraries.
Representative sequences for each OTU were identified as the sequences having the maximum average similarity to the other sequences in the same OTU and were used for a phylogenetic tree reconstruction by neighbor joining (NJ). Phylogenetic trees with 1,000 bootstrap replicates were obtained using the MEGA program (version 4.0) (50). Gene clusters were named according to Purkhold et al. (42) and Stopnišek et al. (49). To compare the AOA or AOB community diversity in the three algae, weighted and unweighted UniFrac analyses were performed (32).
Nucleotide sequence accession numbers.
Partial amoA gene sequences obtained from the AOB and AOA were deposited in GenBank under accession numbers JF715503 to JF715649 and JF715650 to JF715913, respectively.
RESULTS
Abundance of 16S rRNA and amoA genes.
For the bacterial 16S rRNA genes, abundances ranged from 5 × 108 to 7 × 1010 gene copies/g (dry weight [dw]) of algae, whereas for the thaumarchaeota and crenarchaeota, values were significantly lower (P = 0.005) and ranged from 1 × 105 to 3 × 106 (Table 1). Similarly to 16S rRNA genes, amoA abundances were significantly higher for bacteria than for archaea (P < 0.001), with 6 × 106 to 3 × 108 and 4 × 105 to 1 × 107 copies/g dw of algae, respectively (Table 1).
Table 1.
Sampling dates, depths, locations, seascape types, algal species, and abundance of bacterial and thaumarchaeal 16S rRNA genes and bacterial and archaeal amoA genes
| Sampling | Sampling date (day/mo/yr) | Depth (m) | Location | Seascapea | Species | No. of copies/g (dw) of algaeb |
|||
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA gene |
amoA gene |
||||||||
| Bacteria (109) | Archaeac (106) | AOB (107) | AOA (106) | ||||||
| 137 | 09/06/2008 | 72 | Ponent Mallorca (Sóller) | Osmundaria-Phyllophora | P. crispa | 1.70 ± 0.86 | 2.10 ± 0.52 | 1.83 ± 0.68 | 4.50 ± 2.27 |
| 138 | 09/06/2008 | 62 | Ponent Mallorca (Dragonera) | Osmundaria-Phyllophora | P. crispa | 1.84 ± 0.05 | 2.22 ± 0.56 | 2.61 ± 1.52 | 2.37 ± 0.68 |
| 141 | 10/06/2008 | 60 | Migjorn Mallorca (Badia) | Osmundaria-Phyllophora | P. crispa | 1.51 ± 0.17 | 1.55 ± 1.48 | 1.53 ± 1.54 | 1.68 ± 1.20 |
| O. volubilis | 0.57 ± 0.14 | 0.12 ± 0.12 | 1.07 ± 1.05 | 1.24 ± 1.22 | |||||
| L. rodriguezii | 1.27 ± 0.21 | 1.59 ± 1.46 | 1.92 ± 0.16 | 3.21 ± 1.44 | |||||
| 146 | 11/06/2008 | 81 | Canal Menorca | Laminaria | O. volubilis | 1.84 ± 0.92 | 2.61 ± 0.76 | 1.88 ± 0.90 | 12.70 ± 9.21 |
| L. rodriguezii | 0.42 ± 0.20 | 0.54 ± 0.03 | 0.64 ± 0.15 | 1.24 ± 0.67 | |||||
| 147 | 11/06/2008 | 77 | Canal Menorca | Laminaria | O. volubilis | 2.50 ± 1.29 | 2.85 ± 1.21 | 4.55 ± 1.71 | 5.20 ± 3.43 |
| L. rodriguezii | 69.20 ± 11.60 | 6.03 ± 2.17 | 30.50 ± 4.90 | 5.00 ± 3.84 | |||||
| 148 | 11/06/2008 | 57 | Migjorn Menorca (Ciutadella) | Osmundaria-Phyllophora | P. crispa | 3.60 ± 1.99 | 2.26 ± 3.22 | 3.03 ± 3.46 | 1.31 ± 0.64 |
| O. volubilis | 2.35 ± 1.20 | 2.11 ± 0.84 | 2.36 ± 1.02 | 3.72 ± 0.89 | |||||
| L. rodriguezii | 2.22 ± 1.72 | 1.18 ± 0.96 | 2.21 ± 1.61 | 4.25 ± 3.42 | |||||
| 149 | 11/06/2008 | 77 | Migjorn Menorca (Ciutadella) | Osmundaria-Phyllophora | P. crispa | 10.90 ± 2.48 | 2.77 ± 1.57 | 7.39 ± 2.59 | 2.21 ± 0.65 |
| O. volubilis | 37.70 ± 28.70 | 7.98 ± 6.10 | 17.10 ± 10.50 | 7.38 ± 6.80 | |||||
| L. rodriguezii | 44.70 ± 44.10 | 3.17 ± 3.12 | 17.60 ± 15.40 | 1.14 ± 0.99 | |||||
| 161 | 15/06/2008 | 58 | Migjorn Menorca (Maó) | Osmundaria-Phyllophora | P. crispa | 5.86 ± 4.07 | 4.36 ± 2.32 | 4.81 ± 2.48 | 6.57 ± 6.06 |
| O. volubilis | 1.69 ± 0.98 | 2.06 ± 1.67 | 2.23 ± 1.07 | 2.81 ± 2.07 | |||||
| 162 | 15/06/2008 | 65 | Migjorn Menorca (Maó) | Osmundaria-Phyllophora | P. crispa | 3.95 ± 1.66 | 3.55 ± 0.09 | 6.35 ± 2.65 | 3.17 ± 1.02 |
| O. volubilis | 4.54 ± 2.68 | 7.49 ± 3.98 | 6.72 ± 2.19 | 7.45 ± 3.85 | |||||
| 168 | 17/06/2008 | 57 | Llevant Mallorca (Capdepera) | Osmundaria-Phyllophora | P. crispa | 3.93 ± 2.59 | 3.17 ± 2.28 | 5.93 ± 4.14 | 1.28 ± 1.25 |
| O. volubilis | 4.35 ± 1.08 | 2.75 ± 0.80 | 5.32 ± 0.94 | 1.64 ± 0.58 | |||||
| 169 | 17/06/2008 | 75 | Llevant Mallorca (Portocolom) | Osmundaria-Phyllophora | P. crispa | 2.21 ± 0.76 | 2.11 ± 1.04 | 1.79 ± 0.83 | 4.77 ± 2.85 |
| O. volubilis | 3.41 ± 1.71 | 3.85 ± 3.37 | 5.18 ± 3.90 | 6.37 ± 5.05 | |||||
| L. rodriguezii | 2.04 (n = 1) | 1.19 (n = 1) | 2.42 (n = 1) | 1.63 (n = 1) | |||||
| 170 | 17/06/2008 | 52 | Llevant Mallorca (Portocolom) | Osmundaria-Phyllophora | P. crispa | 5.81 ± 5.13 | 3.81 ± 2.51 | 6.59 ± 4.83 | 1.95 ± 1.73 |
| O. volubilis | 6.43 ± 2.13 | 5.87 ± 3.47 | 9.50 ± 3.47 | 4.35 ± 3.28 | |||||
Two seascape types were found: one dominated by Osmundaria-Phyllophora and one dominated by Laminaria.
Means of three replicates and standard deviations are shown, unless stated otherwise.
Thaumarchaeota and crenarchaeota.
Algal seascapes were the only a priori-defined groups displaying significant differences of the abundances according to algal species. AOA amoA gene abundances were significantly lower on the surface of O. volubilis than on the surface of the other two algal species in the Laminaria seascapes (P = 0.005). Pairwise correlations between bacterial 16S rRNA and bacterial amoA gene abundances were highly significant regardless of the algae studied in the Osmundaria-Phyllophora seascapes (see Table S1 in the supplemental material), supporting the idea that the proportion of AOB in relation to total bacteria was stable (about 0.01). Significant correlations between 16 rRNA and amoA genes were also found, with a proportion of 1. In contrast, significant correlations between AOA and AOB amoA genes were found only for O. volubilis in Osmundaria-Phyllophora seascapes, suggesting a clear differentiation between the two groups of ammonia oxidizers (see Table S1 and Fig. S1 in the supplemental material).
Effects of algal species on ammonia-oxidizing community diversity and composition.
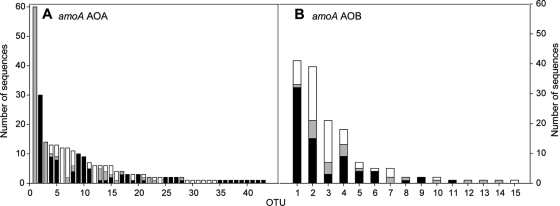
The AOA community was more diverse than the AOB counterpart (see the text and Fig. S2 in the supplemental material). Moreover, the AOA presented higher host specificity, with 29 out of 43 OTUs containing sequences from one algal epiphytic community (Fig. 2A). The three most abundant OTUs were specific for O. volubilis and L. rodriguezii. For bacterial amoA genes, host specificity was less pronounced. Out of the 15 OTUs, only 6 were specific, and all specific OTUs contained only 1 or 2 sequences (Fig. 2B). The observed differences at the OTU level were confirmed by the UniFrac analysis comparing the AOB and AOA communities (Table 2). The weighted analysis showed that the specific selection of the macroalgae was stronger on the AOA than the AOB. For the AOA, significant differences in all pairwise comparisons between algal species were found, but no significant differences for the bacterial communities associated with O. volubilis and P. crispa were detected (Table 2).
Fig 2.
Rank-abundance curves showing the number of OTUs obtained for archaeal (A) and bacterial (B) amoA sequences corresponding to three different algal species. OTUs were obtained using a cutoff distance of 5% for each algal species; gray bars, O. volubilis; white bars, P. crispa; black bars, L. rodriguezii.
Table 2.
UniFrac scores and P values when comparing bacterial and archaeal ammonia oxidizer communities in the three algal species O. volubilis, P. crispa, and L. rodriguezii
| UniFrac test | Groups compared | Score | Pa |
|---|---|---|---|
| Unweighted | Bacterial L. rodriguezii-O. volubilis-P. crispa | 0.54 | * |
| Archaeal L. rodriguezii-O. volubilis-P. crispa | 0.74 | *** | |
| Weighted | Bacterial P. crispa-L. rodriguezii | 1.52 | *** |
| Bacterial O. volubilis-P. crispa | 0.83 | NS | |
| Bacterial L. rodriguezii-O. volubilis | 2.14 | *** | |
| Archaeal P. crispa-L. rodriguezii | 1.72 | *** | |
| Archaeal O. volubilis-P. crispa | 2.00 | *** | |
| Archaeal L. rodriguezii-O. volubilis | 0.76 | *** |
NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
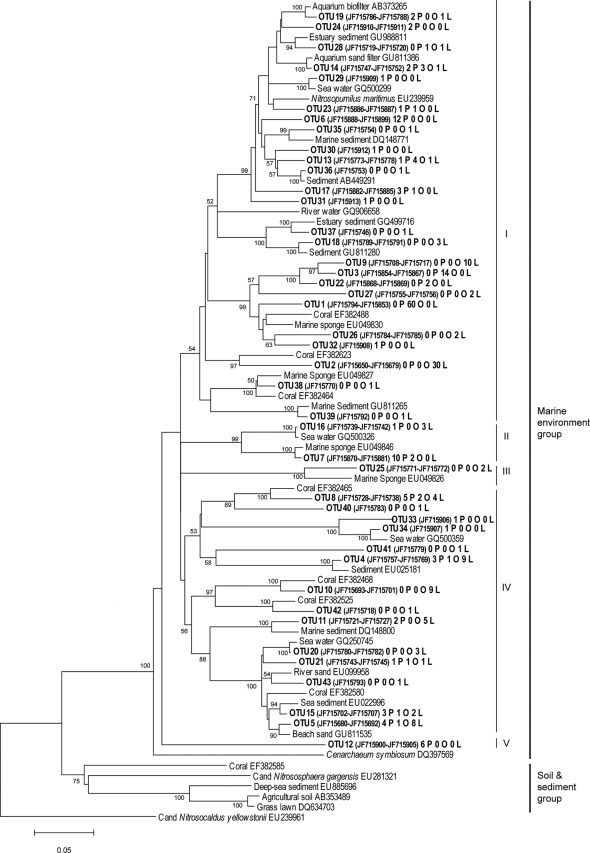
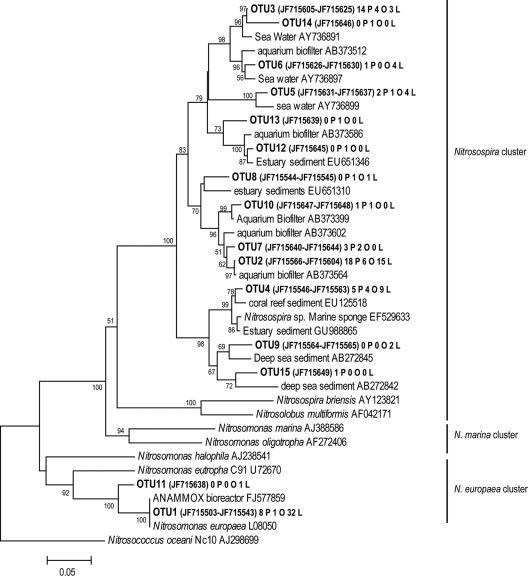
The archaeal amoA sequences showed from 82 to 99% similarity to previously published sequences, with a majority assigned to amoA genes from seawater, marine sediments, and symbionts of corals or marine sponges. All clones distributed in five clusters in the marine environment group 1.1a (41, 49) (Fig. 3). Cluster I contained sequences from 25 OTUs, most of them species specific. Clusters III and V contained sequences specific for L. rodriguezii and P. crispa, respectively, and clusters II and IV contained non-species-specific OTUs. The bacterial amoA sequences showed 93 to 100% similarity to members of the Nitrosomonas europaea and the Nitrosospira clusters (42) (Fig. 4). The majority of sequences belonged to the Nitrosospira cluster and distributed in 13 OTUs. These sequences were closely related to sequences found in seawater and sediments, aquarium filters, and coral reef sediments. The Nitrosospira cluster contained one specific OTU for P. crispa, three for O. volubilis, and one for L. rodriguezii. The Nitrosomonas europaea cluster contained sequences belonging to two OTUs, with one being specific for L. rodriguezii.
Fig 3.
Neighbor-joining phylogenetic tree of archaeal amoA sequences from epiphytic communities of three different algal species. Representative sequences of each OTU (cutoff distance, 5%) are shown in bold, followed by the GenBank accession numbers of all the sequences in the same OTU. Origins of the sequences in each OTU are indicated with letter codes (O, O. volubilis; P, P. crispa; and L, L. rodriguezii). Bootstrap values higher than 50% are indicated at branch points (1,000 replicates). Reference sequences from the GenBank database are added.
Fig 4.
Neighbor-joining phylogenetic tree of bacterial amoA sequences from epiphytic communities of three different algal species. Representative sequences of each OTU (cutoff distance, 5%) are shown in bold, followed by the GenBank accession numbers of all the sequences in the same OTU. Origins of the sequences in each OTU are indicated with letter codes (O, O. volubilis; P, P. crispa; and L, L. rodriguezii). Bootstrap values higher than 50% are indicated at branch points (1,000 replicates). Reference sequences from the GenBank database are added.
DISCUSSION
In all algal samples, AOB amoA genes were about 10 times more abundant than those from the AOA. This contrasts with the majority of marine environments, in which AOA dominate (11, 13), with a few exceptions (8, 36). The relative proportions of AOB and AOA differ significantly between compartments of the sea. For example, on marine sponges, the abundance of AOA assessed by fluorescent in situ hybridization measurements was only twice as high as that of AOB (3), whereas the relative proportion of AOA increased up to 90 to 99% in open ocean waters (55), and AOA completely outnumbered AOB in corals (3). An estimation of the cell number cannot be translated directly from gene abundances since differences in the number of copies of a gene in a genome may exist between phylotypes. However, a mean value of 2.5 amoA gene copies per genome has been proposed for the AOB, whereas the information available for AOA describes a single amoA gene copy per genome (18, 20, 22, 28). Using these estimations for our data, the cell number of the AOB was six times higher than that of the AOA, showing that surfaces of macroalgae favor the presence of AOB instead of AOA. If we compare the estimated number of total bacterial cells (average 16S rRNA gene copy number, 3.6 per cell) to the estimated number of AOB, the bacterial ammonia oxidizers comprise 1% of the total bacterial community. In comparison, only 0.1% of the total bacteria found on marine sponges were Nitrosospira spp. (2). Our results suggest that the three macroalgae studied here are important and selective habitats for AOB in the epiphytic biofilms. However, the reasons for this enrichment remain elusive. Possible explanations could be favorable physicochemical conditions over the algal surface or the fact that enrichment is a result of a combination of specific positive and negative epiphyte-host interactions. Further experiments are needed to determine the local environmental conditions that determine selection for AOB rather than AOA.
In general, the host organism had a greater influence than depth and geographical location on both AOB and AOA community composition. Interestingly, the UniFrac analyses revealed that both AOA and AOB communities specific for L. rodriguezii were significantly different from those specific for the other macroalgal species. L. rodriguezii is a brown seaweed that differs from the red algae O. volubilis and P. crispa in several ways. L. rodriguezii assemblages are found in areas with unidirectional currents and low water temperatures (12, 14) and are normally restricted to deeper areas, those below 60 m (40). At our sampling locations, O. volubilis and P. crispa formed codominant assemblages, and they are known to spread over rocky or detrital bottoms at depths of between 50 to 70 m (1, 19). Moreover, the cell wall composition and the main storage carbohydrates are different between the three algal species considered (10, 43). Availability of organic compounds for mixotrophic nitrification has recently been considered a selective factor for AOA (45, 52). Unfortunately, no information about exudates produced by these macroalgae exists, but the exudates produced could potentially be a key factor that explains the greater host specificity observed among the AOA than the AOB community members. In agreement with our study, others have shown that a core epiphytic bacterial community can be defined at the algal species level, indicating a specific selection for particular microbial epiphytes (25, 26, 53). However, whether this applies to other macroalgal species is not known. All our obtained archaeal amoA sequences grouping in the marine environment group (49, 51) were closely related to sequences retrieved from corals and sponges, indicating that high similarities exist among these epiphytic microbial communities. The AOB communities were mainly represented by Nitrosospira sp.-related sequences, and, in agreement, Nitrosospira spp. have also been described to be the most abundant AOB present in sponges (2, 35). Among our algal species, the differences in the AOA communities in particular indicate host specificity, but larger sample sets are needed to establish if a defined host-specific core community exists.
In summary, we have shown that algal species may exert a specific ecological niche for AOB in the marine environment, traditionally considered to be dominated by AOA. We calculated that AOB account for about 1% of the total bacterial community on the algal surfaces and can be up to 6 times more abundant than the AOA. Similar proportions were found for the three algal species dominating in the mesophotic zone in the western Mediterranean Sea (Balearic Islands). Despite their lower abundance, the AOA showed a higher host specificity, indicating a closer relationship between the algal species and the AOA compared to the AOB. Further studies should aim at deciphering potential interactions between algal species and ammonia-oxidizing bacteria and archaea and determining if the higher abundance of AOB on algal surfaces equals a hot spot for nitrification in marine environments.
Supplementary Material
ACKNOWLEDGMENTS
The work was financially and technically supported by the Spanish Ministerio de Ciencia e Innovación (project CGL2009-08338), the Swedish University of Agricultural Sciences, and the EVADEMED project, cofinanced by the Spanish Institute of Oceanography (IEO) and the European Union.
We also thank everyone who took part in the MEDITS ES05_08 survey and S. Joher, who helped with sampling. S. Ramió-Pujol and A. Vilar-Sanz are acknowledged for helping with DNA extractions.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ballesteros E. 1992. Els fons rocosos profunds amb Osmundaria volubilis (Linné) R. E. Norris a les Balears. Boll. Soc. Hist. Nat. Balears 35:35–50 [Google Scholar]
- 2. Bayer K, Schmitt S, Hentschel U. 2008. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ. Microbiol. 10:2942–2955 [DOI] [PubMed] [Google Scholar]
- 3. Beman JM, Roberts KJ, Wegley L, Rohwer F, Francis CA. 2007. Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl. Environ. Microbiol. 73:5642–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertrand JA, Gil de Sola L, Papaconstantinou C, Relini G, Souplet A. 2002. The general specifications of the MEDITS survey. Sci. Mar. 66:9–17 [Google Scholar]
- 5. Borja A. 2005. The European Water Framework Directive: a challenge for nearshore, coastal and continental shelf research. Cont. Shelf Res. 25:1768–1783 [Google Scholar]
- 6. Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 7. Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S. 2011. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5:590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660–662 [DOI] [PubMed] [Google Scholar]
- 9. Chao A. 1984. Non parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265–270 [Google Scholar]
- 10. Cole KM, Sheath RG. 1990. Biology of red algae. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 11. Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. 2009. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33:855–869 [DOI] [PubMed] [Google Scholar]
- 12. Feldmann J. 1934. Les Laminariaceés de la Méditerranée et leur répartition géographique. Bull. Trav. Stat. Aquic. Péche Castiglione Alg. 2:143–184 [Google Scholar]
- 13. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giaccone G. 1967. Popolamenti a Laminaria rodriguezii Bornet sul Banco Apollo dell'isola di Ustica (Mar Tirreno). Nova Thalassia 3:1–9 [Google Scholar]
- 15. Goecke F, Labes A, Wiese J, Imhoff J. 2010. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409:267–300. [Google Scholar]
- 16. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 17. Hallin S, Jones CM, Schloter M, Philippot L. 2009. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3:597–605 [DOI] [PubMed] [Google Scholar]
- 18. Herrmann M, Saunders AM, Schramm A. 2009. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl. Environ. Microbiol. 75:3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joher S, Ballesteros E, Cebrian E, Sánchez N, Rodríguez-Prieto C. 2010. Algal-dominated seascapes from the continental shelf off Mallorca and Menorca (Balearic Islands, western Mediterranean), p 61–65 Proc. 4th Mediterranean Symposium Mar. Veg. [Google Scholar]
- 20. Junier P, et al. 2010. Phylogenetic and functional marker genes to study ammonia-oxidizing microorganisms (AOM) in the environment. Appl. Microbiol. Biotechnol. 85:425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karner MB, DeLong EF, Karl DM. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510 [DOI] [PubMed] [Google Scholar]
- 22. Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knief C, Frances L, Cantet F, Vorholt JA. 2008. Cultivation-independent characterization of methylobacterium populations in the plant phyllosphere by automated ribosomal intergenic spacer analysis. Appl. Environ. Microbiol. 74:2218–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Könneke M, et al. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 25. Lachnit T, Blumel M, Imhoff JF, Wahl M. 2009. Specific epibacterial communities on macroalgae: phylogeny matters more than habitat. Aquat. Biol. 5:181–186 [Google Scholar]
- 26. Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. 2011. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13:655–665 [DOI] [PubMed] [Google Scholar]
- 27. Lachnit T, Wahl M, Harder T. 2010. Isolated thallus-associated compounds from the macroalga Fucus vesiculosus mediate bacterial surface colonization in the field similar to that on the natural alga. Biofouling 26:247–255 [DOI] [PubMed] [Google Scholar]
- 28. Leininger S, et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed] [Google Scholar]
- 29. Longford SR, et al. 2007. Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat. Microb. Ecol. 48:217–229 [Google Scholar]
- 30. Lopez-Gutierrez JC, et al. 2004. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57:399–407 [DOI] [PubMed] [Google Scholar]
- 31. López-Jurado JL, Jansà J, Aparicio A, Amengual B. 2009. Control de variables químico-biológicas en zonas costeras de las Islas Baleares. Instituto Español de Oceanografía, Ministerio de Ciencia e Innovación, Madrid, Spain [Google Scholar]
- 32. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mills AL, Wassel RA. 1980. Aspects of diversity measurement for microbial communities. Appl. Environ. Microbiol. 40:578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mincer TJ, et al. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162–1175 [DOI] [PubMed] [Google Scholar]
- 35. Mohamed NM, Saito K, Tal Y, Hill RT. 2010. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 4:38–48 [DOI] [PubMed] [Google Scholar]
- 36. Mosier AC, Francis CA. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002–3016 [DOI] [PubMed] [Google Scholar]
- 37. Muyzer G, Teske A, Wirsen CO, Jannasch HW. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165–172 [DOI] [PubMed] [Google Scholar]
- 38. Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787–797 [DOI] [PubMed] [Google Scholar]
- 39. Olson JB, Kellogg CA. 2010. Microbial ecology of corals, sponges, and algae in mesophotic coral environments. FEMS Microbiol. Ecol. 73:17–30 [DOI] [PubMed] [Google Scholar]
- 40. Posidonie UIG. 1990. Livre rouge “Gerard Vuignier” des végétaux, peuplements et paysages marins menacés de Méditerranée. MAP technical reports series no. 43. UNEP, Athens, Greece [Google Scholar]
- 41. Prosser JI, Nicol GW. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931–2941 [DOI] [PubMed] [Google Scholar]
- 42. Purkhold U, et al. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reviers BD. 2003. Biologie et phylogénie des algues, tome 1. Éditions Belin, Paris, France [Google Scholar]
- 44. Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schleper C, Nicol GW. 2010. Ammonia-oxidising archaea—physiology, ecology and evolution. Adv. Microb. Physiol. 57:1–41 [DOI] [PubMed] [Google Scholar]
- 46. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Staufenberger T, Thiel V, Wiese J, Imhoff JF. 2008. Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microbiol. Ecol. 64:65–77 [DOI] [PubMed] [Google Scholar]
- 48. Steger D, et al. 2008. Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ. Microbiol. 10:1087–1094 [DOI] [PubMed] [Google Scholar]
- 49. Stopnišek N, et al. 2010. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 76:7626–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 51. Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357–1364 [DOI] [PubMed] [Google Scholar]
- 52. Tourna M, et al. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tujula NA, et al. 2010. Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 4:301–311 [DOI] [PubMed] [Google Scholar]
- 54. Vargas-Yáñez M. 2010. Cambio climático en el mediterráneo español. Instituto Español de Oceanografía, Ministerio de Ciencia e Innovación, Madrid, Spain [Google Scholar]
- 55. Wuchter C, et al. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zehr JP, Ward BB. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.