Abstract
Water is an important route for human norovirus (HuNoV) transmission. Using magnetic beads conjugated with blood group-like antigens (HuNoV receptors), we developed a simple and rapid receptor-binding capture and magnetic sequestration (RBCMS) method and compared it to the existing negatively charged membrane absorption/elution (NCMAE) method for concentrating HuNoV from sewage effluent. RBCMS required 6-fold-less sample volume than the NCMAE method and also resulted in a significantly higher yield of HuNoV. The NCMAE and RBCMS concentrations of genogroup I (GI) HuNoV measured by quantitative reverse transcription-PCR (qRT-PCR) resulted in average threshold cycle (CT) values of 34.68 (8.68 copies, 252-fold concentration) versus 34.07 (13.05 copies, 477-fold concentration), respectively; the NCMAE and RBCMS concentrations of genogroup II (GII) HuNoV were measured as average CT values of 33.32 (24.7 copies, 239-fold concentration) versus 32.38 (46.9 copies, 333-fold concentration), respectively. The specificity of qRT-PCR was confirmed by traditional RT-PCR and an RNase I protection assay. The qRT-PCR signal from RBCMS-concentrated HuNoV treated with RNase I indicated that it was from encapsidated RNA and, probably, viable virus. In contrast, the qRT-PCR signal from NCMAE-concentrated HuNoV was not protected from RNase I and, likely, degradation. Both GI and GII HuNoV were detected from sewage effluent samples collected between April and July with average concentrations of 7.8 × 103 genomic copies per liter (gc/liter) and 4.3 × 104 gc/liter, respectively. No GI and <2% GII HuNoV were detected in sewage samples stored at room temperature for 4 weeks. We conclude that RBCMS requires less sample volume, has better recovery and sensitivity, and is faster than NCMAE for detection of HuNoV in sewage.
INTRODUCTION
Human noroviruses (HuNoVs) account for approximately 58% of food-borne illnesses in the United States (29). Most food-borne disease outbreaks of HuNoVs are assumed to occur from consumption of food contaminated by food handlers who carry the virus or are ill with the virus. Waterborne outbreaks are often caused by sewage contamination of drinking water or recreational water (19, 24); therefore, water is suspected as a source for contaminating preharvest, and possibly, postharvest produce. The feces of infected humans can contain high concentrations of virus (up to 1011 virus particles/g), and virus shedding can last up to 3 weeks (1). Therefore, septic system and municipal sewage could contain high numbers of HuNoV and be a potential contamination source. Indeed, HuNoVs have been detected in high concentrations in human sewage (7, 14, 36). HuNoV in untreated raw sewage (influent) serves as a source for the development and testing of detection methods and for molecular epidemiological characterization and tracking of HuNoV in the environment that might be related to HuNoV outbreaks. Conventional wastewater treatment, designed mainly for bacterial elimination, is not optimal for viral elimination. Lodder et al. (17) reported enteric virus removal rates ranging between 30 and 99%, which is consistent with a report by van den Berg et al. (36) of average HuNoV in treated-sewage effluents decreasing from 105 to 103 reverse transcription-PCR (RT-PCR) detectable units after treatment. Victoria et al. (37) reported mean concentrations of HuNoV in influent and effluent samples of 7,290 genomic copies per liter (gc/liter) and 3,470 gc/liter for HuNoV genogroup I (GI), respectively, and 2,400 gc/liter and 643 gc/liter for genogroup II (GII), respectively. The removal of virus by treatment was 52% and 79% for GI and GII HuNoV, respectively. These results are representative of the results of numerous studies reporting detection of HuNoV in treated-sewage effluent samples (3, 9, 11, 17, 35, 39). These reports raise concern in public safety, since HuNoV is highly infectious with <10 virus particles capable of causing disease (32). Therefore, inadequate wastewater treatment can result in the persistence of HuNoV in the environment in coastal or surface waters, and possibly, contamination of aquifers and wells supplying irrigation water for produce or other raw commodities. Improved methods for detection and quantification of HuNoV contamination in treated-sewage effluent and the environment are critical for monitoring the sources and transport of the virus and contamination of food. We have developed a rapid and sensitive method to detect and quantitate HuNoV in treated-sewage effluent as a prototype method for future studies of HuNoV in watersheds.
A variety of approaches have been developed to concentrate HuNoV from sewage. HuNoV was detected in sewage influents in concentrations ranging from undetected to a million gc/liter (3, 9, 25, 36). Ueki et al. (35) used a polyethylene glycol (PEG) precipitation method to detect HuNoV in 6 out of 8 samples (75%) of river water, 8 out of 9 samples (89%) of treated wastewater, and all 9 samples (100%) of sewage. Their results indicated that treated wastewater is probably a main source of HuNoV pollution in the study area in the Netherlands. Iwai et al. (9) used both PEG and negatively charged membrane absorption/elution (NCMAE) methods and reported persistence of HuNoV in influent sewage. da Silva et al. used PEG (3) to detect HuNoV in 88% of influent samples and 14% of effluent samples in a study area in France. Sdiri-Loulizi et al. (30) used PEG to detect HuNoV in 2.8% of influent samples and 1.6% of effluent samples of Monastir, Tunisia. In contrast, Lodder et al. (17) reported that HuNoV could be detected in all 5 samples collected with an NCMAE method in both influent and effluent sewage samples.
Histo-blood group antigens (HBGAs) have been characterized recently as receptors for HuNoV, and receptor-based capture assays have been applied to concentrate HuNoV from spiked food samples and spiked concentrated environmental water samples (2, 21, 33). Two-log-unit increases in the sensitivity of HuNoV detection in spiked food samples by a similar method have been reported (33). Cannon and Vinje (2) demonstrated that compounds present in environmental waters that inhibit RT-PCR after concentration by PEG precipitation were removed by magnetic beads conjugated with synthetic HBGAs. However, direct application of this method to concentrate HuNoV from environmental water samples has never been tested. A wide range of viral concentration techniques for water samples have been proposed and include large sample concentration, an approach essential for water samples with low levels of contamination, such as drinking water or seawater (15, 16, 28, 34, 36). However, a recent study by Gregory et al. (6) demonstrated that concentration of viruses from some large volume samples may be counterproductive when inhibitors are coconcentrated. da Silva et al. (3) demonstrated that a small sample size (40 ml) was adequate to concentrate HuNoV in sewage samples. Although our receptor-binding capture and magnetic sequestration (RBCMS) method was designed to work with both large and small volume samples, we initiated our analysis with small volume samples to determine the distribution of HuNoV during a warm season of our region (April through July) and the stability of HuNoV in sewage. This receptor-binding capture amplification type assay relies upon the presence of both viral capsid proteins to bind to HBGA receptors, as well as on viral RNA released from the capsid to be amplified as template in RT-PCR. This method is predicted to correspond to the infectivity of the virus if the concentrated viral RNA is from encapsidated virus. To test this hypothesis, we applied an RNase protection assay to confirm that the concentrated viral RNAs were encapsidated.
MATERIALS AND METHODS
Source of sewage and collection.
Water samples were collected between April and July in 2011 from two sewage treatment plants (STPs) located near the eastern shore of San Pablo Bay in the San Francisco Bay area of California; both STPs used sedimentation and biological treatment with activated sludge as primary and secondary treatment methods, respectively. Samples of influent and effluent water were collected from both STPs. Samples of effluent water were collected on a weekly basis from a single STP to determine differences in the amounts of HuNoV over a period of time. A large sample of effluent water was collected from a STP, and portions stored at either room temperature or 4°C were tested to determine the stability of the virus.
Concentration of HuNoV by the NCMAE method.
HuNoV particles present in sewage samples were concentrated by the methods of Katayama (12) and Fong (5), with minor modifications. Briefly, STP sewage samples were adjusted to pH 3.5 with glacial acetic acid and cleared of large particulates by vacuum filtration through a 1- to 5-μm retention cellulose fiber filter paper (P2 qualitative filter paper; Fisher Scientific, Pittsburgh, PA). A 140-μl sample of the clarified filtrate was saved as preconcentration sample. HuNoV particles were sequestered from 250-ml aliquots of clarified filtrate by vacuum filtration through a 0.45-μm-pore mixed-cellulose HA filter membrane (MF-Millipore HAW; Millipore, Billerica, MA). The HuNoV particles bound to the filter membrane were washed by vacuum filtration with 100 ml of 1 mM H2SO4. The sequestered HuNoV particles were released and eluted from the washed membrane by vacuum filtration with 10 ml of 10 mM NaOH into a clean vacuum flask containing 0.1 ml of 50 mM H2SO4. The eluent was transferred into 50-kDa-NMWL (nominal molecular weight limit) centrifugal filter units (Centriprep YM-50; Millipore, Billerica, MA) and concentrated to approximately 0.5 ml in accordance with the manufacturer's protocol. A 140-μl sample of the concentrate and the saved preconcentration sample were extracted to obtain viral RNA with a viral RNA extraction kit (QIAamp viral RNA minikit; Qiagen, Valencia, CA) in accordance with the manufacturer's protocol.
Concentration of HuNoV by the RBCMS method.
Sewage samples were cleared of large particulates by either vacuum filtration as described above or by centrifugation. For centrifugation, water samples were adjusted to pH 3.5 with glacial acetic acid with or without 1% Tween 80 and then centrifuged at 4,816 relative centrifugal force (RCF) for 60 min. A 40-ml sample of the clarified supernatant was transferred to a new 50-ml tube, and 140 μl of the clarified supernatant was set aside as the preconcentration sample to be extracted later for viral RNA with a viral RNA extraction kit (QIAamp viral RNA minikit; Qiagen, Valencia, CA) as described above. HuNoV particles were concentrated by binding to porcine gastric mucin conjugated to magnetic beads (PGM-MB) as described previously (33) with some procedural modifications. PGM-MB (150 μl) was added to the ~40 ml of clarified supernatant, and any HuNoV present was allowed to bind for 15 min while gently mixing. The HuNoV bound to PGM-MB by placing the HuNoV-PGM-MB suspension in a magnetic separation rack (New England BioLabs, Ipswich, MA) for 60 min. The separated fluid phase was removed by pipetting, the PGM-MB with bound HuNoV was resuspended with 560 μl of viral lysis solution with carrier RNA from the viral RNA extraction kit, and the suspension was transferred to a 1.7-ml microcentrifuge tube. The sample was lysed for 5 min to release the viral RNA into solution. The lysed resuspension was placed in a magnetic separation rack for 5 min to separate the magnetic beads from the fluid phase, and the separated fluid with viral RNA was transferred to a new microcentrifuge tube. The viral RNA was extracted in accordance with the manufacturer's protocol.
qRT-PCR and RT-PCR.
Multiplexed, probe-based quantitative real-time RT-PCR (qRT-PCR) was performed on a qPCR system (MX3000P; Stratagene, La Jolla, CA) using a one-step qRT-PCR kit (Quantitect probe RT-PCR kit; Qiagen, Valencia, CA) in accordance with the manufacturer's protocol. Primers and probe sequences described previously (10) were synthesized with modified fluorophores and quenchers (Integrated DNA Technologies, Inc., San Diego, CA). The primers and probes used for detection of GI HuNoV were COGIF (5′ CGY TGG ATG CGN TTY CAT GA 3′), COGIR (5′ CTT AGA CGC CAT CAT CAT TYA C 3′), GI-P1 (5′ 6-FAM–AGA TYG CGA TCY CCT GTC CA–BHQ-1 3′ where 6-FAM is 6-carboxyfluorescein and BHQ-1 is black hole quencher 1), and GI-P1b (5′ 6-FAM–AGA TCG CGG TCT CCT GTC CA–BHQ-1 3′). The primers and probes for GII HuNoV were COGIIF (5′ CAR GAR BCN ATG TTY AGR TGG ATG AG 3′), COGIIR (5′ TCG ACG CCA TCT TCA TTC ACA 3′), and GII-P (5′ HEX–TGG GAG GGC GAT CGC AAT CT– BHQ-1 3′ where HEX is hexachlorofluorescein). Each 25-μl reaction mixture consisted of 12.5 μl of Quantitect probe RT-PCR master mix, 5.5 μl of RNase-free water, 0.75 μl of each 10 μM primer (COGIF, COGIR, COGIIF, and COGIIR), 0.25 μl of each 10 μM probe (GI-P1 6-FAM, GI-P1b-1 6-FAM, and GII-P HEX), 0.25 μl of Quantitect RT mix, and 3 μl of extracted RNA free water, 0.75 μl of each 10 μM primer (COGIF, COGIR, COGIIF, and COGIIR), 0.25 μl of each 10 μM probe (GI-P1 6-FAM, GI-P1b-1 6-FAM, and GII-P HEX), 0.25 μl of Quantitect RT mix, and 3 μl of extracted RNA. Cycling times and temperatures were 30 min at 50°C and 15 min at 95°C, followed by 45 cycles, with 1 cycle consisting of 15 s at 95°C, 20 s at 53°C, and 50 s at 60°C. Fluorescence was read at the end of each 60°C extension step, and thresholds were determined by MxPro software with amplification-based threshold determination using default settings. qRT-PCR signal specificity was confirmed with RT-PCR with region B, C, and D primer sets for HuNoV (4, 13, 38).
Quantitative analysis.
Plasmids containing GI and GII HuNoV sequences were kindly provided by T. Kageyama (10). The plasmids were diluted in a 10-fold dilution series (ranging from 0.1 to 1,000,000 copies) to generate a standard curve together with positive-control samples for quantitative analysis of HuNoV. In this study, virus copies were defined as copies of virus measured in 3 μl out of 60 μl of RNA extraction output in qRT-PCR. The dilution factor was not considered except in calculation of concentration (gc/liter) of HuNoVs in environmental samples (see Fig. 3) where the dilution factor was 500 [(1,000/40) × 20]. Positive-control reactions were qRT-PCRs of diluted RNA extracted from human fecal samples and containing known amounts of GI and GII HuNoV. The extracted RNA had been divided into single-use aliquots and frozen; new aliquots were thawed for each amplification run to be normalized. The threshold cycle (CT) values of the positive-control reactions were converted to genomic copies using the standard curve constructed from the GI and GII plasmid samples. For each amplification run, positive-control reactions were included and used to normalize all data.
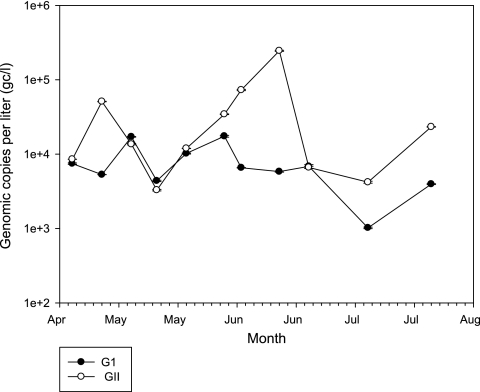
Fig 3.
Distribution of GI and GII HuNoVs in effluent sewage between late April and July in 2011.
RNase protection assay.
The NCMAE and RBCMS procedures were modified to include an RNase I (Epicentre Biotechnologies, Madison, WI) digestion prior to RNA extraction. Concentration of virus for NCMAE was as described in the previous section up to the point after concentration of the sample to approximately 0.5 ml with centrifugal filter units. A 55.6-μl sample of 10× TNE (Tris-NaCl-EDTA) buffer was added to the concentrated sample, and the sample was divided into 155.6-μl reaction volumes. Concentration of virus by RBCMS was as described in the previous section up to the point of viral binding to PGM-MB, magnetic separation, and removal of the fluid phase. The magnetic beads were resuspended with 140 μl of double-distilled H2O (ddH2O) and 15.6 μl of 10× TNE buffer, and the beads were transferred to a 1.7-ml microcentrifuge tube. For both methods, 3 μl of RNase I (1U/μl) was added to each digestion reaction mixture but omitted from control reaction mixtures. All reaction mixtures were incubated at 37°C for 30 min. A 7.5-μl sample of dithiothreitol (DTT) (0.1 M) was added to all reaction mixtures and then heat deactivated at 95°C for 10 min. The viral RNA was extracted from HuNoV-PGM-MB in the reaction mixture as described in the NCMAE and RBCMS methods in the previous sections. Viral RNA extracted previously was digested with RNase I in accordance with the manufacturer's protocol, and in parallel with the test sample assays as a control for RNase I activity.
Statistics.
At least three replicates were performed for each test sample, and at least three independent experiments were performed for each test condition to ensure consistency. One-way analysis of variance (ANOVA) (SigmaStat) or Student's t test was used for statistical analysis.
RESULTS
Quantitation of HuNoV.
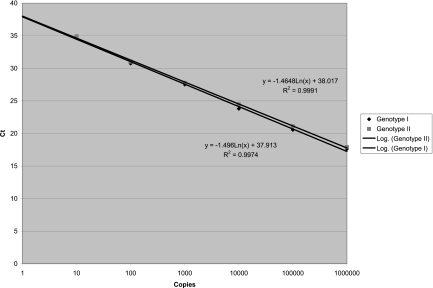
Standard curves were generated with qRT-PCR of samples containing known numbers of viral genomes to convert CT values to copy numbers of GI and GII HuNoVs (Fig. 1). Samples calculated to contain 106, 105, 104, 103, 102, 101, and 100 copies of viral genome in the amplification reaction mixture corresponded to CT values of 17.63, 20.56, 23.8, 27.52, 30.74, 34.89, and 37.77 for GI HuNoV, respectively, and CT values of 17.93, 21.11, 24.49, 27.78, 31.03, 34.93, and 36.66 for GII HuNoV, respectively. Samples calculated to contain a predicted 0.1 copy of GI and GII HuNoV did not amplify. The slope was −1.496 cycles/log 10 for GI HuNoV with an R2 of 0.9974 and was −1.4648 cycles/log 10 for GII HuNoV with an R2 of 0.9991. The positive controls produced an average CT of 32.45 for GI HuNoV, corresponding to 38.54 genomic copies, and an average CT of 29.76 for GII HuNoV, corresponding to 280 genomic copies. The positive controls were used to validate and calibrate the data for each experiment.
Fig 1.
Standard curves for GI and GII HuNoVs.
Pretreatment of sewage samples is required for the RBCMS method with PGM-MB.
In the course of optimizing binding conditions for PGM-MB with sewage samples, we detected trace signals (CT ≥ 42) of both GI and GII HuNoV in sewage by direct qRT-PCR. However, neither GI nor GII HuNoV could be concentrated effectively from these samples by the RBCMS method and, thus, were undetected. Concentration and detection of GI and GII HuNoV by the RBCMS method were achieved only after removing large particulates from the sewage samples by either vacuum filtration through a 1- to 5-μm retention cellulose filter or a low-RCF centrifugation, with or without 1% Tween 80. CT values of 33.6 ± 0.10, 33.14 ± 0.70, and 32.63 ± 0.28, calculated to correspond to 21.84, 24.3, and 34.17 genomic copies of GI HuNoV, respectively, were obtained by filtration, centrifugation, and centrifugation with Tween 80, respectively (Table 1). GI HuNoV yield after clarification by centrifugation with Tween 80 was significantly better than clarification by vacuum filtration (P = 0.005). Although a general improvement in yield was observed with centrifugation with Tween 80, the significance of the difference compared to centrifugation without Tween 80 could not be established, primarily due to high variations in yield for the latter (P = 0.33). For GII HuNoV, CT values of 31.02 ± 0.64, 30.68 ± 0.21, and 30.09 ± 0.22, corresponding to 118.72, 149.8, and 224 genomic copies, respectively, were obtained by filtration, centrifugation, and centrifugation with Tween 80, respectively. GII HuNoV yield after clarification by centrifugation with Tween 80 was significantly better than clarification by vacuum filtration (P = 0.005). In addition, the collected magnetic beads were easily and thoroughly washed from the tube walls when Tween 80 was used. Therefore, centrifugation with Tween 80 was used for clarification of sewage samples in subsequent experiments with the RBCMS method. Clarification of samples by vacuum filtration did not yield a significant improvement in HuNoV yields for the NCMAE method. For GI HuNoV, CT values of 33.55 ± 0.29 and 33.38 ± 0.41, equal to 18.5 and 20.7 genomic copies, respectively, were achieved for clarification filtration and no filtration, respectively. For GII HuNoV, CT values of 30.46 ± 0.27 and 30.49 ± 0.42, equal to 174 and 170 genomic copies, respectively, were achieved for clarification filtration and no filtration, respectively. Despite these results, vacuum filtration was necessary for clarifying the sample to decrease clogging in the HA membrane filtration step, which would otherwise take hours for a 250-ml sample containing particulates. Therefore, vacuum filtration clarification of sewage samples was used in subsequent experiments with the NCMAE method.
Table 1.
Concentrating HuNoV from sewage water by the RBCMS method after pretreatment by filtration or centrifugation
| Treatment | GI HuNoV |
GII HuNoV |
||
|---|---|---|---|---|
| CT | No. of copies | CT | No. of copies | |
| Untreated | >40 | <1 | >40 | <1 |
| P2 filter | 33.60 ± 0.10 | 21.84 | 31.02 ± 0.64 | 118.72 |
| Centrifugation | 33.14 ± 0.70 | 24.30 | 30.68 ± 0.21 | 149.80 |
| Centrifugation with Tween 80 | 32.63 ± 0.28 | 34.17 | 30.09 ± 0.22 | 224.00 |
Comparison of concentration of HuNoV by NCMAE and RBCMS method with PGM-MB.
Nine samples collected from two STPs (marked as P and E) and three samples collected from a local reservoir (R) were tested for HuNoV by NCMAE and RBCMS methods (Table 2). For each sample, both methods were performed in parallel on the same day to maximize the comparability of the methods. The average CT values for GI HuNoV were 34.68 ± 1.47 and 34.07 ± 1.45, equal to 11.48 ± 12.1 and 17.43 ± 11.1 genomic copies, for samples concentrated by NCMAE and RBCMS method, respectively. The average CT values for GII HuNoV were 33.32 ± 2.8 and 32.38 ± 1.76, equal to 57.3 ± 66.5 and 81 ± 27.1 genomic copies, for samples concentrated by NCMAE and RBCMS method, respectively. The concentrated GI HuNoV in sewage samples ranged widely from 0 to 38.3 copies (NCMAE method) and from 1.42 to 34.17 copies (RBCMS method). The concentrated GII HuNoV from sewage ranged from 0 to 174 copies (NCMAE method) and from 7.34 to 222 copies (RBCMS method). The wide range in concentrations of HuNoV in sewage samples necessitated comparing the capability of the two methods to concentrate HuNoV. The mean CT difference (ΔCT) between the starting and concentrated samples for GI was 7.50 ± 1.70 and 8.27 ± 1.87 for NCMAE and RBCMS methods, respectively, and for GII, it was 6.96 ± 1.97 and 7.80 ± 1.47 for NCMAE and RBCMS methods, respectively. After adjusting for the starting volumes, the average concentrating powers by the NCMAE and RBCMS methods were 252-fold and 477-fold for GI, and 239-fold and 333-fold for GII, respectively. However, the average time needed to complete the RBCMS method was significantly less than the time to complete the NCMAE method. For example, it could take more than 8 h to vacuum filter 250 ml of certain samples of clarified sewage through the negatively charged membrane. Among three samples obtained from a nearby reservoir, all samples tested negative (CT > 45) for GI HuNoV by both the NCMAE and RBCMS methods. However, one sample tested positive for GII HuNoV by the RBCMS method. One reaction from triplicates gave a CT value of 40.23, reflecting 0.22 copy of GII. This result was confirmed after the water sample was retested two more times and yielded similar results.
Table 2.
Comparison of concentration of HuNoV by the NCMAE and RBCMS methods with PGM-MB
| STPa | GI HuNoVb |
GII HuNoVb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCMAE |
RBCMS |
NCMAE |
RBCMS |
|||||||||
| CT | ΔCT | No. of copies | CT | ΔCT | No. of copies | CT | ΔCT | No. of copies | CT | ΔCT | No. of copies | |
| P1 | 34.17 | 8.73 | 12.21 | 34.43 | 8.73 | 10.30 | 33.03 | 7.94 | 30.10 | 33.22 | 7.75 | 26.44 |
| P2 | 33.55 | 7.51 | 18.48 | 34.38 | 6.68 | 10.60 | 30.46 | 9.58 | 174.00 | 31.49 | 8.45 | 86.00 |
| P3 | 33.60 | 10.30 | 17.89 | 32.63 | 11.26 | 34.17 | 31.02 | 9.68 | 120.60 | 30.10 | 10.60 | 222.50 |
| P4 | 36.03 | 7.06 | 3.52 | 34.69 | 8.40 | 8.62 | 38.98 | 4.96 | 0.52 | 35.02 | 8.92 | 7.74 |
| P5 | 32.46 | 6.69 | 38.29 | 33.14 | 6.01 | 24.30 | 30.87 | 6.19 | 131.50 | 31.75 | 5.31 | 72.13 |
| P6 | 35.03 | 8.49 | 6.87 | 33.32 | 10.20 | 21.55 | 34.62 | 4.68 | 10.17 | 32.01 | 7.30 | 60.40 |
| P7 | 35.90 | 4.62 | 3.84 | 32.76 | 7.76 | 31.33 | 32.70 | 5.53 | 37.71 | 30.18 | 7.84 | 210.65 |
| E1 | 36.71 | 6.59 | 2.23 | 37.39 | 5.91 | 1.42 | 34.86 | 7.12 | 8.63 | 34.82 | 7.16 | 8.87 |
| E2 | Failed | N/A | 0 | 33.90 | 9.51 | 14.62 | Failed | N/A | 0 | 32.83 | 6.83 | 34.51 |
| Mean | 34.68 | 7.50 | 11.48 | 34.07 | 8.27 | 17.65 | 33.32 | 6.96 | 57.01 | 32.38 | 7.80 | 81.03 |
Nine samples collected from two STPs (P and E STPs) were tested for HuNoV by the NCMAE and RBCMS methods.
ΔCT, CT difference between input and concentrated samples; N/A, not available for detection.
Specificity of the assay.
RNA extraction columns, such as the ones employed in the RBCMS and NCMAE methods, do not exclude DNA, so column outputs are assumed to be a mixture of both RNA and DNA. Therefore, it was necessary to confirm that qRT-PCR was amplifying the RNA template in the sewage sample. We heat deactivated (95°C for 5 min) the reverse transcriptase in the one-step qRT-PCR reaction mixture prior to the addition of template nucleic acids in order to remove the ability to amplify RNA and observe if any DNA amplification occurred by qRT-PCR. No amplification of the nucleic acids extracted from HuNoV signal-producing sewage occurred with the heated deactivated RT reaction mixture (data not shown), confirming that the signals produced from the parallel qRT-PCR reactions were indeed from RNA. This was confirmed further by RNase I digestion of nucleic acids extracted from HuNoV-positive sewage samples and subsequent heat deactivation of RNase I activity prior to their addition to qRT-PCR reaction mixtures, which also resulted in no amplification. Amplicons produced by HuNoV region B, C, and D primer sets were identified as bands of the predicted size in agarose gel electrophoresis. Region D primers resulted in the highest sensitivity and produced a 180-bp amplicon for GI HuNoV and a 250-bp amplicon for GII HuNoV (data not shown).
Stability of HuNoV in sewage.
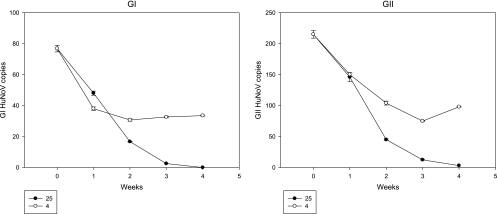
To determine the stability of HuNoV in sewage samples over time, a large-volume sewage sample was collected, and one portion was stored at room temperature while another was kept at 4°C. The samples were analyzed every week for 4 weeks and quantitated for HuNoV (Fig. 2). After 1 week, the copy numbers of GI and GII HuNoV decreased 38% and 32%, respectively, for the room temperature sample, and decreased 51% and 30% for the 4°C sample, respectively. From the 2nd week onwards, copy numbers for both GI and GII HuNoV decreased rapidly in the room temperature sample. At the 4th week, no GI HuNoV could be detected, and GII HuNoV copy numbers decreased 98.6% for the room temperature sample. In contrast, GI and GII HuNoV copy numbers decreased by only 56% and 54%, respectively, for the 4°C sample.
Fig 2.
Stability of GI and GII HuNoVs in sewage water stored at room temperature (25°C) or 4°C.
Detection of HuNoV in sewage samples collected between late April and late July.
Sewage samples were collected from a single STP (P) on a weekly basis for 3 months and tested for HuNoV. The concentrations of virus in the sewage samples were normalized by comparing to values of positive-control reactions with known numbers of copies of virus as described in Materials and Methods. Generally, the average amount of GII HuNoV (42,300 gc/liter) was higher than GI HuNoV (7,825 gc/liter; approximately 5.4-fold) (Fig. 3). The lowest GI virus concentration was measured with the July 7 sample (1,020 gc/liter). High concentrations of GI virus were measured with samples collected May 12 (1.71 × 104 gc/liter) and June 3 (1.74 × 104 gc/liter). The amount of GI virus at other time points was approximately 5,000 gc/liter. Two samples with GII virus lower than 5,000 copies were collected 18 May 2011 (3,300 gc/liter) and 7 July 2011 (4,200 gc/liter). The largest amount of GII HuNoV was observed with the June 16 sample that yielded a copy number of 2.46 × 105 gc/liter. Small amounts of both GI and GII virus were observed with the July 7 sample. It is noteworthy that the peak(s) for high concentrations of GI and GII did not occur at the same time. There were no HuNoV-related illnesses during this period of time reported to the public health department in the same county as the STP (Contra Costa County in California).
RNase protection assay.
Viruses concentrated by the RBCMS or NCMAE method were treated with RNase I or not treated with RNase I to confirm that the RNA signals were from encapsidated viruses. Viral RNA extracted previously (positive control) was also digested with RNase I as a control for the enzyme activity. Amplification was abolished completely for the positive-control viral RNA standard treated with RNase I (data not shown). However, there was no significant difference in virus concentrated by PGM-MB and treated or untreated with RNase I. The CT values for untreated and RNase I-treated samples were 32.93 ± 2.83 and 32.74 ± 0.70 for GII HuNoV (P = 0.687) and 37.45 ± 2.08 and 37.76 ± 0.22 for GI, respectively (P = 0.815). However, there was a significant decrease in CT values after RNase treatment for NCMAE samples. The CT values for untreated and RNase I-treated groups were 34.11 ± 0.37 and 35.81 ± 0.62 for GII HuNoV (P = 0.015) and 39.45 ± 2.47 and 42.12 ± 3.32 for GI, respectively. Indeed, the CT values were undetected in 3 out of 6 samples treated with RNase I (i.e., CT > 45).
DISCUSSION
We developed a rapid, specific, and relatively sensitive method for detection of GI and GII HuNoV in municipal sewage samples and compared our method to a membrane absorption method (NCMAE) for detection of HuNoV in samples collected between April and July, 2011 from two STPs within the San Francisco Bay area of California.
Various approaches have been applied to concentrate HuNoV from sewage. The NCMAE method has been used most frequently and has been reported to provide efficient detection of HuNoV using a standard sample size of 250 ml. However, a recent study by Gregory et al. (6) reported that concentration of viruses from large volumes may be counterproductive for some samples that may contain inhibitors that are coconcentrated with virus. da Silva et al. (3) reported that HuNoV could be detected from a smaller sample size (e.g., 40 ml) by a PEG precipitation method. However, the detection rate for effluent samples (14%) was much lower than for the influent samples (88%), indicating either low sensitivity of the method, possibly due to PCR inhibitors in the effluent, or inadequate concentration due to complex effluent sample particulates. The RBCMS method we developed provides improved recovery and concentration of HuNoV, and most importantly, improved sensitivity. The average CT of concentrated samples was lower for the RBCMS method than for the NCMAE method, corresponding to improved sensitivity of the RBCMS method. The average concentrations of HuNoV by the NCMAE and RBCMS methods were 252-fold and 477-fold for GI, respectively, and 239-fold and 333-fold for GII, respectively. The average time needed to complete the RBCMS method was significantly less than for the NCMAE method and due mostly to the complexity of the samples; NCMAE required more than 8 h to filter 250 ml of some samples. In contrast, the total concentration time for the RBCMS method was less than 2 h. It is possible to decrease the low-speed centrifugation step to as short as 15 min, since no significant difference in virus recovery between 15 and 60 min of centrifugation was observed (data not shown).
HuNoV is detected in surface waters less frequently than in wastewaters concentrated from liters or gallons of water. However, we identified a low concentration of GII HuNoV contamination by the RBCMS method from one 40-ml sample collected at a nearby reservoir open to public recreation. The signal was minimal in that only one reaction of three samples tested resulted in a CT value of 40.23, equal to 0.22 copies of GII HuNoV. This particular reservoir sample was collected from water close to a recreation area in the reservoir, suggesting that the contamination might be from an infected individual(s) in the recreation area. The same sample was tested two additional times, producing similar results. Although the results were ambiguous, the results are intriguing and support additional testing of surface water samples in produce production regions.
HuNoV continues to circulate seasonally, causing disease peaking in the winter season. Currently, there is no direct evidence of the existence of a disease reservoir for reintroduction into the human population. Our data suggest that treated-sewage samples may contain high concentrations of HuNoV in nonepidemic seasons. We are about to initiate a year-long survey of concentrations in sewage in selected STPs. The persistance of HuNoV in treated sewage throughout the year would obviate another source/reservoir of the virus other than humans.
GI strains were identified as the most frequent cause of waterborne outbreaks associated with HuNoV, whereas GII strains have been reported to be more common in HuNoV-related food-borne outbreaks and in health care settings (18, 19, 26). The reason for this association is not known, but it has been hypothesized that GI strains could be more stable in water than GII strains due to differences in their capsid proteins (18, 19). However, both GI and GII could be detected in all samples tested simultaneously in our improved detection method, with GI concentrations lower than GII concentrations in all samples tested. The ambiguous results obtained for the detection of HuNoV in sewage samples could be the result of differences in concentration methods and locations. Relative to other locations, the temperature variations are low throught the year in the unusually mild climate of the San Francisco Bay area.
The effluent sewage samples were all collected prior to their release into coastal waters. The effluents were treated by multiple steps, including pretreatment, primary treatment, secondary treatment with activated sludge, and chlorination with up to 12 ppm of chlorine. However, it is worth noting that the presence of a qRT-PCR-detected signal did not confirm the presence of infectious viral particles. Chlorine is a highly oxidizing agent, and hypochloric acid is considered to be the active moiety (20). Nuanualsuwan and Cliver (22, 23) reported in their studies that hypochlorite affected both the RNA and capsid of poliovirus, feline calicivirus, and hepatitis A virus. The minimal chlorine concentration needed to inactivate HuNoV has not been established, because no tissue culture system is available for testing interventions with HuNoV and comparison of results to inactivation methods for other viruses.
Histoblood group antigens (HBGAs) have been identified as receptors of HuNoV and are associated with genetic susceptibility of some humans to HuNoV infection, if they are exposed to the virus (27, 31). Several studies demonstrated that a HBGA receptor-based capture method could be used to concentrate HuNoV from food samples (21, 33) and facilitate the removal of PCR inhibitors from concentrated water samples (2). Recently, Huhti et al. (8) reported using an HBGA-binding assay together with electron microscropy (EM) to confirm morphological integrity, antigenicity, and functionality of HuNoV GII-4 viral like particles. However, application of this kind of assay to the characterization of biological functions of the virus has not been well evaluated. Since an intact viral capsid (for attachment) and genome (for replication) are required for a biologically viable virus, a receptor-based capture assay quantitated by qRT-PCR can facilitate measurement of the presence of both capsid and genome. Our method captures HuNoV with HBGA conjugated to magnetic beads, and the viral genomes are then extracted and amplified by qRT-PCR; only encapsidated viral RNA is amplified. The results of the RNase protection assay are consistent with HuNoV signal by the RBCMS method being the result of captured encapsidated viral RNA. In contrast, not all of the HuNoV signal produced by NCMAE was protected from RNase digestion. Two-thirds of the HuNoV signal measured by NCMAE was eliminated by RNase treatment. We speculate that capsid proteins might be damaged by the harsh treatment of a strong basic buffer wash, or alternatively, that free viral RNA (in addition to intact viral particles) also binds to the negatively charged membrane. We conclude that the results of measurement of HuNoV concentrations in complex samples by the RBCMS assay are more biologically relevant than the NCMAE assay. The RBCMS method is faster and requires less sample compared to the NCMAE method, resulting in better recovery of HuNoV and improved sensitivity. Since correlations between positive results measured by this method and HuNoV infectivity in humans cannot be determined due to the lack of in vitro tissue culture methods and an animal model, sensitive and specific assays for accurately measuring HuNoV in the environment may be the best strategy for identifying reservoirs and source tracking relevant to epidemiological studies of HuNoV related to contamination of food and illness.
ACKNOWLEDGMENTS
We thank T. Kageyama (Section of Infectious Disease, R&D Center, BML, Saitama, Japan) for providing HuNoV plasmids used to construct the standard curve.
This work was supported by USDA Agricultural Research Service CRIS projects 5325-42000-021-044 and 5325-42000-021-045.
Footnotes
Published ahead of print 18 November 2011
REFERENCES
- 1. Atmar RL, et al. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14:1553–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cannon JL, Vinje J. 2008. Histo-blood group antigen assay for detecting noroviruses in water. Appl. Environ. Microbiol. 74:6818–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. da Silva AK, et al. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 73:7891–7897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fankhauser RL, et al. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1–7 [DOI] [PubMed] [Google Scholar]
- 5. Fong TT, Griffin DW, Lipp EK. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gregory JB, Webster LF, Griffith JF, Stewart JR. 2011. Improved detection and quantitation of norovirus from water. J. Virol. Methods 172:38–45 [DOI] [PubMed] [Google Scholar]
- 7. Haramoto E, et al. 2006. Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci. Technol. 54:301–308 [DOI] [PubMed] [Google Scholar]
- 8. Huhti L, et al. 2010. A comparison of methods for purification and concentration of norovirus GII-4 capsid virus-like particles. Arch. Virol. 155:1855–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwai M, et al. 2009. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyama, Japan (2006 to 2008). Appl. Environ. Microbiol. 75:1264–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kageyama T, et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katayama H, et al. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42:1441–1448 [DOI] [PubMed] [Google Scholar]
- 12. Katayama H, Shimasaki A, Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kitajima M, et al. 2011. Genetic diversity of genogroup IV noroviruses in wastewater in Japan. Lett. Appl. Microbiol. 52:181–184 [DOI] [PubMed] [Google Scholar]
- 14. Kitajima M, et al. 2010. Seasonal distribution and genetic diversity of genogroups I, II, and IV noroviruses in the Tamagawa River, Japan. Environ. Sci. Technol. 44:7116–7122 [DOI] [PubMed] [Google Scholar]
- 15. La Rosa G, et al. 2007. Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription-PCR assays. Appl. Environ. Microbiol. 73:4152–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laverick MA, Wyn-Jones AP, Carter MJ. 2004. Quantitative RT-PCR for the enumeration of noroviruses (Norwalk-like viruses) in water and sewage. Lett. Appl. Microbiol. 39:127–136 [DOI] [PubMed] [Google Scholar]
- 17. Lodder WJ, van den Berg HH, Rutjes SA, de Roda Husman AM. 2010. Presence of enteric viruses in source waters for drinking water production in The Netherlands. Appl. Environ. Microbiol. 76:5965–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lysen M, et al. 2009. Genetic diversity among food-borne and waterborne norovirus strains causing outbreaks in Sweden. J. Clin. Microbiol. 47:2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maunula L, Miettinen IT, von Bonsdorff CH. 2005. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 11:1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morton V, Jean J, Farber J, Mattison K. 2009. Detection of noroviruses in ready-to-eat foods by using carbohydrate-coated magnetic beads. Appl. Environ. Microbiol. 75:4641–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nuanualsuwan S, Cliver DO. 2003. Capsid functions of inactivated human picornaviruses and feline calicivirus. Appl. Environ. Microbiol. 69:350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nuanualsuwan S, Cliver DO. 2003. Infectivity of RNA from inactivated poliovirus. Appl. Environ. Microbiol. 69:1629–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parshionikar SU, et al. 2003. Waterborne outbreak of gastroenteritis associated with a norovirus. Appl. Environ. Microbiol. 69:5263–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pusch D, et al. 2005. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch. Virol. 150:929–947 [DOI] [PubMed] [Google Scholar]
- 26. Riera-Montes M, et al. 2011. Waterborne norovirus outbreak in a municipal drinking-water supply in Sweden. Epidemiol. Infect. 139:1928–1935 [DOI] [PubMed] [Google Scholar]
- 27. Rockx BH, Vennema H, Hoebe CJ, Duizer E, Koopmans MP. 2005. Association of histo-blood group antigens and susceptibility to norovirus infections. J. Infect. Dis. 191:749–754 [DOI] [PubMed] [Google Scholar]
- 28. Rutjes SA, van den Berg HH, Lodder WJ, de Roda Husman AM. 2006. Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl. Environ. Microbiol. 72:5349–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sdiri-Loulizi K, et al. 2010. Detection and molecular characterization of enteric viruses in environmental samples in Monastir, Tunisia between January 2003 and April 2007. J. Appl. Microbiol. 109:1093–1104 [DOI] [PubMed] [Google Scholar]
- 31. Tan M, Jiang X. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 13:285–293 [DOI] [PubMed] [Google Scholar]
- 32. Teunis PF, et al. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 [DOI] [PubMed] [Google Scholar]
- 33. Tian P, Engelbrektson A, Mandrell R. 2008. Two-log increase in sensitivity for detection of norovirus in complex samples by concentration with porcine gastric mucin conjugated to magnetic beads. Appl. Environ. Microbiol. 74:4271–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trujillo AA, et al. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ueki Y, Sano D, Watanabe T, Akiyama K, Omura T. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 39:4271–4280 [DOI] [PubMed] [Google Scholar]
- 36. van den Berg H, Lodder W, van der Poel W, Vennema H, de Roda Husman AM. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Res. Microbiol. 156:532–540 [DOI] [PubMed] [Google Scholar]
- 37. Victoria M, et al. 2010. One year monitoring of norovirus in a sewage treatment plant in Rio de Janeiro, Brazil. J. Water Health 8:158–165 [DOI] [PubMed] [Google Scholar]
- 38. Vinje J, Hamidjaja RA, Sobsey MD. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116:109–117 [DOI] [PubMed] [Google Scholar]
- 39. Zakhour M, et al. 2010. Bovine norovirus: carbohydrate ligand, environmental contamination, and potential cross-species transmission via oysters. Appl. Environ. Microbiol. 76:6404–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]