Abstract
Bacillus subtilis induces expression of the gene ytnP in the presence of the antimicrobial streptomycin, produced by the Gram-positive bacterium Streptomyces griseus. ytnP encodes a lactonase-homologous protein that is able to inhibit the signaling pathway required for the streptomycin production and development of aerial mycelium in S. griseus.
TEXT
Bacterial communities display diverse defensive strategies in response to the presence of secreted natural products from coexisting microorganisms. These molecules act as signals to trigger changes in gene expression that allow bacteria to adapt to particular conditions (7, 22, 46, 53). Adaptation sometimes involves the activation of a defensive strategy against the presence of these natural products. For instance, the soil-dwelling organism Bacillus subtilis forms a biofilm in response to certain antimicrobials (30). Alternatively, several Bacillus species produce hydrolytic enzymes that are able to degrade the quorum-sensing signals from competing bacteria, a phenomenon known as quorum quenching (10–12). Some of these hydrolytic enzymes are metallolactamases (4, 5), which are able to degrade the lactone ring of the acyl-homoserine lactone signaling molecules (28, 35). Metallolactamases share a similar molecular structure, and they conserve important residues in their active sites (5, 6, 50), despite the low protein identity among the different subgroups (approximately 25%).
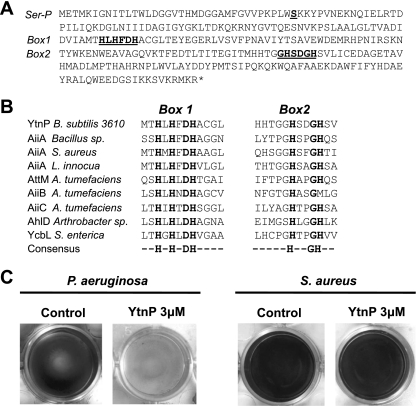
Bioinformatic analysis carried out in the model organism Bacillus subtilis NCIB3610 led to the identification of YtnP, a protein that shares a similar deduced domain to proteins described as quorum-quenching lactonases (Fig. 1A). Using ClustalW, a comparative alignment of the following proteins was performed: YtnP and AiiA from Bacillus sp. (10–12); AiiA homologues from Staphylococcus aureus and Listeria innocua (6); AttM, AiiB, and AiiC from Agrobacterium tumefaciens (6, 17, 29); AhlD from Arthrobacter sp. (39); and glyoxalase II (YcbL) from Salmonella enterica (45). AiiM from Microbacterium testaceum (52) and AidH from Ochrobactrum sp. (33) were excluded from this comparative analysis because of their nature of unusual lactonases. Results of the alignment confirmed that the important residues in the active sites of metallohydrolases are present in YtnP (Fig. 1B) (12, 24, 26, 27). Subsequently, overexpression of a 6×His-tagged version of the YtnP protein was carried out with Escherichia coli. 6×His-YtnP was purified using Ni+2 exchange chromatography (see Fig. S1 in the supplemental material). Inhibition of the quorum-sensing activity of YtnP was assayed based on the ability of Pseudomonas aeruginosa to form a biofilm in a quorum-sensing-dependent manner (3, 8, 32). Small concentrations of purified YtnP added to the biofilm formation assay of the cultures of P. aeruginosa inhibited biofilm formation, while similar concentrations of YtnP did not affect biofilm formation in cultures of S. aureus (Fig. 1D). This might be because the quorum-sensing molecule that controls biofilm formation in S. aureus has a peptidic nature and is not affected by the lactonase activity of YtnP (36, 38, 49).
Fig 1.
YtnP is a lactonase-homologous protein. (A) Amino acid sequence of YtnP from B. subtilis strain 3610. Conserved motifs present in other metallolactamases are marked as Box 1 and Box 2 and are underlined and bold. Also an additional serine (Ser36) important for YtnP activity is marked as Ser-P. (B) Alignment of the conserved motifs of the active site of the following: YtnP and other metallolactamases; AiiA from Bacillus sp. and homologues from S. aureus and L. innocua; AttM, AiiB, and AiiC from A. tumefaciens; AhlD from Arthrobacter sp.; and glyoxalase II (YcbL) from S. enterica. The last lane of the alignment shows the consensus sequence. (C) Addition of YtnP to the biofilm formation assays inhibited biofilm formation in P. aeruginosa but not in S. aureus. Biofilm formation is observed as a pellicle attached to the submerged surfaces (at the bottom of the well plate). Crystal violet staining was used in the assay for better visualization, according to the protocol described by O'Toole and Kolter (37). For the quantification of biofilm formation, the crystal violet associated with biofilms was dissolved in acetic acid (33%) and was measured spectrophotometrically with an optical density at 595 nm (37).
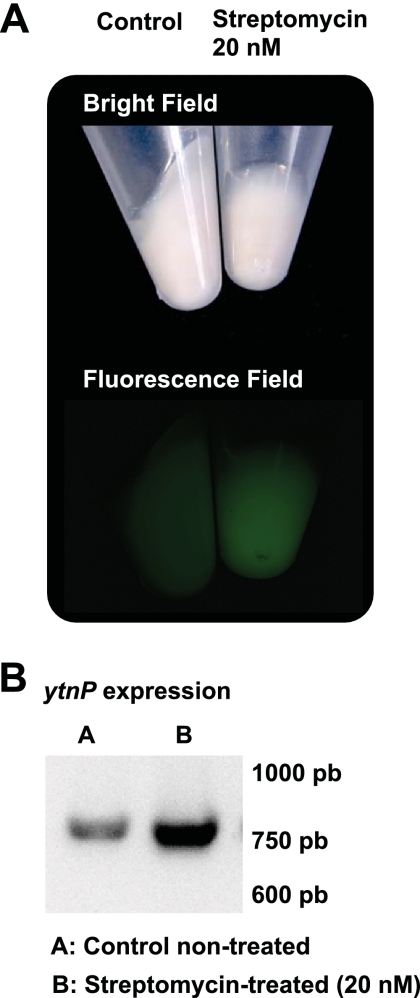
To determine whether the expression of ytnP in B. subtilis serves as a defensive strategy against competing bacteria, an experiment was carried out to test the activation of ytnP expression in the presence of several acylhomoserine lactone (AHL) molecules. To do this, B. subtilis strain 3610 was labeled with a transcriptional fusion, PytnP-yfp (yfp encodes the yellow fluorescence protein [YFP]), and the emission of fluorescence was monitored when small concentrations of AHL (purchased from Sigma-Aldrich) were added to cultures of B. subtilis growing in MSgg (2). We did not observe any change in ytnP expression associated with the addition of AHL molecules to the cultures of B. subtilis. Then, other natural products were tested for their abilities to induce ytnP expression in B. subtilis. This idea was based on the results published in a previous report that describes an increase in the expression of a protein similar to YtnP in S. aureus when certain antimicrobials were added to the cultures (1). Consequently, a battery of antibiotics was collected, and they were tested for their abilities to induce the expression of PytnP-YFP when added at sub-growth-inhibitory concentrations (<50 nM) (see Table S1 in the supplemental material). We found that streptomycin, an antimicrobial produced by the soil-dwelling bacterium Streptomyces griseus, increased the expression of the reporter PytnP-YFP approximately 5-fold when added at sub-growth-inhibitory concentrations to B. subtilis cultures (Fig. 2A) (41, 51). Reverse transcription-PCR (RT-PCR) experiments confirmed that cultures that were previously treated with sub-growth-inhibitory concentrations of streptomycin showed an increase in the expression of the ytnP gene (Fig. 2B).
Fig 2.
Streptomycin induces the expression of ytnP. (A) Fluorescence emitted by the expression of the reporter PytnP-yfp in B. subtilis when incubated in the presence or absence of streptomycin. Pellets of nontreated (control) and YtnP-treated (20 nM) cultures of B. subtilis PytnP-yfp are presented in bright field (top) and fluorescence field (bottom). (B) RT-PCR analysis of ytnP expression from nontreated and streptomycin-treated (20 nM) cultures of B. subtilis.
A search in the DBTBS software was performed to study the mechanisms responsible for the activation of ytnP expression in response to streptomycin. This search identified the CodY repressor as a regulatory protein involved in the regulation on ytnP expression (43). CodY loses activity and target genes are transcribed when the growth rate of B. subtilis slows down in response to adverse conditions (42, 44). Given that antimicrobials affect bacterial growth rate, B. subtilis probably responds to a decrease in growth rate with an induction of ytnP expression. Hence, a ΔcodY strain labeled with the PytnP-yfp reporter was constructed and the expression of the reporter in this strain was compared to the expression observed in the wild-type strain. The absence of codY increased the expression of ytnP 3- to 5-fold in B. subtilis cultures, similar to the addition of streptomycin (Fig. 2D). Activation of ytnP in response to cellular stress is consistent with results from a previous publication, which shows that the expression of AttM in A. tumefaciens increases in response to starvation and to other cellular stresses (54).
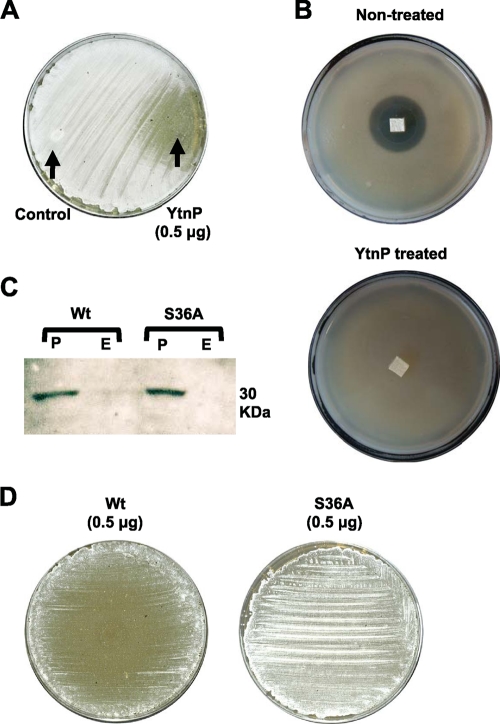
Although the aminoglycoside antibiotic streptomycin is not a lactone, the quorum-sensing signal that positively regulates streptomycin production and development of aerial hyphae in S. griseus is a γ-butyrolactone (termed A factor) (15, 18, 19, 25). We predicted that this molecule could be a putative target for YtnP. Thus, experiments were conducted to elucidate whether YtnP interferes with the development of S. griseus by assaying the ability of YtnP to inhibit the formation of aerial hyphae during S. griseus development. The 6×His-YtnP-tagged protein was overexpressed and purified in Bacillus subtilis PY79. A sample of purified YntP was spotted on an agar plate that had previously been planted with S. griseus spores. The aerial mycelium of S. griseus arose from the surfaces of the colonies, giving a white, hairy appearance to the agar plate. After 3 days of growth, the area subjected to the action of YtnP did not develop any aerial mycelium, lacking the characteristic fuzzy colony morphology of the wild type (Fig. 3A). Interestingly, YtnP did not inhibit the development of aerial hyphae on S. coelicolor (see Fig. S2 in the supplemental material). This might be due to the fact that the projection of the aerial mycelium in S. coelicolor is not regulated by the A factor (14, 16, 20, 47). Development of the aerial mycelium in S. griseus is intimately related to streptomycin production, since both processes are positively regulated by the A factor. Consequently, the inhibition of streptomycin production was monitored in cultures of S. griseus that were previously exposed to the activity of YtnP. Detection of streptomycin produced by S. griseus was based on the inhibitory halo that the secreted streptomycin caused in the soft agar that was previously planted with B. subtilis strain ATCC 6633 (9, 18, 21, 34). An agar section from a plate of S. griseus that was exposed to the activity of YtnP was excised and placed on the surface of the soft agar previously inoculated with B. subtilis ATCC 6633. After 24 h of incubation at 37°C, the agar section from YtnP-treated cultures of S. griseus showed no growth-inhibitory halo on B. subtilis ATCC 6633 (Fig. 3B), indicating that YtnP-treated cultures of S. griseus has decreased in streptomycin production.
Fig 3.
YtnP inhibits development of aerial mycelium in Streptomyces griseus. (A) Inhibition of the formation of the aerial mycelium in S. griseus when grown on R2YE agar medium (23). Purified YtnP was spotted in the agar (spotted on the right side of the plate). Negative control was solution buffer with bovine serum albumin (BSA) (spotted on the left side of the plate). The plate was incubated for 3 days at 30°C. (B) Quantification of streptomycin production from nontreated and YtnP-treated agar cultures of S. griseus by the formation of an inhibitory halo on a lawn of B. subtilis ATCC 6633 grown on soft agar. (C) Detection of YtnP (Wt) and YtnPS36A (S36A) alleles by Western blot analysis. Anti-6×His antibodies were used to detect the proteins. Signals exclusively occurred in the cell pellet (P), and no signal was found in the extracellular protein fraction (E). The band has a molecular mass of 30 kDa. (D) Inhibition of formation of the aerial mycelium in S. griseus caused by the two alleles YtnP (Wt) and YtnPS36A (S36A). An agar plate of R2YE medium was planted with S. griseus (23). Purified Wt and S36A alleles were spotted in the center of the agar. Plates were incubated for 3 days at 30°C. Development of the aerial mycelium could be observed by the white, hairy appearance that the formation of the aerial mycelium gives to the lawn of S. griseus growing on the plate. Development of the aerial mycelium occurred only in the plate complemented with the S36A allele.
Inhibition of the aerial mycelium in S. griseus was used to assay the activity of YtnP. A previous study of the phosphoproteome of B. subtilis identified a serine phosphorylation (Ser36) in YtnP, a widely spread mechanism of posttranslational regulation (31, 40). To investigate the effect that phosphorylation of Ser36 causes on the activity of YtnP, an allele of YtnP-6XHis was constructed harboring a Ser36Ala replacement (YtnPS36A). The YtnPS36A allele was overexpressed and purified in B. subtilis PY79. Immunological studies (via Western blot analysis) using antibodies against the 6×His tag showed similar expression of YtnPS36A and wild-type alleles (Fig. 3C). Purified YtnPS36A was assayed for inhibition of aerial mycelium of S. griseus by spotting 0.5 μg YtnPS36A on a plate previously planted with spores of S. griseus. After 3 days of growth, the purified YtnPS36A protein did not abrogate the development of aerial mycelium in S. griseus (Fig. 3D). This might indicate the importance of the phosphorylation of Ser36 for the activity of YtnP, albeit other indirect effects that the substitution of Ser36 might cause in the structure and functional activity of the enzyme YtnP should not be discarded.
Taken altogether, data from this report suggest that the expression of YtnP from B. subtilis is induced in the presence of streptomycin and other cellular stresses. The prior expression of YtnP interferes with the signaling pathway that leads to the development of the aerial mycelium and streptomycin production in S. griseus. B. subtilis might activate ytnP expression in response to the presence of certain antimicrobials, serving this as a defensive strategy against threatening bacteria because it would allow B. subtilis to selectively inhibit the quorum-sensing system of harmful microbial communities. The selective inhibition of quorum sensing would not occur if B. subtilis triggered the expression of ytnP in response to the presence of lactone molecules. Lactones are signaling molecules broadly found among prokaryotes, and the activation of YtnP expression in response to the presence of these signaling molecules will not allow B. subtilis to discriminate between threatening or potentially benign microbial communities that can generate beneficial interactions to B. subtilis. To ensure that the quorum-quenching activity of YtnP acts only in the presence of antimicrobials or other stressing factors that compromise the physiology of B. subtilis cells, YtnP is expressed as a cytoplasmic enzyme, as well as the rest of the quorum-quenching enzymes (11–13). Hence, the release of YtnP to the extracellular space occurs concomitantly with cell lysis. This might resemble other cases in microbiology, in which the release of proteins with extracellular activities requires cell lysis, as is the case with the secretion of TcdA and TcdB toxins in Clostridium difficile facilitated by TcdE, a holin-like protein responsible for cell lysis (48).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Young Investigator Research Program, from the Institute for Molecular Infection Biology (University of Würzburg). J. C. Garcia-Betancur is recipient of a Ph.D. fellowship from the Graduate School of Life Sciences (University of Würzburg).
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Balibar CJ, Shen X, Tao J. 2009. The mevalonate pathway of Staphylococcus aureus. J. Bacteriol. 191:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carfi A, Duee E, Galleni M, Frere JM, Dideberg O. 1998. 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus. Acta Crystallogr. D Biol. Crystallogr. 54:313–323 [DOI] [PubMed] [Google Scholar]
- 5. Carfi A, et al. 1995. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14:4914–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlier A, et al. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-Acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34:171–198 [DOI] [PubMed] [Google Scholar]
- 8. Davies DG, et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
- 9. Distler J, Mansouri K, Mayer G, Stockmann M, Piepersberg W. 1992. Streptomycin biosynthesis and its regulation in streptomycetes. Gene 115:105–111 [DOI] [PubMed] [Google Scholar]
- 10. Dong YH, Gusti AR, Zhang Q, Xu JL, Zhang LH. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong YH, et al. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813–817 [DOI] [PubMed] [Google Scholar]
- 12. Dong YH, Xu JL, Li XZ, Zhang LH. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U. S. A. 97:3526–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dong YH, Zhang LH. 2005. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43:101–109 [PubMed] [Google Scholar]
- 14. Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 15. Hara O, Beppu T. 1982. Mutants blocked in streptomycin production in Streptomyces griseus—the role of A-factor. J. Antibiot. (Tokyo) 35:349–358 [DOI] [PubMed] [Google Scholar]
- 16. Hara O, Horinouchi S, Uozumi T, Beppu T. 1983. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J. Gen. Microbiol. 129:2939–2944 [DOI] [PubMed] [Google Scholar]
- 17. Haudecoeur E, et al. 2009. Different regulation and roles of lactonases AiiB and AttM in Agrobacterium tumefaciens C58. Mol. Plant Microbe Interact. 22:529–537 [DOI] [PubMed] [Google Scholar]
- 18. Horinouchi S, Beppu T. 1993. A-factor and streptomycin biosynthesis in Streptomyces griseus. Antonie Van Leeuwenhoek 64:177–186 [DOI] [PubMed] [Google Scholar]
- 19. Horinouchi S, Beppu T. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859–864 [DOI] [PubMed] [Google Scholar]
- 20. Horinouchi S, Hara O, Beppu T. 1983. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J. Bacteriol. 155:1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horinouchi S, Kumada Y, Beppu T. 1984. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J. Bacteriol. 158:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller L, Surette MG. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4:249–258 [DOI] [PubMed] [Google Scholar]
- 23. Kieser T, Buttner BMJMJ, Chater K F, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England [Google Scholar]
- 24. Kim MH, et al. 2005. The molecular structure and catalytic mechanism of a quorum-quenching N-acyl-L-homoserine lactone hydrolase. Proc. Natl. Acad. Sci. U. S. A. 102:17606–17611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kudo N, Kimura M, Beppu T, Horinouchi S. 1995. Cloning and characterization of a gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 177:6401–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee SJ, et al. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li de la Sierra-Gallay I, Pellegrini O, Condon C. 2005. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433:657–661 [DOI] [PubMed] [Google Scholar]
- 28. Liu D, et al. 2008. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 47:7706–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu D, et al. 2007. Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry 46:11789–11799 [DOI] [PubMed] [Google Scholar]
- 30. López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Macek B, et al. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell Proteomics 6:697–707 [DOI] [PubMed] [Google Scholar]
- 32. McLean RJ, Whiteley M, Stickler DJ, Fuqua WC. 1997. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154:259–263 [DOI] [PubMed] [Google Scholar]
- 33. Mei GY, Yan XX, Turak A, Luo ZQ, Zhang LQ. 2010. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl. Environ. Microbiol. 76:4933–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyake K, Kuzuyama T, Horinouchi S, Beppu T. 1990. The A-factor-binding protein of Streptomyces griseus negatively controls streptomycin production and sporulation. J. Bacteriol. 172:3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Momb J, et al. 2008. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 2. Substrate modeling and active site mutations. Biochemistry 47:7715–7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Gara JP. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179–188 [DOI] [PubMed] [Google Scholar]
- 37. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 38. Otto M. 2008. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322:207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park SY, et al. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541–1550 [DOI] [PubMed] [Google Scholar]
- 40. Pereira SF, Goss L, Dworkin J. 2011. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75:192–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rake G, Donovick R. 1946. Studies on the nutritional requirements of Streptomyces griseus for the formation of streptomycin. J. Bacteriol. 52:223–226 [DOI] [PubMed] [Google Scholar]
- 42. Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sierro N, Makita Y, de Hoon M, Nakai K. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36:D93–D96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927 [DOI] [PubMed] [Google Scholar]
- 45. Stamp AL, et al. 2010. Structural and functional characterization of Salmonella enterica serovar Typhimurium YcbL: an unusual Type II glyoxalase. Protein Sci. 19:1897–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Straight PD, Kolter R. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99–118 [DOI] [PubMed] [Google Scholar]
- 47. Takano E. 2006. Gamma-butyrolactones: streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9:287–294 [DOI] [PubMed] [Google Scholar]
- 48. Tan KS, Wee BY, Song KP. 2001. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J. Med. Microbiol. 50:613–619 [DOI] [PubMed] [Google Scholar]
- 49. Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem. Rev. 111:117–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomas PW, Stone EM, Costello AL, Tierney DL, Fast W. 2005. The quorum-quenching lactonase from Bacillus thuringiensis is a metalloprotein. Biochemistry 44:7559–7569 [DOI] [PubMed] [Google Scholar]
- 51. Waksman SA, Reilly HC, Johnstone DB. 1946. Isolation of streptomycin-producing strains of Streptomyces griseus. J. Bacteriol. 52:393–397 [DOI] [PubMed] [Google Scholar]
- 52. Wang WZ, Morohoshi T, Ikenoya M, Someya N, Ikeda T. 2010. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 76:2524–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yim G, Wang HH, Davies J. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296:163–170 [DOI] [PubMed] [Google Scholar]
- 54. Zhang HB, Wang C, Zhang LH. 2004. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol. Microbiol. 52:1389–1401 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.