Abstract
Indole production by Escherichia coli, discovered in the early 20th century, has been used as a diagnostic marker for distinguishing E. coli from other enteric bacteria. By using transcriptional profiling and competition studies with defined mutants, we show that cyclic AMP (cAMP)-regulated indole formation is a major factor that enables E. coli growth in mixed biofilm and planktonic populations with Pseudomonas aeruginosa. Mutants deficient in cAMP production (cyaA) or the cAMP receptor gene (crp), as well as indole production (tnaA), were not competitive in coculture with P. aeruginosa but could be restored to wild-type competitiveness by supplementation with a physiologically relevant indole concentration. E. coli sdiA mutants, which lacked the receptor for both indole and N-acyl-homoserine lactones (AHLs), showed no change in competitive fitness, suggesting that indole acted directly on P. aeruginosa. An E. coli tnaA mutant strain regained wild-type competiveness if grown with P. aeruginosa AHL synthase (rhlI and rhlI lasI) mutants. In contrast to the wild type, P. aeruginosa AHL synthase mutants were unable to degrade indole. Indole produced during mixed-culture growth inhibited pyocyanin production and other AHL-regulated virulence factors in P. aeruginosa. Mixed-culture growth with P. aeruginosa stimulated indole formation in E. coli cpdA, which is unable to regulate cAMP levels, suggesting the potential for mixed-culture gene activation via cAMP. These findings illustrate how indole, an early described feature of E. coli central metabolism, can play a significant role in mixed-culture survival by inhibiting quorum-regulated competition factors in P. aeruginosa.
INTRODUCTION
In nature, bacteria normally occur in polymicrobial communities. Interactions between community members typically involve several mechanisms, including responses to antimicrobial compounds, nutritional interactions, and signaling (9, 12, 42). Chemical signaling is widespread in bacteria, and in Gram-negative bacteria it involves several compounds, including N-acyl derivatives of homoserine lactone (AHLs), furans, small peptides, quinolones, and indole (37). In Pseudomonas aeruginosa, many genes involved with virulence and competition are regulated by AHL- and quinolone-based quorum signaling (35, 50). Quorum signal disruption (quenching) has been shown to alter bacterial competition and reduce virulence (18). In one animal model study of P. aeruginosa lung infections, administration of a ginseng extract caused a reduction in AHL levels and AHL-regulated elastase without affecting bacterial growth (44). Microbial nutrition is also important in species' interactions. Central metabolism, long regarded as reflecting a number of routine housekeeping functions, is now being reexamined for its role in mixed-culture interactions within biofilm and planktonic populations (9).
One component of central metabolism in Escherichia coli involves purine and pyrimidine nucleic acid synthesis (reviewed in reference 36). Pathways for de novo synthesis and salvage pathways exist for both nucleotides. During purine synthesis, ribose-5-phosphate, an intermediate in the pentose phosphate cycle, is converted through a series of intermediates to the end products AMP and GMP. Aside from being nucleic acid components, purines serve other important functions in bacteria. These functions include energy transfer (ATP and GTP) and cell signaling [cyclic AMP (cAMP), bis-(3′-5′)-cyclic di-GMP (c-di-GMP), and guanosine tetraphosphate (ppGpp)] (11, 17). The secondary signal molecule, cAMP, is synthesized from AMP by adenylate cyclase, which is encoded by cyaA (21) and is broken down to 5′-AMP by cAMP phosphodiesterase, encoded by cpdA (22). The receptor for cAMP is the cAMP receptor protein, encoded by crp (21). The second messenger cAMP has been linked to a number of cellular functions in E. coli, most notably regulation of carbon catabolism (8) and more recently a number of stress responses (14).
One E. coli metabolite, indole, is produced from the amino acid tryptophan by tryptophanase (encoded by tnaA), which is regulated by cAMP (23). Since the discovery of its formation from tryptophan in the early 20th century (19), indole production has been employed as a biochemical test for distinguishing E. coli from other members of the Enterobacteriaceae (7). Here we show that cAMP-regulated indole production, traditionally associated with central metabolism, quenches the production of AHL-regulated pyocyanin production in P. aeruginosa (29) and facilitates E. coli growth in mixed culture.
MATERIALS AND METHODS
Bacterial strains and media.
The strains used in this study are listed in Table 1 and were maintained in Luria-Bertani (LB) medium supplemented with kanamycin (50 μg/ml) for E. coli mutants. Prior to experimentation, cultures were revived from frozen stocks and cultured overnight in LB agar. The cultures were then subcultured in LB broth and incubated overnight at 37°C with aeration (100 rpm). Unless stated otherwise, during experimentation the turbidity of the overnight cultures was adjusted to an optical density at 600 nm (OD600) of 0.1 ([1.08 ± 0.15] × 105 CFU/ml [mean ± standard error of the mean]) using sterile LB broth. During this study, we used several strains of E. coli, including ZK126 and MG1655 (for transcriptional profiling) and BW25113 (for the majority of the gene investigations, due to the availability of a mutant collection for it [3]). All three strains are E. coli K-12 derivatives (3, 6). During preliminary experiments, we tested several media to differentiate E. coli and P. aeruginosa during dilution plating. Strain ZK126 showed the most promise in exhibiting different colony morphologies from P. aeruginosa on chromogenic agar (Hi-Chrome ECC agar; Fluka) and so was used initially. However, due to concern about colony appearance being affected by mixed growth, we developed selective media with LB plus ampicillin (100 μg/ml) to select for P. aeruginosa and LB plus cefsulodin (20 μg/ml) to select for E. coli (27). In preliminary experiments all three E. coli parent strains showed similar competition profiles in mixed culture. Although spontaneous E. coli ampicillin-resistant and P. aeruginosa cefsulodin-resistant mutants did occur, the estimated frequency was 10−6, so we do not consider spontaneous mutations to be a source of error.
Table 1.
Bacterial strains used
| Species and strain | Description | Source and/or reference(s)a |
|---|---|---|
| E. coli BW25113 | wt | 3 |
| E. coli BW25113 cyaA::kan | Lacks adenylate cyclase gene | 3, 48 |
| E. coli BW25113 cpd::kan | Lacks cAMP-phosphodiesterase gene | 3, 22 |
| E. coli BW25113 crp::kan | Lacks cAMP receptor protein | 3, 14 |
| E. coli BW25113 sdiA::kan | Lacks luxR homologue (receptor for AHLs and indole) | 3, 30, 43 |
| E. coli BW25113 tnaA::kan | Lacks tryptophanase | 3, 23 |
| E. coli MG1655 | wt | D. A. Siegele (6) |
| E. coli MG1655 purH | Lacks IMP cyclohydrolase (purine synthesis) | F. R. Blattner (25) |
| E. coli MG1655 purD | Lacks phosphoribosyltransferase (purine synthesis) | F. R. Blattner (25) |
| E. coli ZK126 | wt | D. A. Siegele (1) |
| Pseudomonas aeruginosa PAO1 | wt | V. Deretic (45) |
| P. aeruginosa PDO100 ΔrhlI::Tn501 | Lacks AHL synthase for C4-HSL | E. P. Greenberg (50) |
| P. aeruginosa PAO-MW1 ΔrhlI ΔlasI | Lacks AHL synthases for C4-HSL and 3-o-C12-HSL | E. P. Greenberg (50) |
E. coli BW25113 strains were obtained from the Genome Analysis Project in Japan.
Microarray.
P. aeruginosa PAO1 and E. coli ZK126 were grown in LB at 37°C with a shaking speed of 200 rpm to an OD600 of 0.3. The cultures were mixed 1:1 and subsequently incubated for an additional 45 min at 37°C. All cultures were mixed 1:1 with RNAlater (Ambion) and stored at 4°C for a maximum of 2 weeks until processed. RNA extraction, purification, processing for cDNA, and microarray analysis using E. coli and P. aeruginosa Gene Chip arrays (Affymetrix) were performed as described previously (34). During RNA purification, we monitored genomic DNA contamination by multiplex PCR using lacZ as an indicator of E. coli (primers Ec-LacZ-F, 5′-ACT ATC CCG ACC GCC TTA CT-3′; Ec-LacZ-R, 5′-TAG CGG CTG ATG TTG AAC TG-3′) and rplU as an indicator of P. aeruginosa genomic contamination (primers and PCR conditions were as described in reference 20).
Competition experiments.
For mixed-culture experiments, cells were inoculated into 50 ml LB as described above. The 125-ml culture flasks contained eight sterile silicon discs (7-mm diameter) (49) as biofilm colonization substrata. Competition experiments were performed for up to 7 days. Samples were removed periodically for dilution plating (27) and chemical analysis. Pure culture experiments for each strain were also conducted as controls. Each experiment was replicated a minimum of three times.
Exogenous addition of cAMP and indole.
Stock solutions of cAMP and indole were prepared in distilled and dimethyl formamide, respectively, and sterilized by filtration (0.2-μm pore size) prior to use. To investigate whether exogenous addition of cAMP or indole could restore competitiveness of the mutant strains of E. coli in mixed culture growth with PAO1, these compounds were added to LB prior to experimentation at different concentrations (0.2, 0.5, and 1 mM), and the competition experiments were conducted as described above. We also investigated if indole could inhibit growth of P. aeruginosa or promote growth of E. coli and thus account for a change in E. coli competitiveness. Here, E. coli BW25113 and P. aeruginosa PAO1 were grown (37°C, 100 rpm) as monocultures in LB broth in 125-ml sidearm flasks, in the presence (1 mM) or absence of indole, and growth was monitored by turbidity (OD600).
Indole assay.
Bacterial cultures were tested for indole production in pure and mixed cultures by using the protocol described by Kawamura-Sato et al. (26).
P. aeruginosa virulence factor assays.
Elastase activity was determined by using the Congo red elastin protocol (51). Colorimetric pyocyanin analysis was conducted using the chloroform extraction and HCl acidification protocol (13).
Data analysis.
Data calculations were conducted using Excel (Microsoft Office 2007). Statistical analysis and graphing were performed using Sigma Plot v. 11 (Systat Software Inc.).
Microarray sequence accession number.
The gene array results have been deposited in Gene Expression Omnibus (GEO) under accession number GSE26931.
RESULTS
Microarray.
We conducted microarray analysis on planktonic cocultures of P. aeruginosa PAO1 and E. coli ZK126 (Table 1) grown in LB. As shown in Table S1 of the supplemental material, a total of 162 genes (approximately 3% of the genome) were differentially expressed in the E. coli mixed culture, whereas only 10 genes (approximately 0.1%) were differentially expressed in the P. aeruginosa coculture. Interestingly, many genes upregulated in E. coli mixed cultures were involved in de novo purine biosynthesis (see Table S1). In contrast, pyrimidine synthesis genes were unaffected, which rules out a requirement for DNA or RNA synthesis. Similar upregulation in de novo purine synthesis genes was seen in microarray analysis of mixed culture E. coli MG1655 and P. aeruginosa PAO1 grown for 5 h in a chemically defined minimal medium (38) with vitamin supplementation (10) containing 1 mM N-acetylglucosamine as a carbon source (see Table S2 in the supplemental material) (GEO accession number GSE26932), suggesting that E coli purine upregulation in coculture is not strain or medium dependent. Nevertheless, we cannot rule out cross-hybridization of E. coli cDNA on P. aeruginosa gene chips and vice versa.
Competitiveness of purine mutants.
We tested the competitiveness of E. coli strains lacking two different genes in de novo purine synthesis (purD and purH) in mixed culture but observed no significant changes in planktonic or biofilm cultures of either E. coli or P. aeruginosa (see Fig. S1 in the supplemental material). It is conceivable that the salvage pathway for purine synthesis (36) allowed E. coli purD and purH strains to compensate for these mutations.
cAMP.
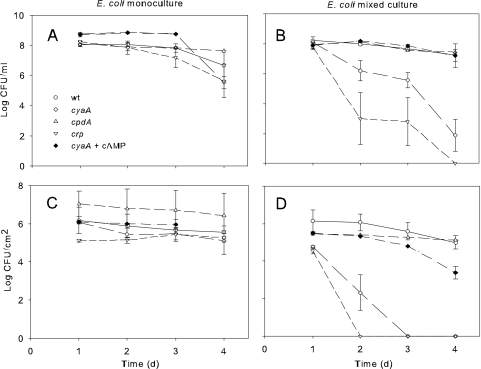
Aside from being a nucleic acid component, the purine adenine is a component of the signal molecule cAMP. To explore this possibility, we explored the competitiveness of E. coli in mixed culture by using strains deficient in cAMP production (cyaA), reception (crp), or intracellular regulation (cpdA). As shown in Fig. 1, E. coli cAMP mutants were not affected in monoculture planktonic (Fig. 1A) or biofilm (Fig. 1C) cultures. In coculture, significant loss of E. coli competitiveness occurred in both planktonic (Fig. 1B) and biofilm (Fig. 1D) populations in mutants unable to produce cAMP (cyaA) or lacking the cAMP receptor gene (crp) (14). Competitiveness in the cyaA but not crp mutants could be restored to wild-type (wt) levels for 24 h by addition of 1 mM cAMP (Fig. 1C and D); however, after 48 h these effects had disappeared, likely due to cAMP depletion. Surprisingly, there was no change in the cAMP degradation (cpdA) mutant (22) due to cAMP supplementation in monoculture (Fig. 1A and C) or mixed culture (Fig. 1B and D), suggesting that the levels of cAMP present were not inhibitory to E. coli under our growth conditions. P. aeruginosa planktonic and biofilm populations were unaffected by cAMP disruption in E. coli (see Fig. S2 in the supplemental material). Based on these results, we conclude that cAMP plays a key role in E. coli competitiveness in mixed culture.
Fig 1.
Influence of mutations in E. coli cAMP production (adenylate cyclase [cyaA]), cAMP receptor gene ([crp]), and cAMP phosphodiesterase, encoded by cpdA on E. coli monoculture planktonic (A) and biofilm (C) cultures and E. coli populations in mixed-planktonic (B) and biofilm cultures (D) with P. aeruginosa. The growth decline in E. coli cyaA mixed cultures was reversed by the addition of 1 mM cAMP. The growth medium in all figures is LB. Error bars in all figures represent standard errors of the means (n ≥ 3).
Indole.
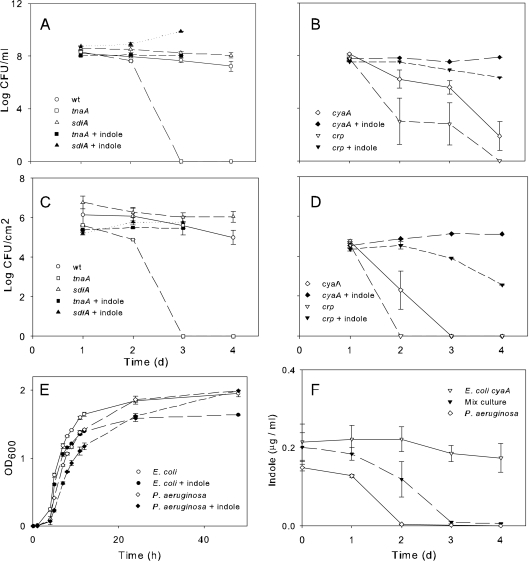
As shown previously (23), we found that indole production was absent in E. coli crp and reduced in cyaA mutants. We tested mutants deficient in indole production (tnaA) and the indole receptor (sdiA) (30). As shown in Fig. 2, the E. coli tnaA mutant was notably less competitive than its wild-type counterparts in both planktonic (Fig. 2A) and biofilm (Fig. 2C) mixed cultures. In both cases, supplementation with 1 mM indole restored mutant populations to wild-type levels for 24 h (Fig. 2A and C). Population levels of the sdiA mutant were not significantly different from the wild type, but its competitive ability was enhanced by indole supplementation (Fig. 2A and C). P. aeruginosa populations were not affected (see Fig. S2 in the supplemental material). Indole supplementation also restored the competitiveness of the cAMP mutants cyaA and crp (Fig. 2B and D). In order to determine whether indole altered growth, we grew E. coli and P. aeruginosa monocultures in the presence and absence of 1 mM indole and observed a slight indole-mediated inhibition of growth of both cultures (Fig. 2E). The most prominent effects were seen at physiologically relevant concentrations between 500 μM and 1 mM (47) (Fig. 2E shows the effects in 1 mM indole). At higher concentrations (≥5 mM), indole was toxic to both E. coli and P. aeruginosa and no growth occurred. With P. aeruginosa, the growth inhibition caused by indole disappeared between 24 and 48 h. As shown in Fig. 2F, indole was degraded by P. aeruginosa after 24 to 48 h in pure or mixed cultures and not by E. coli, explaining the transient effects of indole supplementation seen in Fig. 2A to E.
Fig 2.
Influence of mutations in E. coli indole production (tryptophanase [tnaA]) and receptor gene (sdiA) on growth in mixed culture with P. aeruginosa in planktonic (A) and biofilm (C) cultures. Addition of 1 mM indole restored competitiveness of E. coli tryptophanase mutants (tnaA) to wt levels (A and C) as well as strains lacking adenylate cyclase (cyaA) and the cAMP receptor gene (crp) (B and D). (E) Monoculture growth of E. coli and P. aeruginosa in the presence and absence of 1 mM indole. Supplemented indole was metabolized by P. aeruginosa over 48 h in pure or mixed culture with an E. coli strain unable to produce indole (such as E. coli cyaA [panel F]).
Coculture stimulates indole formation.
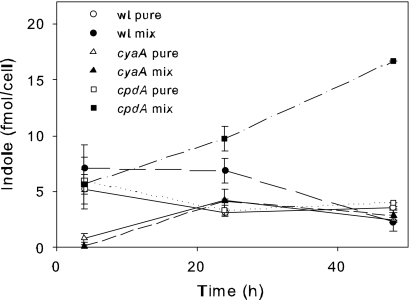
We investigated whether coculture influenced indole production in E. coli. As shown in Fig. 3, mixed-culture growth induced indole production in E. coli; however, this effect was only seen with the E. coli cpdA strain. These findings suggest that mixed culture growth can enhance cAMP, which in turn promotes indole production. However, this effect did not occur in E. coli populations in which cAMP levels were moderated through cpdA-mediated homeostasis (22).
Fig 3.
Influence of pure and mixed-culture growth on indole production in wt and cyaA and cpdA mutants. Indole concentrations were calculated per E. coli cell.
Mixed-culture growth inhibits pyocyanin and other AHL-regulated virulence factors.
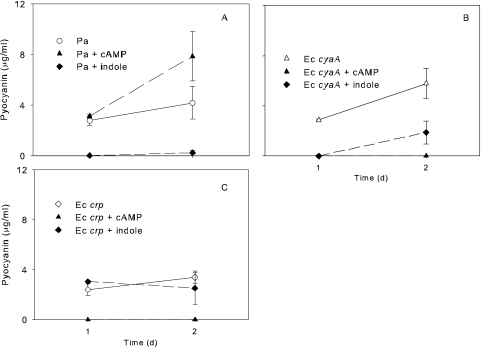
One consistent observation was that P. aeruginosa cultures became nonpigmented during mixed-culture growth. We measured pyocyanin production by P. aeruginosa in pure and mixed cultures (Fig. 4). Under all conditions tested, pyocyanin production was abolished in the presence of indole. In pure culture, addition of cAMP enhanced pyocyanin production (Fig. 4A). However, in mixed culture with E. coli cyaA (Fig. 4B), addition of cAMP decreased pyocyanin, likely due to indole formation. In mixed culture with the E. coli crp mutant (Fig. 4C), there was no significant change in pyocyanin formation by P. aeruginosa regardless of the presence or absence of cAMP. As pyocyanin production is controlled by quorum signaling (41), we investigated the influence of indole on other AHL-regulated virulence factors of P. aeruginosa as well as P. aeruginosa strains deficient in AHL production. As measured colorimetrically by dye release with the Congo red elastin protocol (51), elastase production in 24-h P. aeruginosa cultures, normalized to P. aeruginosa cell density in monoculture, dropped from an OD495 of 0.744 ± 0.027 in pure cultures to a 0.023 ± 0.011 (P < 0.001) when P. aeruginosa was grown in mixed culture with indole-producing E. coli strains. In the same time frame, mixed-culture growth with wt E. coli caused a small reduction in P. aeruginosa cell density (log CFU/ml for pure culture, 9.87 ± 0.35, compared with log CFU/ml of the mixed culture, 8.64 ± 0.21; P = 0.04). There was a statistically insignificant reduction in P. aeruginosa cell density when it was grown with the E. coli tnaA strain (log CFU/ml, 9.61 ± 0.07; P = 0.57). While the lower P. aeruginosa cell density in mixed culture could certainly contribute to a reduction in quorum-regulated elastase, two other studies by Lee et al. (29, 33) have shown indole to inhibit P. aeruginosa quorum-regulated virulence factors, including elastase and pyocyanin. On the basis of work reported by Lee et al. (29, 33) and our own studies, we attribute the reductions in elastase and pyocyanin to E. coli indole production.
Fig 4.
Effects of 1 mM cAMP and 1 mM indole supplementation on pyocyanin production by P. aeruginosa in pure culture (A) and mixed culture with E. coli cyaA (B) or E. coli crp (C). The two E. coli strains are unable to produce indole.
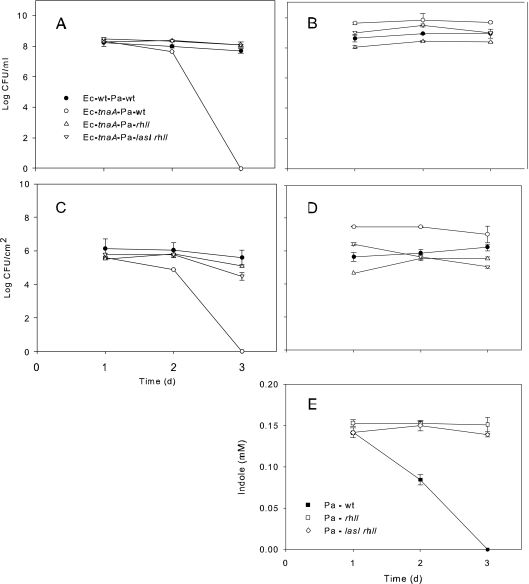
In order to further explore the connection between E. coli indole production and P. aeruginosa AHL production, we tested the competitiveness of E. coli wt and tnaA strains with P. aeruginosa strains defective in one (rhlI) or both (lasI rhlI) AHL synthase genes (50). As shown in Fig. 5, E. coli competitiveness in planktonic (Fig. 5A) and biofilm (Fig. 5C) cultures was restored in tnaA mutants when cocultured against P. aeruginosa strains deficient in one (rhlI) or both (lasI rhlI) AHL synthase genes. In comparison to coculture with wt E. coli, P. aeruginosa cell numbers increased in mixed planktonic (Fig. 5B) and biofilm (Fig. 5D) populations during coculture with E. coli tnaA. Interestingly, there was a small but significant decline in the P. aeruginosa rhlI mutant, but not in the lasI rhlI mutant, in biofilm populations (Fig. 5D) during coculture with E. coli. One explanation may relate to the inability of P. aeruginosa AHL synthesis mutants to degrade indole (Fig. 5E) and reduce its growth inhibition (Fig. 2E). Based on our results, we conclude that the major protective effect of indole during E. coli mixed-culture growth is due to direct inhibition of pyocyanin and other quorum-regulated competition factors of P. aeruginosa.
Fig 5.
Interaction of E. coli tryptophanase (tnaA) and P. aeruginosa AHL synthesis (lasI and rhlI) on E. coli (A [planktonic] and C [biofilm]) and P. aeruginosa (B [planktonic] and D [biofilm]) growth in mixed culture. Symbols for panels A to D are shown in panel A. Data for growth of E. coli wt with P. aeruginosa wt and of E. coli tnaA with P. aeruginosa wt from Fig. 2 are shown again for comparison. (E) In contrast to the wild type, P. aeruginosa AHL synthesis mutants were unable to degrade indole.
DISCUSSION
In nature most bacteria exist within mixed populations. In terms of broad-based genetic studies (involving transcriptomic or proteomic approaches), most mixed-culture investigations have explored a Gram-positive and a Gram-negative organism grown in coculture. Because of fundamental differences in their cell wall architecture (5), one can selectively lyse one member of the consortium and extract desired cellular materials for analysis. These studies have been useful in highlighting some of the novel aspects of mixed-culture interactions (10, 34, 40). When the Gram-negative dental pathogen Aggregatibacter actinomycetemcomitans is grown in mixed culture with the Gram-positive commensal Streptococcus gordonii, it shifts its energy source to lactate from glucose (10), but it also gains enhanced serum protection that can enable it to enter the circulatory system (40). In another example, coculture of P. aeruginosa and Staphylococcus aureus resulted in P. aeruginosa lysing S. aureus as an iron source (34), a process mediated by outer membrane vesicles produced in P. aeruginosa through the action of the Pseudomonas quinolone signal (PQS) (35). PQS is a quorum signal in P. aeruginosa under the control of the las and rhl AHL quorum system (39). In one study involving two Gram-negative organisms, An et al. (2) showed motility genes, associated with biofilm formation and quorum signal regulation, enhanced P. aeruginosa competition with Agrobacterium tumefaciens in this mixed-culture system. Of relevance to the current study, P. aeruginosa mutants deficient in AHL-mediated quorum signaling were less competitive against A. tumefaciens than the wild type. While A. tumefaciens also has an AHL-regulated quorum-signaling system (2), this system did not appear to play a role during its interactions with P. aeruginosa. In the An et al. study (2), the enhanced growth rate of P. aeruginosa relative to A. tumefaciens also provided a competitive advantage to P. aeruginosa. In our study, the growth rates of P. aeruginosa and E. coli, at least in monoculture, were similar (Fig. 2E), so we do not consider the growth rate differences to be important. One common theme from these aforementioned mixed-culture studies involving P. aeruginosa (2, 35) appears to be a role of quorum-regulated competition factors. We also found P. aeruginosa AHL-regulated quorum signaling to be important in its interactions with E. coli (Fig. 5), and we address this issue below. In general, microorganisms appear to adopt several strategies for mixed-culture survival, including nutritional and metabolic flexibility, mixed-culture-induced gene (mcg) expression, and signaling. These strategies must be considered in context with previously identified mechanisms, including bacteriocins, resource competition, and generation of harmful metabolites, that are employed during competition (9, 12).
Based on transcriptional profiling results showing elevated E. coli purine synthesis in mixed culture (see Table S1 in the supplemental material), we investigated the growth of E. coli purD and purH strains in mixed culture and found no loss of competitiveness (see Fig. S1 in the supplemental material), possibly due to the action of the purine salvage pathway (36). Pyrimidine synthesis genes were unaffected by mixed culture (see Table S1), which discounts a requirement for enhanced nucleic acid synthesis. Although ATP and GTP levels were not measured, we saw no evidence that E. coli growth was inhibited in purine synthesis mutants (purD and purH) either during growth in pure culture or in mixed culture (see Fig. S1), as might be expected from energy depletion. Aside from cAMP, purines are also involved in other signaling systems in E. coli, notably c-di-GMP and ppGpp (17). In contrast to wt, E. coli relA and relA spoT stringent response mutants that are defective in ppGpp synthesis show inhibited growth in pure culture (4) and so were not investigated in coculture, as growth inhibition would mask any competitive disadvantage. The effect of c-di-GMP on mixed-culture growth remains to be investigated. However, the most significant changes in E. coli competition were due to cAMP disruption (Fig. 1), which became the focus of this study.
In E. coli, cAMP and its receptor protein, Crp, have been traditionally associated with glucose-mediated catabolite repression (8). Transcriptome analyses (15) have shown that approximately 200 operons are regulated directly or indirectly by cAMP. Aside from carbon catabolism, many other cell functions are now recognized as components of the Crp regulon, including some genes associated with stress responses, cell division, and amino acid metabolism, including the tryptophanase gene tnaA (23). During mixed-culture growth, E. coli would need to be flexible for nutrient utilization as individual carbon sources are depleted and metabolites accumulate, including some that may induce stress. From both a nutrition and stress response perspective, a central role of cAMP in E. coli mixed-culture growth is logical (Fig. 1).
During competition with other species, P. aeruginosa produces a number of toxic compounds that are regulated by quorum signaling. These include pyocyanin and other phenazine compounds (16), rhamnolipids (24), and membrane vesicles (35). Pyocyanin enhances oxidative stress for competing organisms (16). Pyocyanin and membrane vesicles are toxic against Staphylococcus aureus during pulmonary infections in cystic fibrosis (35), and rhamnolipids have been shown to be toxic against eukaryotic cells (24). As well, rhamnolipids enhance swarming motility in P. aeruginosa (28). It was quite notable that coculture with wild-type E. coli caused a decrease in several AHL-regulated features in P. aeruginosa. These included pyocyanin production (Fig. 4) and elastase (described above). Inhibition of protease and pyocyanin was not present in the cAMP mutants cyaA and cpdA or in the tnaA mutant (data not shown), but it could be restored upon supplementation with 1 mM indole. Our observations of indole-based inhibition of P. aeruginosa AHL-regulated elastase and pyocyanin are in agreement with those reported by Lee et al. (29). E. coli tnaA strains regained competitive fitness during coculture with P. aeruginosa rhlI and lasI rhlI AHL synthesis mutants (Fig. 5A and C). As well, P. aeruginosa AHL synthesis mutants were unable to degrade exogenously supplied indole (Fig. 5E), giving further support to the interactions between AHL signaling and indole. The mechanism of indole degradation is not known but is under investigation. Based on our results and those of Lee et al. (29), it would appear that rhl-regulated genes are involved in indole degradation (Fig. 5E), but both las- and rhl-regulated genes in P. aeruginosa are affected by indole. In contrast to the culture data, we did not observe any alterations in AHL-mediated gene expression during transcriptome studies. The changes in competition (Fig. 1 and 2) occurred after 24 h, whereas the transcriptome analyses were done in early-log-phase cultures (45 min in LB and 3 to 4 h in defined medium). Quorum signaling typically begins in late-log-phase and stationary-phase cultures (37), which occurred after 12 h (Fig. 2E), and was therefore not detected. Indole and related compounds have been shown to inhibit pyocyanin and other quorum-regulated virulence factors in P. aeruginosa (29), although the role of indole in microbial competition has not been previously described.
Indole has been shown to be a cell signal molecule (30, 47) in E. coli. Functions associated with indole regulation in E. coli include amino acid catabolism (47), acid resistance, biofilm inhibition, and motility (30). In this context, it was important to determine whether any competition-enhancing effect was due to a direct influence of indole on P. aeruginosa or whether it was due to an indirect effect via indole-mediated gene expression in E. coli. Deletion of the E. coli indole receptor, sdiA (30), had no effect on E. coli competition in either planktonic (Fig. 2A) or biofilm (Fig. 2C) populations, which discounts an indirect effect. Although E. coli does not produce AHLs (it lacks a luxI homologue), it has a gene, sdiA, that functions as a luxR homologue (30, 31) and could conceivably use and deplete AHLs produced by other organisms. Our results (Fig. 2A and C) do not support sdiA-mediated quorum quenching as a factor in E. coli competitiveness under our experimental conditions. With respect to indole signaling, our investigations were performed at 37°C, whereas E. coli indole signaling is only prominent at 25 to 30°C and largely absent at 37°C (30, 32). Interestingly, the loss of competition of cAMP mutants cyaA and crp could be reversed by supplementation with physiologically relevant concentrations of indole (47) (Fig. 2B and D). As shown in Fig. 3, we saw that mixed-culture growth had the potential to enhance indole production via increasing cAMP levels in E. coli cpdA cells but not in other strains. As cpdA-encoded phosphodiesterase removes excess intracellular cAMP levels in E. coli (22), this observation shows that exposure to P. aeruginosa has the potential to increase indole production via enhanced cAMP levels, but any stimulation is buffered by cAMP reduction by the cpdA gene product (22). P. aeruginosa produces a number of metabolites during growth (46), and further investigation is needed to identify the metabolites that may impact E. coli indole production.
In summary, we conclude that cAMP and indole enhance E. coli during mixed-culture growth with P. aeruginosa through the indole-based inhibition of several las- and rhl-regulated P. aeruginosa virulence factors, notably pyocyanin. P. aeruginosa indole degradation appears to require rhl-mediated quorum signaling. Similar beneficial effects of indole on E. coli competition were seen in both biofilm and planktonic populations, and so we interpreted this phenomenon as a global effect, rather than a planktonic- or biofilm-specific effect, as reported in other studies (27, 49). While indole production by bacteria has been known for over a century (19), this study shows that this metabolite provides a key mechanism to explain the natural ecological success of E. coli in mixed communities.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grants from the Norman Hackerman Advanced Research Program of the Texas Higher Education Coordinating Board to R.J.C.M. and M.W. (award 003615-0037-2007).
We thank Matt Ramsey and Qun Ma for assistance, G. Eagleson for advice, and F. R. Blattner, V. Deretic, C. Fuqua, E. P. Greenberg, M. J. Schurr, and D. A. Siegele for cultures.
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adams JL, McLean RJC. 1999. The impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An D, Danhorn T, Fuqua C, Parsek MR. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. U. S. A. 103:3828–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balzer GJ, McLean RJC. 2002. The stringent response genes relA and spoT are important for Escherichia coli biofilms under slow-growth conditions. Can. J. Microbiol. 48:675–680 [DOI] [PubMed] [Google Scholar]
- 5. Beveridge TJ. 1981. Ultrastructure, chemistry, and function of the bacterial wall. Int. Rev. Cytol. 72:229–317 [DOI] [PubMed] [Google Scholar]
- 6. Blattner FR, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1474 [DOI] [PubMed] [Google Scholar]
- 7. Blazevic DJ, Ederer GM. 1975. Principals of biochemical tests in diagnostic microbiology. John Wiley and Sons, New York, NY [Google Scholar]
- 8. Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol. Mol. Biol. Rev. 56:100–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown SA, Palmer KL, Whiteley M. 2008. Revisiting the host as a growth medium. Nat. Rev. Microbiol. 6:657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown SA, Whiteley M. 2007. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189:6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cashel M, Gentry DR, Hernandez VJ, Vinella D. 1996. The stringent response, p. 1458–1496 In Niedhardt FC, et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 12. Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Essar DW, Eberly L, Hadero A, Crawford IP. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186:3516–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gutierrez-Ríos RM, et al. 2007. Identification of regulatory network topological units coordinating the genome-wide transcriptional response to glucose in Escherichia coli. BMC Microbiol. 7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hassan HM, Fridovich I. 1980. Mechanism of the antibiotic action pyocyanine. J. Bacteriol. 141:156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 18. Hentzer M, et al. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hopkins FG, Cole SW. 1903. A contribution to the chemistry of proteids. Part II. The constitution of tryptophane, and the action of bacteria upon it. J. Physiol. 29:451–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huse HK, et al. 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. mBio 1:e00199–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Husnain SI, Busby SJ, Thomas MS. 2009. Downregulation of the Escherichia coli guaB promoter by upstream-bound cyclic AMP receptor protein. J. Bacteriol. 191:6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imamura R, et al. 1996. Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 271:25423–25429 [DOI] [PubMed] [Google Scholar]
- 23. Isaacs H, Chao D, Yanofsky C, Saier MH. 1994. Mechanism of catabolite repression of tryptophanase synthesis in Escherichia coli. Microbiology 140:2125–2134 [DOI] [PubMed] [Google Scholar]
- 24. Jensen PO, et al. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329–1338 [DOI] [PubMed] [Google Scholar]
- 25. Kang Y, et al. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kawamura-Sato K, et al. 1999. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179:345–352 [DOI] [PubMed] [Google Scholar]
- 27. Kay MK, Erwin TC, McLean RJC, Aron GM. 2011. Bacteriophage ecology in Escherichia coli and Pseudomonas aeruginosa mixed biofilm communities. Appl. Environ. Microbiol. 77:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Köhler T, Kocjancic Curty L, Barja F, Van Delden C, Pechère JC. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee J, Attila C, Cirillo SL, Cirillo JD, Wood TK. 2009. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2:75–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee J, Maeda T, Hong SH, Wood TK. 2009. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75:1703–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J, et al. 2008. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2:1007–1023 [DOI] [PubMed] [Google Scholar]
- 33. Lee JH, Cho MH, Lee J. 2011. 3-Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ. Microbiol. 13:62–73 [DOI] [PubMed] [Google Scholar]
- 34. Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425 [DOI] [PubMed] [Google Scholar]
- 36. Neuhard J, Nygaard P. 1987. Purines and pyrimidines, p. 445–473 In Niedhardt FC, et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC [Google Scholar]
- 37. Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palmer KL, Mashburn LM, Singh PK, Whiteley M. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pesci EC, et al. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramsey MM, Whiteley M. 2009. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U. S. A. 106:1578–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reimmann C, et al. 2002. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148:923–932 [DOI] [PubMed] [Google Scholar]
- 42. Russell AB, et al. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sitnikov DM, Schineller JB, Baldwin TO. 1996. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. U. S. A. 93:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song Z, et al. 2010. Panax ginseng has anti-infective activity against opportunistic pathogen Pseudomonas aeruginosa by inhibiting quorum sensing, a bacterial communication process critical for establishing infection. Phytomedicine 17:1040–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 46. Vinayavekhin N, Saghatelian A. 2009. Regulation of alkyl-dihydrothiazole-carboxylates (ATCs) by iron and the pyochelin gene cluster in Pseudomonas aeruginosa. ACS Chem. Biol. 4:617–623 [DOI] [PubMed] [Google Scholar]
- 47. Wang D, Ding X, Rather PN. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wayne PK, Rosen OM. 1974. Cyclic 3′:5′-adenosine monophosphate in Escherichia coli during transient and catabolite repression. Proc. Natl. Acad. Sci. U. S. A. 71:1436–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weber MM, French CL, Barnes MB, Siegele DA, McLean RJC. 2010. A previously uncharacterized gene, yjfO (bsmA), influences Escherichia coli biofilm formation and stress response. Microbiology 156:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winson MK, et al. 1995. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92:9427–9431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.