Abstract
Pseudomonas putida KT2440 is capable of producing medium-chain-length polyhydroxyalkanoates (MCL-PHAs) when grown on unrelated carbon sources during nutrient limitation. Transcription levels of genes putatively involved in PHA biosynthesis were assessed by quantitative real-time PCR (qRT-PCR) in P. putida grown on glycerol as a sole carbon source. The results showed that two genes, phaG and the PP0763 gene, were highly upregulated among genes potentially involved in the biosynthesis of MCL-PHAs from unrelated carbon sources. Previous studies have described phaG as a 3-hydroxyacyl-acyl carrier protein (ACP)-coenzyme A (CoA) transferase, and based on homology, the PP0763 gene was predicted to encode a medium-chain-fatty-acid CoA ligase. High expression levels of these genes during PHA production in P. putida led to the hypothesis that these two genes are involved in PHA biosynthesis from non-fatty acid carbon sources, such as glucose and glycerol. The phaGpp and PP0763 genes from P. putida were cloned and coexpressed with the engineered Pseudomonas sp. 61-3 PHA synthase gene phaCl (STQK)ps in recombinant Escherichia coli. Up to 400 mg liter−1 MCL-PHAs was successfully produced from glucose. This study has produced the largest amount of MCL-PHAs reported from non-fatty acid carbon sources in recombinant E. coli to date and opens up the possibility of using inexpensive feedstocks to produce MCL-PHA polymers.
INTRODUCTION
Polyhydroxyalkanoates (PHAs) are carbon and energy storage materials synthesized by a variety of bacteria when grown under nutrient limitation in the presence of excess carbon (10, 11). These polymers have attracted extensive interest as environmentally friendly, biodegradable alternatives to petroleum-based plastics. Unlike petroleum-based plastics, PHAs can be obtained from renewable resources and are completely degraded to CO2 and H2O (25). PHAs are generally grouped into three categories: short-chain-length PHAs (SCL-PHAs) that contain repeating units of 3 to 5 carbon atoms, medium-chain-length PHAs (MCL-PHAs) that contain repeating units of 6 to 14 carbon atoms, and SCL-co-MCL PHAs, which consist of both SCL and MCL repeating units. MCL-PHAs show promise as thermoelastomers and are attractive for biomedical applications, such as drug delivery (16) and tissue engineering (28).
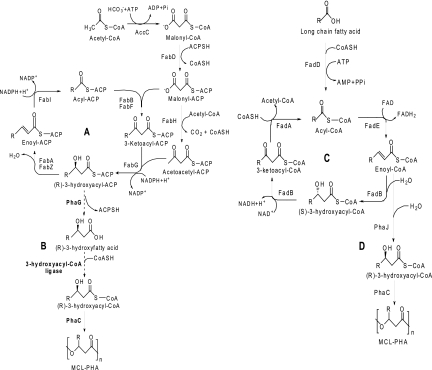
Pseudomonas strains are known to produce large amounts of MCL-PHAs (23). The genome of Pseudomonas putida KT2440 is fully sequenced, and several genes involved with PHA synthesis have been identified (1, 14). Two PHA synthase genes, phaC1 and phaC2, which are separated by the PHA depolymerase gene phaZ, are characterized as class II PHA synthases (3, 17). These synthases generally prefer 3-hydroxyacyl-coenzyme A (CoA) with chain lengths of 6 to 14 carbon atoms as substrates to produce MCL-PHA polymers (20). As shown in Fig. 1, precursors for PHA synthesis are thought to be derived from fatty acid biosynthesis when the microorganism is grown on unrelated carbon sources, such as glucose and glycerol, and β-oxidation when the microorganism is grown on related carbon sources, such as fatty acids (5, 11). The (R)-specific enoyl-CoA hydratase PhaJ is an enzyme that could potentially provide 3-hydroxyacyl-CoA precursors for PHA synthesis derived via the fatty acid β-oxidation pathway when the organism is grown on fatty acids (Fig. 1D) (4). For PHA production from unrelated carbon sources, an enzyme encoded by phaG was identified to be a key link between fatty acid biosynthesis and MCL-PHA biosynthesis (18). PhaG was first reported as a 3-hydroxyacyl- acyl carrier protein (ACP)-CoA transferase and was thought to transfer the 3-hydroxyacyl group from the ACP moiety to the CoA moiety (18). However, our results suggest that PhaG functions as a 3-hydroxyacyl-ACP thioesterase to produce 3-hydroxy fatty acids. Therefore, at least one 3-hydroxyacyl-CoA ligase is hypothesized to be necessary to provide the PHA precursor (R)-3-hydroxyacyl-CoA (11), as shown in Fig. 1B. It has been previously shown that the AlkK enzyme from Pseudomonas oleovorans has 3-hydroxyacyl-CoA ligase activity and can catalyze conversion of 3-hydroxyalkanoate substrates into 3-hydroxyalkanoate-CoA thioester PHA precursors in vitro (21). Therefore, several P. putida KT2440 genes encoding proteins with homology to AlkK from P. oleovorans (AlkKPo) were investigated in this study to find candidates for the 3-hydroxyacyl-CoA ligase putatively involved in PHA biosynthesis.
Fig 1.
Proposed metabolic pathways for PHA biosynthesis. (A) Fatty acid biosynthesis; (B) PHA biosynthesis via fatty acid biosynthesis; (C) β-oxidation; (D) PHA biosynthesis via β-oxidation.
In this study, transcription levels of genes putatively involved in PHA synthesis and genes encoding putative proteins homologous to AlkKPo were monitored by quantitative real-time PCR (qRT-PCR). Hypothetical MCL-PHA biosynthesis pathways were proposed by comparing the differences in gene transcription. A putative medium-chain-fatty-acid ligase gene (the PP0763 gene) displayed upregulated transcription levels when cells were grown under PHA-producing conditions. This gene was cloned and coexpressed with phaG from P. putida (phaGPp) and the PHA synthase gene phaC1(STQK)Ps in recombinant Escherichia coli. PhaC1(STQK) is an engineered PHA synthase originally derived from Pseudomonas sp. 61-3 that has two point mutations and has previously been shown to have increased activity toward 3-hydroxydecanoate (3HD)-CoA substrates in an in vitro assay (24). By coexpressing these genes, the production of MCL-PHAs in recombinant E. coli from non-fatty acid carbon sources was about 4-fold higher than that in P. putida KT2440. The results of this study have identified a new PHA biosynthesis pathway with the highest production of MCL-PHAs achieved to date in recombinant E. coli from inexpensive non-fatty acid carbon sources such as glucose and glycerol.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
P. putida KT2440 was grown in 100-ml batches of a defined mineral salt (MS) medium (7) with high nitrogen (high N) (7.2 g liter−1) or low nitrogen (low N) (0.072 g liter−1) and glycerol as the carbon source at 20 mM. Growth was monitored by measuring the optical densities at 540 nm (OD540) at 0 h, 4 h, 8 h, 12 h, 24 h, and 48 h. E. coli JM109 was used as a host for plasmid construction and was grown on Luria-Bertani (LB) medium. When necessary, ampicillin (100 μg ml−1) and/or kanamycin (50 μg ml−1) were added to the medium. For PHA production, recombinant E. coli BL21(DE3) and E. coli LS5218 strains were grown on LB medium with ampicillin (100 μg ml−1) and kanamycin (50 μg ml−1). IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM at 5 h when the OD540 was ∼1.0, and 20 g liter−1 glucose was added at 8 h when the OD540 was ∼2.0. For measuring 3-hydroxy fatty acids in the medium, recombinant E. coli LS5218 cells were grown on high N MS medium with 20 g liter−1 glycerol as the sole carbon source. All the experiments were performed in triplicate, and data are presented as the average ± the standard deviation for each sample point.
Determination of gene transcription levels by qRT-PCR.
qRT-PCR was performed in a manner similar to that previously described (27). Briefly, a total volume of 500 μl of P. putida KT2440 cell culture was harvested from MS medium when the OD540 reached 1.0. RNAs were stabilized using RNAprotect bacteria reagent (Qiagen, Valencia, CA). The total RNA was isolated using the RNeasy minikit (Qiagen, Valencia, CA). Prior to reverse transcription, the trace DNA remaining in RNA samples was removed by digestion with RQ1 RNase-free DNase (Promega, Madison, WI). The cDNA libraries were synthesized with a SuperScript VILO cDNA synthesis kit (Invitrogen, Carlsbad, CA). Total RNA (1 μg) was added to the reaction mixture as the template for each sample. All procedures using kits were performed according to the manufacturer's instructions. The qRT-PCRs were performed using the iQ SYBR green supermix kit with the iQ5 multicolor real-time PCR detection system (Bio-Rad, Hercules, CA). The primers used in qRT-PCR are listed in Table 2. The gene expression levels were assessed by comparing the ratios of the housekeeping gene rpoD threshold cycle (CT) values to target gene CT values using the following equation: ratio of rpoD/target = 2CT(rpoD) − CT(target).
Table 2.
Primers used in this study
| Primers | Sequence(s)a |
|---|---|
| Primers used in qRT-PCR | |
| phaC1 | F, 5′-CAGGTTGCTTTGTTTGTC-3′; R, 5′-TTGTTTACCCAGTAGTTCC-3′ |
| phaZ | F, 5′-AGTTTGCTCACGATTAC-3′; R, 5′-CACCTTGGGCTTGC-3′ |
| phaC2 | F, 5′-ATGAGCAGACCATCG-3′; R, 5′-GTTTACCCAGTAGTTCC-3′ |
| phaG | F, 5′-TTCAAACGCTTCAAC-3′; R, 5′-CGGTCTTGTTCTCC-3 |
| phaJ | F, 5′-TGTCCCAGGTCAC-3′; R, 5′-GGAACATGCTCTTG-3′ |
| phaJ3 | F, 5′-CCGAGGCTGAAGATGGTATC-3′; R, 5′-ACGGCACTGAGATGGATAGG-3′ |
| phaJ4 | F, 5′-ATGGTTTCCTGACCTTGTCG-3′; R, 5′-AAAACAGAGCGACAGCGACT-3′ |
| PP0763 | F, 5′-CCAAGGGCGTGTATTTCAGT-3′; R, 5′-CCTGGGTACACCTGCTTCAT-3′ |
| PP3553 | F, 5′-ACACGGTGATGAAGGGCTAC-3′; R, 5′-CTGGCTTTAAGGCAACGAAG-3′ |
| PP2795 | F, 5′-GAAAGGCTACCTGCACAACC-3′; R, 5′-ATTTCCACATAGCCGTCCAG-3′ |
| PP3458 | F, 5′-CCGAAGTGCAGATTGCCTAT-3′; R, 5′-GCTTTGGGGTTGTTCCAGTA-3′ |
| PP2038 | F, 5′-CTGACCGAACAGGACCATTT-3′; R, 5′-AGTAGCCCTGCATGATGTCC-3′ |
| PP4063 | F, 5′-GCCACGCTCAGTCACTACAA-3′; R, 5′-GAAAGCATCGTTGGGGTAGA-3′ |
| PP4549 | F, 5′-ACGTATACCCCAACGAGCTG-3′; R, 5′-CGGTACCTTGTAGCCGGTAA-3′ |
| PP4550 | F, 5′-GCAGAAGGCTGGTTCAAGAC-3′; R, 5′-GGGTACTTTGTAGCCGGTGA-3′ |
| PP4487 | F, 5′-GAGCACCGCAACATCACTTA-3′; R, 5′-ACCACCTTGGATTTGCAGTC-3′ |
| PP2213 | F, 5′-GATCAAGGCGTTCGTGGTAT-3′; R, 5′-ATTCGATTTCCCTGGGGTAG-3′ |
| PP2351 | F, 5′-ACCACGGTGTTCTACGAAGG-3′; R, 5′-AGCGAGCTGAGGTCATGTTT-3′ |
| PP4702 | F, 5′-CGGATGGTCGACACCTACTT-3′; R, 5′-CCAGTGCACTTTCCACTTCA-3′ |
| PP3071 | F, 5′-CCGTTACCACGATGCCTACT-3′; R, 5′-CCACAACCTGTTCGACCTTT-3′ |
| PP2709 | F, 5′-CGCTTGTGTGCAGCTCTATC-3′; R, 5′-ATCAGGCTCTGAGCACCACT-3′ |
| Primers used for plasmid construction and knockout mutant | |
| phaG F_KpnI | 5′-GGTACCGGCAAATGCCTACCTGTCAT-3′ |
| phaG R_XbaI | 5′-TCTAGACTGCGGGTAT TGGCTACAAG-3′ |
| PP0763 F_SacI | 5′-GAGCTCTTTTCAGAAAAGGGATCCCC-3′ |
| PP0763 R_KpnI | 5′-GGTACCTTACAACGTGGAAAGGAACG-3′ |
| ΔfadE forward | 5′-CTCGCGTGCTGCAAAAACTCTCCGGCGTGAGCGGGATCCTGGCGATTACCGTGTAGGCTGGAGCTGCTTC-3′ |
| ΔfadE reverse | 5′-CGTTGACGTCATTGTTCTTCGCCGCTTCCATCTCTTCCAGCACGTACGGAATTCCGGGGATCCGTCGACC-3′ |
| fadE Check | F, 5′-GGAGTTGTGGGCGTACCTTA-3′; R, 5′-AGCCTGATACAGCGCATCTT-3′ |
F, forward; R, reverse. Underlining indicates 50-bp regions homologous to the fadE gene; restriction sites are indicated in bold.
The n-fold induction was determined by the expression levels of genes of cells grown on low N MS medium versus those of cells grown on high N MS medium.
Plasmid construction.
The putative 3-hydroxyacyl-ACP thioesterase gene phaG was amplified by PCR from Pseudomonas putida KT2440 genomic DNA using the primers F_KpnI and R_Xbal (see Table 2). The putative medium-chain-fatty-acid CoA ligase gene (the PP0763 gene) was also amplified by PCR from P. putida KT2440 genomic DNA using the primers F_SacI and R_KpnI (see Table 2). The phaG and PP0763 PCR products were cloned into the vector pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA) to produce pTOPOG and pTOPOK. The orientation and overall correctness were confirmed by restriction digest and DNA sequencing. The 1-kb fragment including phaG was removed by restriction digest with KpnI and XbaI from pTOPOG and subcloned into pTrc99A to construct the plasmid pTrcG. The 1.7-kb fragment including the PP0763 gene was removed by restriction digest with SacI and KpnI from pTOPOK, and the fragment was subcloned into the plasmids pTrc99A and pTrcG, generating the constructs pTrcK and pTrcKG, respectively. The plasmids pTrcG, pTrcK, and pTrcKG were cotransferred with pBBRSTQK (15) into E. coli BL21(DE3) and E. coli LS5218 (Table 1). The gene PP2795 from P. putida KT2440 was also amplified and inserted into pTrcG to generate the plasmid pTrc2795G using the same strategy as described above and cotransferred with pBBRSTQK into E. coli BL21(DE3).
Table 1.
Plasmids and bacterial strains used in this study
| Plasmid or strain | Relevant characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR-Blunt II-TOPO | TA cloning vector, Kmr | Invitrogen |
| pBBRSTQK | pBBR1-MCS2 derivative, Pseudomonas sp. 61-3 phaC1(STQK) | 15 |
| pTOPOG | pCR-Blunt II-TOPO derivative, P. putida KT2440 phaG | This study |
| pTOPOK | pCR-Blunt II-TOPO derivative, P. putida KT2440 PP0763 | This study |
| pTrc99A | Expression vector, Ampr PtrcrrnB ori (pBR322) lacIq | Amersham |
| pTrcG | pTrc99A derivative, P. putida KT2440 phaG | This study |
| pTrcK | pTrc99A derivative, P. putida KT2440 PP0763 | This study |
| pTrcGK | pTrc99A derivative, P. putida KT2440 phaG PP0736 | This study |
| Strains | ||
| P. putida KT2440 | Wild type | |
| E. coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 lac [F′ proAB lacIq ZΔM15] | Takara |
| E. coli BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | 9 |
| E. coli LS5218 | fadR601 atoC2(con) | 6 |
| E. coli LS11 | E. coli LS5218 derivative, ΔfadE | This study |
Gene knockouts in E. coli LS5218.
The method for chromosomal gene inactivation in E. coli developed by Datsenko and Wanner (2) was used to inactivate the fadE gene in E. coli LS5218. The kanamycin-resistant cassette from the plasmid pKD13 was amplified by the primers (Table 2) containing 50-bp regions homologous to the fadE gene. The PCR product was transformed via electroporation into E. coli LS5218 that had expressed the λ-Red system from the pKD46 plasmid. Transformants were selected on LB agar plates with 50 μg ml−1 kanamycin. Complete elimination of the gene from the chromosome was confirmed by PCR with fadE check primers (Table 2). The pKD46 plasmid was removed by growth at 37°C. After fadE was disrupted, the kanamycin-resistant cassette was removed by expression of FLP recombinase from the pCP20 plasmid. The pCP20 plasmid was removed by growth at 37°C. The fadE mutant strain E. coli LS11 was confirmed by failure to grow on fatty acids as the sole carbon source.
Analytical procedures.
For measuring glycerol concentrations in the supernatant, free glycerol reagent (Sigma, St. Louis, MO) was used according to the manufacturer's instructions. Briefly, 10 μl of NANOpure water (blank), glycerol standard (2.5 mg ml−1), or supernatant from medium was mixed with 0.8 ml of the free glycerol reagent and incubated for 5 min at 37°C. The absorbance (A) was measured at 540 nm. The glycerol concentration in supernatant was calculated by the following equation: glycerol content = (Asupernatant − Ablank)/(Astandard − Ablank) × 2.5 mg ml−1.
For analysis of polymer production and composition, liquid cultures (100 ml) were centrifuged at 5,000 × g for 15 min at 4°C, washed with 15 ml NANOpure water, and lyophilized for a minimum of 24 h, and ∼15 mg dry cells per sample was treated with 2 ml of methanol-sulfuric acid (85:15) solution and 2 ml chloroform at 100°C for 140 min and assayed using a GC 2010 gas chromatograph (Shimadzu Scientific Instruments, Inc., Marlborough, MA) (7). Cell materials for extraction and purification of PHA polymer were prepared and lyophilized as described above. The lyophilized cells were added to 100 ml chloroform and were shaken at 60°C, 100 rpm overnight, to extract polymer produced in the cells. The chloroform-polymer solution was vacuum filtered through Whatman filter paper 1, and polymers were precipitated by pouring the filtrate into a 10× volume of methanol. Precipitates were separated from the liquid by centrifugation at 5,000 × g for 15 min. The supernatant was poured off, and the pellet containing the polymers was allowed to dry at room temperature. Once dry, the polymers were dissolved again in chloroform, and the solution was cast in a glass petri dish. The chloroform was removed by evaporation at room temperature for 48 h. The purified polymers were analyzed by 1H nuclear magnetic resonance (NMR) spectroscopy and 13C NMR using a Bruker BioSpin AVANCE 600 (Bruker BioSpin Corp., Billerica, MA) operating at 600 MHz (31). For analyzing extracellular 3-hydroxy fatty acids, 10 ml supernatant of each sample was lyophilized for at least 24 h, and the lyophilized materials were subjected to methanolysis and assayed by GC (7).
RESULTS AND DISCUSSION
Growth of P. putida KT2440 on low-nitrogen or high-nitrogen MS medium.
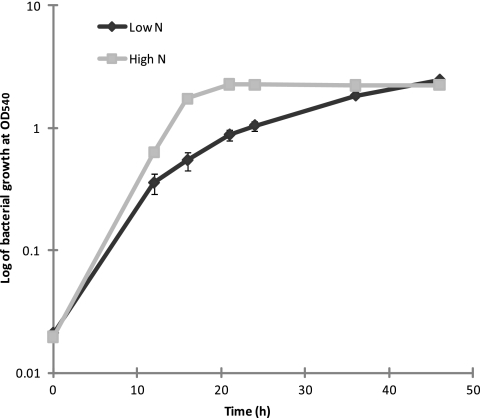
The OD540 of P. putida KT2440 grown on either high-nitrogen (high N) or low-nitrogen (low N) MS medium was measured at different time points (Fig. 2). The doubling time of cells grown on high-nitrogen MS medium (2.5 h) was considerably shorter than that of cells grown on low-nitrogen MS medium (3.4 h) during the exponential phase (from 0 h to 16 h). However, the cells grown on high N MS medium stopped growing after 24 h, while the cells grown on low N medium grew until 48 h. The concentration of glycerol in the supernatant of high N MS medium dropped from 20 mM to zero at 48 h, but the concentration of glycerol in low N MS medium decreased from 20 mM to 3 mM. Therefore, depletion of the carbon source was the main reason that the cells stopped growing in high N MS medium.
Fig 2.
Differences in growth rates of P. putida KT2440 grown on mineral salt (MS) medium with different nitrogen concentrations. Growth curves of P. putida KT2440 grown on MS medium with either high N (0.072 g liter−1), represented by squares, or low N (0.072 g liter−1), represented by diamonds. Points represent the average absorbance of three independent data points (OD540) ± standard deviation.
Transcription levels of genes involved in PHA synthesis in cells grown on low N MS medium versus cells grown on high N MS medium.
In order to determine the best conditions for sampling cells to determine the expression levels of genes potentially involved in PHA biosynthesis from non-fatty acid carbon feedstocks, P. putida was grown in two sets of media with different nitrogen (N) concentrations to induce PHA production and get RNA for qRT-PCR analysis. The PHA production of P. putida KT2440 on low N medium was 71-fold higher than that on high N medium (Table 3). In order to identify genes upregulated in cells grown under PHA-producing conditions, candidate gene transcription levels for cells grown on low N medium versus transcription levels of cells grown on high N medium were assayed by qRT-PCR as described in Materials and Methods. Specifically, the transcription levels of genes involved in PHA synthesis, which were homologous to genes known in other Pseudomonas species to be involved in PHA production, and homologs of the alkK gene from P. oleovorans that encodes an acyl-CoA ligase (26) were investigated. The transcription of the rpoD gene encoding the housekeeping sigma70 factor was used as the housekeeping gene for qRT-PCR since it has already been identified as an ideal housekeeping gene for this task due to its stable expression levels in Pseudomonas aeruginosa (22). The ratios of transcription levels of genes from cells grown on low N MS medium versus those of cells grown on high N MS medium that were >1 were defined as induced by low-nitrogen conditions. The transcription levels of all three genes in the PHA synthesis cluster (phaC1, phaZ, and phaC2) were higher when cells were grown on low N MS than on high N MS medium. The phaJ4 gene had the highest transcription level among all three phaJ genes, and this result was consistent with previous research showing that PhaJ4 was the major enzyme channeling β-oxidation for PHA biosynthesis (27). Surprisingly, the phaG gene had significantly higher transcription levels (∼220-fold) when cells were grown on low N MS medium than on high N MS medium (Table 4). This result suggested that the PhaG enzyme plays an important role in PHA biosynthesis from unrelated carbon sources, and it was most likely to be a critical step for PHA biosynthesis in P. putida. Growing cells on a non-fatty acid carbon source, such as glycerol, under nitrogen limitation led to PHA production. PHA biosynthesis under these conditions is highly likely to be derived from fatty acid biosynthesis rather than β-oxidation. Results indicated that phaG was highly upregulated when the cells were grown under nitrogen limitation conditions in P. putida KT2440 and suggest that phaG expression and PHA production are stress induced.
Table 3.
PHA production in the P. putida KT2440 strain on MS medium with high N or low Na
| Cultureb | CDW (g liter−1) | PHA produced (mg liter−1) | % PHA | Composition (mol%) |
|||
|---|---|---|---|---|---|---|---|
| 3HHx | 3HO | 3HD | 3HDD | ||||
| High N | 0.6 ± 0.04 | 2.0 ± 0.1 | 0.4 ± 0.02 | ND | ND | 56.8 ± 4.4 | 44.2 ± 4.4 |
| Low N | 0.4 ± 0.08 | 109 ± 21 | 28.4 ± 1.0 | 2.6 ± 0.02 | 26.0 ± 0.05 | 67.5 ± 0.1 | 3.9 ± 0.2 |
CDW, cell dry weight; 3HHx, 3-hydroxyhexanoate; 3HO, 3-hydroxyoctanoate; 3HD, 3-hydroxydecanoate; 3HDD, 3-hydroxydodecanoate; ND, not determined.
The cells were grown on MS medium with either high N (7.2 g liter−1) or low N (0.072 g liter−1).
Table 4.
Expression levels of genes involved in PHA biosynthesis pathways in P. putida KT2440 cells grown on low N MS medium and those grown on high N MS medium
| Locus name | Gene symbol | Putative identification | Inductiona (n-fold) |
|---|---|---|---|
| PP5003 | phaC1 | Poly(3-hydroxyalkanoate) polymerase 1 | 2.6 ± 0.5 |
| PP5004 | phaZ | Poly(3-hydroxyalkanoate) depolymerase | 4.5 ± 0.8 |
| PP5005 | phaC2 | Poly(3-hydroxyalkanoate) polymerase 2 | 4.3 ± 0.5 |
| PP4552 | phaJ1 | (R)-specific enoyl-CoA hydratase 1 | 1.1 ± 0.2 |
| PP0580 | phaJ3 | (R)-specific enoyl-CoA hydratase 2 | 0.9 ± 0.2 |
| PP4817 | phaJ4 | (R)-specific enoyl-CoA hydratase 3 | 3.8 ± 1.3 |
| PP1048 | phaG | 3-Hydroxyacyl-ACP thioesterase | 220 ± 31 |
Induction is defined as the ratio of expression level of genes in cells grown on low N MS medium versus that in cells grown on high N MS medium.
Transcription levels of all of the genes homologous to alkK in P. oleovorans (alkKpo) (26) were also analyzed by qRT-PCR (Table 5). Transcription levels of most of the genes were upregulated during growth in medium with low N concentrations. Based on the transcription levels and the hypothesis that a CoA ligation step is necessary for PHA production from unrelated carbon sources, this result showed that multiple enzymes might be involved in this reaction. Among the P. putida homologs to the AlkK protein from P. oleovorans, the gene encoding the putative medium-chain-fatty-acid CoA ligase (the PP0763 gene), which shares 36% identity and 54% similarity to AlkKPo, showed the highest transcription level; therefore, it was hypothesized to play an important role in the production of (R)-3-hydroxyacyl-CoA for PHA production from unrelated carbon sources.
Table 5.
Expression levels of genes encoding acyl-CoA ligase homologs in P. putida KT2440 cells growing on low N MS medium and on high N MS medium
| Locus name | Gene symbol | Putative identification | Induction (n-fold) |
|---|---|---|---|
| PP0763 | Medium-chain-fatty-acid CoA ligase | 10.5 ± 1.0 | |
| PP3553 | AMP-binding domain protein | 3.2 ± 1.8 | |
| PP2795 | AMP-binding domain protein | 6.1 ± 0.2 | |
| PP3458 | Long-chain-fatty-acid CoA ligase, putative | 2.1 ± 1.3 | |
| PP2038 | Long-chain-fatty-acid CoA ligase, putative | 2.6 ± 0.6 | |
| PP4063 | Long-chain-fatty-acid CoA ligase, putative | 0.9 ± 0.2 | |
| PP4549 | fadD | Long-chain-fatty-acid CoA ligase | 1.5 ± 0.03 |
| PP4550 | fadD2 | Long-chain-fatty-acid CoA ligase | 1.3 ± 0.1 |
| PP4487 | acsA | Acetyl-CoA synthetase | 0.6 ± 0.02 |
| PP2213 | fadDx | Acyl-CoA ligase | 1.8 ± 0.2 |
| PP2351 | Acetyl-CoA synthetase, putative | 1.2 ± 0.1 | |
| PP4702 | acsB | Acetyl-CoA synthetase | 1.3 ± 0.5 |
| PP3071 | Acetoacetyl-CoA synthetase, putative | 4.2 ± 0.7 | |
| PP2709 | Long-chain-fatty-acid CoA ligase, putative | 3.3 ± 0.5 |
MCL-PHA production in recombinant E. coli strains.
Previous studies demonstrated that phaG is important for PHA biosynthesis from unrelated carbon sources in pseudomonads (18). However, based on phaG expression experiments in E. coli, phaG alone is unable to provide 3-hydroxyacyl-CoA substrates for PHA production from unrelated carbon sources (18, 30). In order to investigate if the gene encoding the putative medium-chain-fatty-acid CoA ligase (the PP0763 gene) was involved in PHA biosynthesis from unrelated carbon sources, the phaG and PP0763 genes were amplified by PCR from P. putida KT2440 and inserted into the expression vector pTrc99A to construct the plasmid pTrcGK. For construction of the control plasmids, phaG and the PP0763 gene were also cloned into pTrc99A separately to make the plasmids pTrcG and pTrcK.
The P. putida PP0763 gene encoding the AlkK-like protein and phaG genes encoding the synthetic pathway to produce MCL-PHA precursors were coexpressed from pTrcKG with pBBRSTQK (Table 1) carrying the PHA synthase gene phaC1(STQK)Ps in E. coli to assess their ability to produce MCL-PHA from non-fatty acid carbon sources. The initial strain used for PHA production was E. coli BL21(DE3). This strain was chosen because it has been used in previous studies for overproducing recombinant proteins, ethanol, and other biomolecules (29). The results of this coexpression are shown in Table 6. PHA was accumulated to ∼4% cell dry weight in recombinant E. coli BL21(DE3) harboring the plasmids pTrcGK and pBBRSTQK. The major repeating units in the PHA polymers produced were 3-hydroxyoctanoate (3HO) (∼51 mol%) and 3HD (∼44 mol%), which was slightly different in composition compared to PHA polymer produced in the wild-type P. putida strain (Table 3). A recombinant E. coli BL21(DE3) strain containing plasmid pTrcK (PP0763) and pBBRSTQK was examined as a negative control and failed to produce any polymer, while another control strain harboring pTrcG (phaGPp) and pBBRSTQK produced trace amounts of 3HD (∼0.2% per cell dry weight) based on GC. In addition to the PP0763 gene, another open reading frame encoding an AlkKPo homolog, the PP2795 gene, which was transcribed at the second highest level (∼6-fold) among genes encoding AlkK homologs in cells grown under PHA-producing conditions (Table 5), was also coexpressed with phaGPp and phaC1(STQK)Psin E. coli. However, unlike the PP0763 gene, no detectable MCL-PHAs were produced in the recombinant cells. Therefore, the PP2795 gene is unlikely to be involved in PHA biosynthesis.
Table 6.
PHA production in recombinant E. coli strainsa
| Relevant plasmidsb | CDW (g liter−1) | PHA produced (mg liter−1) | % PHA | Composition (mol%) |
|||
|---|---|---|---|---|---|---|---|
| 3HHx | 3HO | 3HD | 3HDD | ||||
| E. coli BL21(DE3) | |||||||
| pTrcG, pBBRSTQK | 1.2 ± 0.1 | 3.1 ± 0.7 | 0.2 ± 0.05 | ND | ND | 100 | ND |
| pTrcK, pBBRSTQK | 0.8 ± 0.1 | ND | ND | ND | ND | ND | ND |
| pTrcGK, pBBRSTQK | 1.7 ± 0.1 | 69 ± 21 | 4.1 ± 1.3 | 3.3 ± 0.4 | 51.0 ± 1.1 | 44.3 ± 1.6 | 1.4 ± 0.3 |
| E. coli LS5218 | |||||||
| pTrcGK, pBBRSTQK | 3.4 ± 0.2 | 392 ± 61 | 11.6 ± 1.3 | 1.3 ± 0.1 | 39.2 ± 0.8 | 56.9 ± 0.8 | 2.6 ± 0.1 |
| E. coli LS11 | |||||||
| pTrcGK, pBBRSTQK | 1.7 ± 0.1 | 123 ± 30 | 7.2 ± 1.5 | N.D. | 32.9 ± 0.9 | 65.1 ± 2.0 | 2.0 ± 0.1 |
CDW, cell dry weight; 3HHx, 3-hydroxyhexanoate; 3HO, 3-hydroxyoctanoate; 3HD, 3-hydroxydecanoate; 3HDD, 3-hydroxydodecanoate; ND, not determined.
The cells were grown on LB medium with 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin. IPTG was added to a final concentration of 1 mM at 5 h, and 20 g liter−1 glucose was added at 8 h.
The plasmids pTrcGK and pBBRSTQK were also transformed into E. coli LS5218 [fadR601 atoC2(con)], which allowed for constitutive expression of enzymes involved in utilization of fatty acids (6). PHA content increased to 11.6% per cell dry weight, and the cell dry weight was also about 2-fold higher than that of the recombinant BL21(DE3) strain. The PHA production reached ∼400 mg liter−1, which was ∼4-fold higher than that of wild-type Pseudomonas putida KT2440 (109 mg liter−1). The composition of PHAs was 1.3% 3-hydroxyhexanoate (3HHx), 32.9% 3HO, 56.9% 3HD, and 2.6% 3-hydroxydodecanoate (3HDD), which was similar to that of wild-type Pseudomonas putida KT2440 (Table 3). Prior to the current study, MCL-PHA production from unrelated carbon sources had been problematic due to the difficulty of drawing PHA precursors from fatty acid biosynthesis. In a previous study, MCL-PHAs were produced at 2.3% per cell dry weight in the presence of gluconate and overexpression of E. coli thioesterase I (8). By adding the fatty acid biosynthesis inhibitor triclosan into the medium, another study claimed that coexpression of phaC1 and phaGPp led to accumulation of 2 to 3% PHA per cell dry weight in recombinant E. coli (19). However, since the PHA content was measured only by GC, the 3HD detected in that study might have been extracellular 3-hydroxydecanoic acids rather than PHA polymers, based on a previous study (30) and our current study.
Since β-oxidation was constitutively expressed in E. coli LS5218, the higher MCL-PHA production in E. coli LS5218 than in E. coli BL21(DE3) might be due to MCL-PHA substrates contributed from intermediates of β-oxidation. In order to exclude the possibility that the PHA production was derived from fatty acid β-oxidation, the fadE gene encoding an acyl-CoA dehydrogenase was inactivated in E. coli 5218 to produce the mutant strain E. coli LS11. This strain was unable to grow on fatty acids as the sole carbon sources (data not shown). The two plasmids pTrcGK and pBBRSTQK were also coexpressed in E. coli LS11, and the PHA yield was 7.2% per cell dry weight, which was still higher than that from E. coli BL21(DE3). The PHA composition was similar to that of recombinant E. coli LS5218. This result suggested that the PHA production in recombinant E. coli strains was derived mainly from fatty acid biosynthesis. The higher production in E. coli LS5218 is likely due to better transportation of the free 3-hydroxy fatty acids generated by PhaG back into the cells to make PHAs.
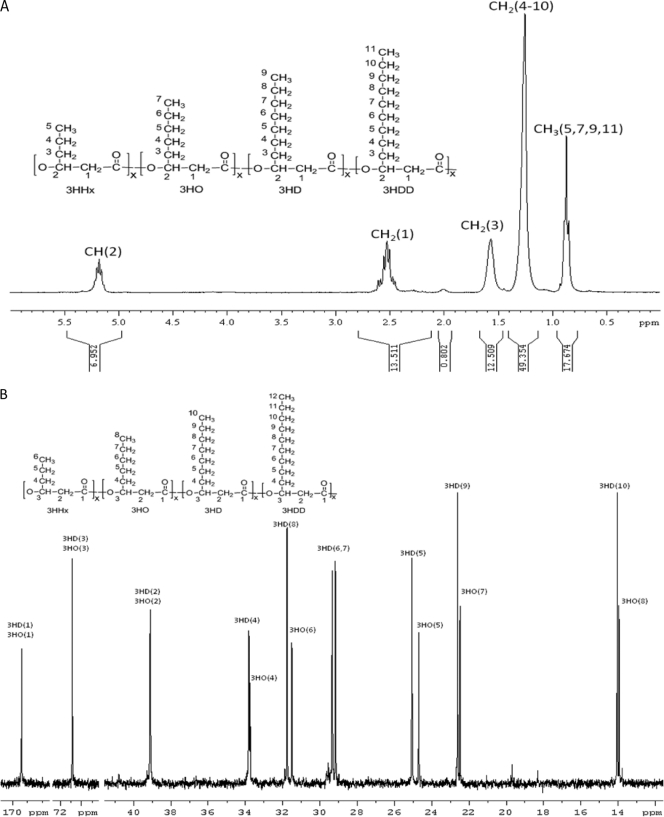
For further confirmation, the PHA polymer produced by recombinant E. coli LS5218 was purified and analyzed by 1H and 13C NMR spectroscopy (Fig. 3). The chemical shifts for the 1H NMR were assigned based on NMR spectroscopy performed in the study by Huijberts et al. (5), and the chemical shifts for 13C NMR spectroscopy were assigned based on the study by Matsusaki et al. (13). The results confirmed that MCL-PHAs were produced and the major repeating units were 3HD and 3HO. The ratios of 3HHx and 3HDD were too low to be detected by 13C NMR.
Fig 3.
NMR spectrum of polymers isolated from E. coli LS5218 harboring the pTrcGK and pBBRSTQK plasmids grown in the presence of glucose. (A) 1H NMR spectrum. Protons in the polymers are numbered and assigned to the peaks in the spectrum. (B) 13C NMR spectrum. Carbon atoms in polymers are numbered and assigned to peaks in the spectrum.
Production of free 3-hydroxy fatty acids in recombinant E. coil expressing the phaG gene.
PhaG was previously identified as an (R)-3-hydroxyacyl-ACP-CoA transferase based on the ability of the partially purified enzyme to convert 3-hydroxydecanoyl-CoA to 3-hydroxydecanoyl-ACP in the presence of ACP (18). However, there was no PHA produced when the phaG and PHA synthase genes were heterogeneously expressed in recombinant E. coli (12, 18). On the other hand, overexpression of the phaG gene in E. coli led to the extracellular production of 3-hydroxydecanoic acid as demonstrated in the current study and a previous study (30). These results suggest that PhaG functions as a 3-hydroxyacyl-ACP thioesterase to remove the ACP from the 3-hydroxyacyl-ACP to form free 3-hydroxy fatty acids rather than as an (R)-3-hydroxyacyl-ACP-CoA transferase.
In order to further verify that PhaG is a 3-hydroxy-acyl-ACP thioesterase rather than a 3-hydroxyacyl-ACP-CoA transacylase, recombinant E. coli LS5218 harboring pTrcG (expressing only the phaG gene) was grown under the same conditions as those described in Table 6. Results indicate that expression of phaG alone led to the production of ∼1.6 g liter−1 free 3-hydroxy fatty acids consisting of 45% 3-hydroxyoctanoate and 55% 3-hydroxydecanoate as detected in the supernatant and no detectable PHAs in the cells. This result is consistent with previous research showing (30).
In order to demonstrate that the substrates for PHA synthesis were most likely derived from the fatty acid biosynthesis pathway and to further prove that PhaG is a 3-hydroxy-acyl-ACP thioesterase, E. coli LS5218 harboring pTrcG and pBBRSTQK was also grown on MS medium using glycerol as the sole carbon source. E. coli LS5218 harboring pTrcGK and pBBRSTQK grown under the same conditions was used as a control. The concentration of free fatty acids was measured in the supernatant. The E. coli strain harboring pTrcG and pBBRSTQK produced about 30 mg liter−1 3-hydroxy fatty acids, comprised of 3-hydroxydecanoic acid (58%) and 3-hydroxyoctanoic acid (42%) in the supernatant. These results indicate that that PhaG functions as a 3-hydroxyacyl-ACP thioesterase rather than a 3-hydroxyacyl-ACP-CoA transferase.
Meanwhile, the control strain did not produce any detectable 3-hydroxy fatty acids in the supernatant but produced PHA polymers in the cell. The E. coli strain harboring pTrcG and pBBRSTQK produced about 0.9 mg liter−1 PHAs as detected by GC, while the strain harboring pTrcGK and pBBRSTQK produced about 25 mg liter−1 PHAs. This result was consistent with the PHA production data described in Table 6. Since glycerol was the only carbon source for these experiments, this result further proved that PP0763 is essential to establish a link to PHA production from non-fatty acid carbon sources in recombinant E. coli.
Conclusions.
Our study demonstrates for the first time that MCL-PHA polymers could be successfully synthesized from the non-fatty acid carbon sources glucose and glycerol in recombinant E. coli, with production levels as high as 400 mg liter−1 and 11.6% per cell dry weight. This study has identified and characterized a novel pathway to make MCL-PHAs from non-fatty acid carbon sources. Glucose can be derived from cellulosic feedstocks, and glycerol is a by-product of biodiesel production. It is expected that the cost of these feedstocks will decrease as the technologies for their production mature. Due to the ubiquity of fatty acid biosynthesis in organisms and because the biosynthetic pathway developed by our study derives 3-hydoxyacyl-CoA substrates from fatty acid biosynthetic pathways, it offers great flexibility in carbon source utilization for MCL-PHA production. This could have great potential to expand the number of inexpensive carbon sources to be used for the production of MCL-PHA polymers.
ACKNOWLEDGMENTS
We thank D. Kiemle and SUNY-ESF Analytical and Technical Services for assistance with NMR studies.
This work was supported by NSF DMR 0907085 awarded to C. T. Nomura.
Footnotes
Published ahead of print 18 November 2011
REFERENCES
- 1. Ballerstedt H, et al. 2007. Genomotyping of Pseudomonas putida strains using P. putida KT2440-based high-density DNA microarrays: implications for transcriptomics studies. Appl. Microbiol. Biotechnol. 75:1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Eugenio LI, et al. 2007. Biochemical evidence that phaZ gene encodes a specific intracellular medium chain length polyhydroxyalkanoate depolymerase in Pseudomonas putida KT2442: characterization of a paradigmatic enzyme. J. Biol. Chem. 282:4951–4962 [DOI] [PubMed] [Google Scholar]
- 4. Fiedler S, Steinbuchel A, Rehm BH. 2002. The role of the fatty acid beta-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans and of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch. Microbiol. 178:149–160 [DOI] [PubMed] [Google Scholar]
- 5. Huijberts GN, Eggink G, de Waard P, Huisman GW, Witholt B. 1992. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl. Environ. Microbiol. 58:536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jenkins LS, Nunn WD. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kato M, Bao HJ, Kang CK, Fukui T, Doi Y. 1996. Production of a novel copolymer of 3-hydroxybutyric acid and medium-chain-length-3-hydroxyalkanoaic acids by Pseudomonas sp. 61-3 from sugar. Appl. Microbiol. Biotechnol. 45:363–370 [Google Scholar]
- 8. Klinke S, Ren Q, Witholt B, Kessler B. 1999. Production of medium-chain-length poly(3-hydroxyalkanoates) from gluconate by recombinant Escherichia coli. Appl. Environ. Microbiol. 65:540–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee SY. 2009. Systems biology and biotechnology of Escherichia coli. Springer, Dordrecht, The Netherlands [Google Scholar]
- 10. Li R, Zhang H, Qi Q. 2007. The production of polyhydroxyalkanoates in recombinant Escherichia coli. Bioresour. Technol. 98:2313–2320 [DOI] [PubMed] [Google Scholar]
- 11. Lu J, Tappel RC, Nomura CT. 2009. Minireview: biosynthesis of poly(hydroxyalkanoates). Polym. Rev. 49:226–248 [Google Scholar]
- 12. Matsumoto K, et al. 2001. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) copolymer from sugars by recombinant Ralstonia eutropha harboring the phaC1Ps and the phaGPs genes of Pseudomonas sp. 61-3. Biomacromolecules 2:934–939 [DOI] [PubMed] [Google Scholar]
- 13. Matsusaki H, Abe H, Taguchi K, Fukui T, Doi Y. 2000. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant bacteria expressing the PHA synthase gene phaC1 from Pseudomonas sp. 61-3. Appl. Microbiol. Biotechnol. 53:401–409 [DOI] [PubMed] [Google Scholar]
- 14. Nelson KE, et al. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799–808 [DOI] [PubMed] [Google Scholar]
- 15. Nomura CT, et al. 2005. Expression of 3-ketoacyl-acyl carrier protein reductase (fabG) genes enhances production of polyhydroxyalkanoate copolymer from glucose in recombinant Escherichia coli JM109. Appl. Environ. Microbiol. 71:4297–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pouton CW, Akhtar S. 1996. Biosynthetic polyhydroxyalkanoates and their potential in drug delivery. Adv. Drug Deliv. Rev. 18:133–162 [Google Scholar]
- 17. Rehm BH. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rehm BH, Kruger N, Steinbüchel A. 1998. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The PHAG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme A transferase. J. Biol. Chem. 273:24044–24051 [DOI] [PubMed] [Google Scholar]
- 19. Rehm BH, Mitsky TA, Steinbüchel A. 2001. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 67:3102–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rehm BH, Steinbüchel A. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 25:3–19 [DOI] [PubMed] [Google Scholar]
- 21. Satoh Y, Murakami F, Tajima K, Munekata M. 2005. Enzymatic synthesis of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) with CoA recycling using polyhydroxyalkanoate synthase and acyl-CoA synthetase. J. Biosci. Bioeng. 99:508–511 [DOI] [PubMed] [Google Scholar]
- 22. Savli H, et al. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403–408 [DOI] [PubMed] [Google Scholar]
- 23. Steinbüchel A, Fuchtenbusch B. 1998. Bacterial and other biological systems for polyester production. Trends Biotechnol. 16:419–427 [DOI] [PubMed] [Google Scholar]
- 24. Takase K, Matsumoto K, Taguchi S, Doi Y. 2004. Alteration of substrate chain-length specificity of type II synthase for polyhydroxyalkanoate biosynthesis by in vitro evolution: in vivo and in vitro enzyme assays. Biomacromolecules 5:480–485 [DOI] [PubMed] [Google Scholar]
- 25. Tokiwa Y, Calabia BP, Ugwu CU, Aiba S. 2009. Biodegradability of plastics. Int. J. Mol. Sci. 10:3722–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Beilen JB, Eggink G, Enequist H, Bos R, Witholt B. 1992. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol. Microbiol. 6:3121–3136 [DOI] [PubMed] [Google Scholar]
- 27. Wang Q, Nomura CT. 2010. Monitoring differences in gene expression levels and polyhydroxyalkanoate (PHA) production in Pseudomonas putida KT2440 grown on different carbon sources. J. Biosci. Bioeng. 110:653–659 [DOI] [PubMed] [Google Scholar]
- 28. Williams SF, Martin DP, Horowitz DM, Peoples OP. 1999. PHA applications: addressing the price performance issue. I. Tissue engineering. Int. J. Biol. Macromol. 25:111–121. [DOI] [PubMed] [Google Scholar]
- 29. Yoon SH, Jeong H, Kwon S-K, Kim JF. 2009. Genomics, biological features, and biotechnology applications of Escherichia coli B: “Is B for better?!” In Lee SY. (ed), System biology and biotechnology of Escherichia coli. Springer, Dordrecht, The Netherlands [Google Scholar]
- 30. Zheng Z, Zhang MJ, Zhang G, Chen GQ. 2004. Production of 3-hydroxydecanoic acid by recombinant Escherichia coli HB101 harboring phaG gene. Antonie Van Leeuwenhoek 85:93–101 [DOI] [PubMed] [Google Scholar]
- 31. Zhu C, Nomura CT, Perrotta JA, Stipanovic AJ, Nakas JP. 2010. Production and characterization of poly-3-hydroxybutyrate from biodiesel-glycerol by Burkholderia cepacia ATCC 17759. Biotechnol. Prog. 26:424–430 [DOI] [PubMed] [Google Scholar]