Abstract
Experimental and in silico PCR analysis targeting ISAba11 and TnAbaR islands in 196 epidemiologically unrelated Acinetobacter strains representative of ≥19 species were performed. The first two Acinetobacter baumannii ISAba11 elements identified had been found to map to the same site on TnAbaR transposons. However, no further evidence of physical linkage between the two elements was demonstrated. Indeed, examination of 25 definite or putative insertion sites suggested limited sequence specificity. Importantly, an aacC1-tagged version of ISAba11 was shown to actively transpose in A. baumannii. Similarity searches identified nine iso-ISAba11 elements in Acinetobacter and one in Enhydrobacter and single representatives of four distant homologs in bacteria belonging to the phyla “Cyanobacteria” and Proteobacteria. Phylogenetic, sequence, and structural analyses of ISAba11 and/or its associated transposase (TnpISAba11) suggested that these elements be assigned to a new family. All five homologs encode transposases with a shared extended signature comprising 16 invariant residues within the N2, N3, and C1 regions, four of which constituted the cardinal ISAba11 family HHEK motif that is substituted for the YREK DNA binding motif conserved in the IS4 family. Additionally, ISAba11 family members were associated with either no flanking direct repeat (DR) or an ISAba11-typical 5-bp DR and possessed variable-length terminal inverted repeats that exhibited extensive intrafamily sequence identity. Given the limited pairwise identity among TnpISAba11 homologs and the observed restricted distribution of ISAba11, we propose that substantial gaps persist in the evolutionary record of ISAba11 and that this element represents a recent though potentially highly significant entrant into the A. baumannii gene pool.
INTRODUCTION
Insertion sequences (IS) are small, typically <2.5-kb, self-transposable genetic elements present in all domains of life. More than 3,145 distinct IS elements have been identified in bacteria (http://www-is.biotoul.fr/), but only a minority have been demonstrated to actively transpose (13). Many IS families have been shown to be disseminated widely among multiple bacterial families, genera, and/or species (29).
Individual IS elements can be grouped into IS families based on characteristics such as transposase homology, transposase motifs, terminal inverted repeat (TIR) length, and composition, target specificity, and direct repeat (DR) length (6, 13). The majority of IS elements are flanked by DR sequences that arise from staggered DNA cuts at the target site, followed by pasting of the IS elements into the insertion locus. The highly dominant IS4 family consists of members distributed throughout numerous organisms belonging to at least eight bacterial phyla and one archaeal phylum (6). In 2008, this large and broad IS4 family was divided into seven recognizable subgroups. Additionally, 74 IS elements with variant forms of a transposase motif conserved in the IS4 family were reclassified into three distinct novel IS families (6).
Only a few IS4 family elements have been experimentally demonstrated to actively transpose in vivo. These include IS231A in Bacillus thuringiensis (9), IS4Bsu1 in Bacillus subtilis (18) and ISAba1 in Acinetobacter baumannii (16). However, molecular mechanisms of transposition have been extensively studied for Tn5 (26) and Tn10 (10), both of which are resistance-encoding composite transposons with a central fragment and flanking terminal IS4 family elements. In addition, crystal structures have been resolved for the Tn5 transposase (TnpTn5) (4). Tn5 transposition is achieved by binding of transposase monomers to the target site, followed by transposase dimerization to form a highly ordered protein-DNA structure. Symmetrical contact of the left and right Tn5 TIRs with both molecules of this dimer results in transposition (4).
The A. baumannii ISAba11 element encodes a predicted transposase that is identical to a hypothetical protein encoded by A. baumannii ATCC 17978 (31). The wider associated ATCC 17978 DNA sequence was subsequently recognized to constitute an insertion sequence designated ISAba11 and deposited in ISfinder (30) as a member of the IS701 family (http://www-is.biotoul.fr/is.html). A. Rose, working in our laboratory, identified an identical IS element in A. baumannii strain A473 (27). His analysis revealed that ISAba11 contained matching 13-bp TIRs and was flanked by identical, target site-generated, 5-bp DRs (27). Remarkably, both of these ISAba11 elements had been found to be located in a location with the same sequence and in the same orientation within the orf1 gene of TnAbaR resistance islands. The orf1 gene is the first of five predicted, tandemly orientated, transposition-associated genes in these Tn7-related transposons (27). The ∼86-kb TnAbaR1 transposon, commonly designated the AbaR1 island, was first identified in A. baumannii strain AYE and found to carry 45 putative resistance genes (8). Subsequently, 18 other elements with features similar to those of TnAbaR1 were reported (12), although unlike counterparts in strains ATCC 17978 and A473, none carried ISAba11 elements. Most recently, Moffatt et al. (15) reported the presence and transposition of ISAba11 in A. baumannii ATCC 19606 and demonstrated a role for this element in the acquisition of colistin resistance through interruption of particular lipid A biosynthesis-associated genes (15).
The Acinetobacter genus comprises over 30 species. A. baumannii is the major human pathogenic species and accounts for an increasing burden of hospital-associated infections frequently caused by multiantibiotic-resistant clones circulating in hospitals worldwide (7). As of October 2011, 37 distinct IS elements had been identified in this genus (http://www-is.biotoul.fr/). Importantly, eight of these IS elements, ISAba1, ISAba2, IS18, an IS3-like element, ISAba3, ISAba125, ISAba825, and ISAba11, have been shown in A. baumannii to be associated with enhanced resistance to penicillins, cephalosporins, carbapenems, and colistin in natural clinical isolates and/or experimentally derived strains (15, 17, 23, 24).
In this study, we examined the distribution of ISAba11 within the Acinetobacter genus and demonstrated that this element actively transposes in A. baumannii. Sequence analysis and modeling of the ISAba11 transposase supported the hypothesis that ISAba11 belongs to a novel family that has diverged from the much broader and highly successful IS4 superfamily.
MATERIALS AND METHODS
Bacterial strains, plasmids, genome sequences, and growth media.
Details of the bacterial strains, plasmids, and genome sequences used in this study are listed in Tables S1 to S3 in the supplemental material. All strains were grown at 37°C in Luria-Bertani (LB) medium. A. baumannii colonies were selected following conjugation on Simmons citrate agar (Oxoid) supplemented with 80 μg/ml gentamicin.
DNA procedures.
Acinetobacter DNA was isolated using the GenElute Bacterial Genomic DNA kit (Sigma) or the ArchivePure DNA Cell/Tissue and Tissue kit (5 Prime), while plasmid DNA was isolated using E.Z.N.A. Plasmid Miniprep Kit I (Omega Bio-Tec). Restriction enzymes (New England BioLabs) and T4 DNA ligase (Promega) were used as indicated by the suppliers, and chemical transformation and electrotransformation performed as previously described (28). For colony PCR, a single bacterial colony was resuspended in 30 μl of ultrapure water, heated at 94°C for 5 min, and centrifuged, and 2 μl of supernatant was used as the template. PCR assays were performed with GoTaq DNA polymerase (Promega) according to the manufacturer's instructions, except that dimethyl sulfoxide was added to a final concentration of 5%. The temperature cycling protocol used was 25 cycles of 95°C for 30 s, the annealing temperature for 30 s, and 72°C for 30 s; the annealing temperature was set at the melting temperature (Tm) minus 1°C for the primer with the lower of the two Tms. Genomic walking was achieved by a two-step gene walking method (21). Finnzymes Phire Hot Start DNA polymerase (Fisher) was used with annealing performed at ≥60°C at a Tm as calculated at https://www.finnzymes.fi/tm_determination.html. Alternatively, inverse-PCR analysis was performed with outwardly facing ISAba11 primers and genomic DNA that had been first restricted and then religated to form single restriction fragment circular template molecules (19). Amplicons were purified using the E.Z.N.A. Cycle Pure kit (Omega Bio-Tec) and sequenced by Eurofins MWG Operon or GATC Biotech. For Southern blotting, 1 μg EcoRI-digested genomic DNA was resolved by agarose gel (0.8%) electrophoresis and capillary blotted onto positively charged nylon membrane (Bio-Rad). Hybridization and detection were performed as specified by the digoxigenin (DIG) High Prime DNA Labeling and Detection Starter kit (Roche).
Construction of a suicide vector carrying a tagged version of ISAba11.
An ∼1.8-kb fragment containing an intact ISAba11 element was amplified from A. baumannii A473 DNA with primers orf1-F/orf1-R and cloned into pGEM-T Easy (Promega) to generate pISAba11. Next, a BamHI restriction site was introduced between the inverted repeat right (IRR) and the 3′ terminus of tnpISAba11 by inverse PCR with primers ISAba11-InF/ISAba11-InR. A 1,052-bp aacC1 cassette from pUC18R6KminiTn7T-Gm was amplified with primers Gm-F-BamHI/Gm-R-BamHI and inserted into this BamHI site to generate pISAba11Gm. The 2.9-kb SacI/SphI aacC1-tagged ISAba11 fragment of pISAba11Gm was then ligated into the same sites in pDS132 (see Table S1 in the supplemental material) to construct pDSISAba11Gm. For details of the primers used in this study, see Table S4 in the supplemental material.
Bacterial conjugation.
Overnight cultures of the donor strain KR1459 (Escherichia coli S17.1 λ pir/pDSISAba11Gm) and the A. baumannii A424 recipient strain were used to inoculate fresh LB medium that was then cultured to an optical density at 600 nm of 0.3 to 0.4. Equal volumes of exponential cultures of the donor and recipient strains were mixed and centrifuged at 16,000 × g (Sigma 1-14 microcentrifuge). Pellets were resuspended in 10% glycerol and spread onto LB agar plates, which were incubated at 37°C for overnight mating. The following day, bacteria were recovered by scraping and resuspended in 10% glycerol prior to plating of suitable dilutions onto Simmons citrate agar (Oxoid) supplemented with gentamicin (80 μg/ml). Plates were incubated at 37°C until colonies appeared.
Homology modeling.
To compare structural features between the transposases of Tn5 and ISAba11, a homology model was built using Modeler (14). HHPred (32) and InterProScan (25) were used to identify and confirm TnpTn5 structures (HHPred E value for Protein Data Bank [PDB] entry 1MUS [33], 9.1e-22) as suitable templates for modeling of TnpISAba11 and to provide a starting point for the target template alignment. As pairwise sequence identity between the TnpISAba11 sequence and the 1MUS sequence is only 11%, a multistep strategy was employed to generate a satisfactory target-template alignment. First, multiple-sequence alignments for both Tn5 and TnpISAba11 homologs were generated by a series of BLAST searches, followed by sequence retrieval, alignment, and manual editing in Jalview (39). The target-template alignment was then derived from a profile-profile alignment between the two multiple-sequence alignments. Secondary structure information for TnpTn5 and secondary structure prediction for TnpISAba11 were also taken into account during the alignment process. The best model out of 20 was selected based on the Modeler scoring function and manual inspection (14). Taking into account the low pairwise sequence identity, the model of TnpISAba11 should be considered a low-resolution model with errors in structural details expected. The PyMOL Molecular Graphics System (version 1.2r3pre; Schrödinger, LLC) was used for visualization of the TnpISAba11 model.
Nucleotide sequence accession numbers.
Sequence data obtained by genomic walking have been deposited in GenBank under accession numbers JN819186 to JN819201. Further details are provided in Table S5 in the supplemental material.
RESULTS
PCR-based survey of ISAba11 and TnAbaR carriage in Acinetobacter.
A total of 148 strains of Acinetobacter species originating as epidemiologically unrelated clinical isolates were screened by PCR for the presence of ISAba11 and TnAbaR elements. A further 48 Acinetobacter strains with complete or near-complete genome sequences deposited in GenBank were investigated by BlastN analysis and in silico PCR. Collectively, 19 or more Acinetobacter species were investigated (Table 1). Screening for ISAba11 was performed with internal tnpISAba11-directed primers (tnp-F/tnp-R) and an outer primer pair (ISAba11-F/ISAba11-R) amplifying the entire element, as we had previously noted evidence of truncated ISAba11 elements (B. Rieck and K. Rajakumar, unpublished data) (Fig. 1A). Combining both experimental and in-silico-derived data, only 4 of the 99 A. baumannii strains (A473, AS42, ATCC 17978, ATCC 19606) were conclusively shown to possess an intact ISAba11 element. However, our PCR, Southern hybridization, and genomic walking analyses of an ATCC 19606 isolate obtained in 2009 from the Salmonella Genetic Stock Centre suggested that our clone possessed only two partial copies of ISAba11, one of which was truncated by 15 bp at the IRR terminus (see below for details). Nevertheless, as an intact ISAba11 sequence was recently reported for A. baumannii ATCC 19606 we have included these latter data in Table 1 and excluded our ATCC 19606 findings to avoid duplication (15). A fifth A. baumannii strain (A479) harbored a remnant of ISAba11 that was missing 110 and 35 bp of the IRR and inverted repeat left (IRL) termini, respectively (Fig. 1B). A sixth A. baumannii strain (6013113) possessed an identical 35-bp deletion at the IRL alone, while the extent of the likely partial ISAba11 element in A. baumannii strain AL7 remains to be determined. However, none of the remaining 92 A. baumannii strains revealed evidence of ISAba11 carriage by either PCR assay. In contrast, ISAba11 was very much more prevalent in several of the non-A. baumannii Acinetobacter species investigated, including A. haemolyticus (present in 4/5 strains examined), A. lwoffii/Acinetobacter genomospecies 9 (28/31), A. johnsonii (6/8), and A. junii (8/9) (Table 1).
Table 1.
Distribution of ISAba11 and TnAbaR elements in Acinetobacter strains
| Acinetobacter speciesa | No. of strains analyzed |
|||
|---|---|---|---|---|
| Totalb | ISAba11 positivee | tniA positivef | Dually positive | |
| A. baumannii | 64 + 35 = 99 | 7 (4) | 56 | 2 |
| A. baylyi/Acinetobacter genomospecies 11c | 2 + 0 = 2 | 0 (0) | 0 | 0 |
| A. berezinae | 2 + 0 = 2 | 0 (0) | 0 | 0 |
| A. beijerinckii | 1 + 0 = 1 | 1 (1) | 0 | 0 |
| A. calcoaceticus | 1 + 2 = 3 | 0 (0) | 0 | 0 |
| Acinetobacter genomospecies 13 | 2 + 0 = 2 | 1 (1) | 0 | 0 |
| Acinetobacter genomospecies 16 | 1 + 0 = 1 | 0 (0) | 0 | 0 |
| Acinetobacter genomospecies 15TU | 3 + 0 = 3 | 3 (3) | 1 | 1 |
| A. gyllenbergii | 1 + 0 = 1 | 1 (1) | 0 | 0 |
| A. haemolyticus | 3 + 2 = 5 | 4 (0) | 0 | 0 |
| A. johnsonii | 6 + 2 = 8 | 6 (5) | 0 | 0 |
| A. junii | 8 + 1 = 9 | 8 (8) | 2 | 2 |
| A. lwoffii/Acinetobacter genomospecies 9c | 29 + 2 = 31 | 28 (26) | 6 | 6 |
| A. nosocomialis sp. nov. | 2 + 1 = 3 | 0 (0) | 0 | 0 |
| A. parvus | 3 + 0 = 3 | 2 (2) | 0 | 0 |
| A. pittii sp. nov. | 7 + 1 = 8 | 0 (0) | 1 | 0 |
| A. radioresistens | 3 + 2 = 5 | 1 (1) | 1 | 0 |
| A. schindleri | 3 + 0 = 3 | 1 (0) | 1 | 1 |
| A. ursingii | 4 + 0 = 4 | 1 (1) | 0 | 0 |
| A. ursingii groupd | 3 + 0 = 3 | 0 (0) | 1 | 0 |
One hundred eighteen strains were identified to the species level by rpoB gene sequencing, while a further 27 of 64 A. baumannii strains were identified to the species level by API NE biochemical testing and blaOXA-51-like PCR analysis. A final three A. lwoffii/Acinetobacter genomospecies 9 strains that were rpoB PCR negative were identified by API NE biochemical testing alone. Species designations corresponding to the 48 sequenced genomes analyzed were as identified in the GenBank database, with the exception of the strains currently unidentified to the species level, ATCC 27244, DR1, ADP1, RUH2624, and SH024, which were identified by in silico rpoB sequence analysis as A. haemolyticus, A. baumannii, A. johnsonii, A. nosocomialis sp. nov., and A. pittii sp. nov., respectively. Full details of strains and PCR results are shown in Tables S2 and S3 in the supplemental material.
The numbers of experimentally and in-silico-analyzed sequenced genomes (underlined values) are shown; the latter were investigated by BlastN and in silico PCR analyses alone. The sequenced genomes analyzed comprised the 48 complete and near-complete Acinetobacter genome sequences available in GenBank as of 30 September 2011, except for the A. baumannii ATCC 19606 and 6013150 sequences, which were excluded to avoid duplication.
rpoB sequence analysis could not reliably differentiate between these species.
The rpoB sequences of the six strains assigned to this group exhibited the best match but nonidentity to the A. ursingii rpoB sequence.
Total numbers of strains harboring intact and/or partial ISAba11 elements are shown; values in parentheses indicate those possessing intact ISAba11 elements only.
tniA is a TnAbaR-specific gene.
Fig 1.
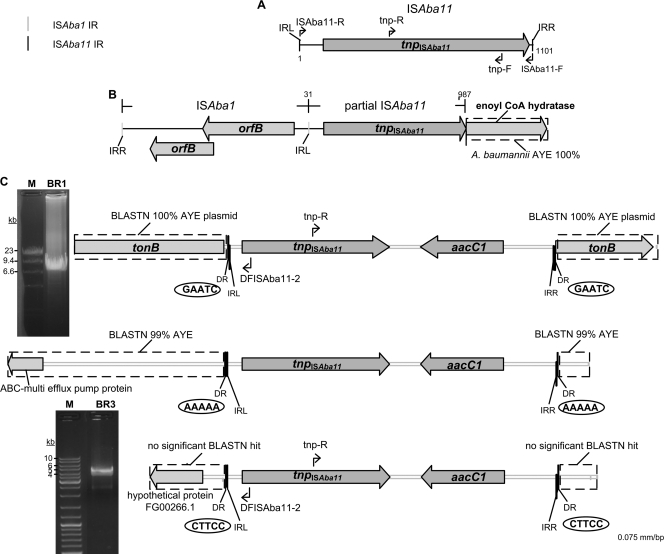
Locations of ISAba11 and ISAba11-aacC1 in four A. baumannii strains. (A) Schematic of ISAba11 showing locations of TIRs, primer binding sites, and tnpISAba11. (B) Overview of the partial ISAba11 element in A. baumannii A479 highlighting its “fusion” to an intact ISAba1 element. (C) Genetic contexts of ISAba11-aacC1 in the A. baumannii A424 derivatives BR1 (top), BR2 (middle), and BR3 (bottom). Identified 5-bp DRs are displayed in ellipses. Gel images show inverse-PCR amplicons (primers: DFISAba11-2/tnp-R) with BR1 (top) and BR3 (bottom) template DNA. Lane M contains a DNA molecular size ladder. Genes are shown as gray arrows and labeled with gene or protein descriptors. Numbers, where shown, are ISAba11 coordinates.
The PCR survey for the presence of TnAbaR elements utilized primers targeting the TnAbaR-specific tniA gene (tniAF/tniAR) (27). The first two A. baumannii ISAba11 elements identified had been found to map to the same location within TnAbaR elements. However, we observed a marked discordance in the prevalence of the two elements. Fifty-six of the 99 A. baumannii strains were positive for tniA and, by inference, for a TnAbaR element, suggesting an A. baumannii TnAbaR carriage rate of ∼57%, in contrast to the 7% of A. baumannii strains that harbored intact and/or remnant ISAba11 elements (Table 1). Examination of data for the non-A. baumannii strains also failed to reveal obvious evidence of the “colocalization” of these two elements to a common host strain and/or Acinetobacter species.
Locations of ISAba11 in selected Acinetobacter sp. strains.
As ISAba11 was known to have been inserted twice within the same site on the TnAbaR orf1 gene (27, 31), we sought further evidence of physical linkage between orf1 and ISAba11. However, amplification across orf1 using primers orf1-F/orf1-R in the 12 Acinetobacter strains testing positive for both ISAba11 and TnAbaR revealed no other examples of an interrupted orf1 gene (see Fig. S1A in the supplemental material). PCR-based genome walking analysis of two A. baumannii and six non-A. baumannii strains led to the localization of ISAba11 elements in these empirically selected, ISAba11-positive strains. As examples, in A. lwoffii/Acinetobacter genomospecies 9 AL5 and A. junii AJ11, ISAba11 inter-rupted genes coding for an esterase (GenBank accession no. ZP_06069171; 98% amino acid identity) and a membrane protein (GenBank accession no. ZP_06066150; 98% amino acid identity), respectively. One of the partial copies of ISAba11 in our clone of A. baumannii ATCC 19606 was found to lie immediately downstream of a putative phospholipase C gene (GenBank accession no. EEX02177; 98% amino acid identity), and the partial copy in A. baumannii A479 interrupted a putative enoyl coenzyme A hydratase/isomerase gene (GenBank accession no. YP_001712458; 97% amino acid identity) and was directly “fused” to an ISAba1 element (Fig. 1B). The insertion site of ISAba11 in the incompletely assembled ATCC 19606 genome sequence could not be determined. Available insertion site data for the eight characterized strains are shown in Table S5 in the supplemental material; no duplicate target sites were detected.
Identification and characterization of de novo transposition of ISAba11 in A. baumannii.
ISAba11 was tagged with an aacC1 gentamicin resistance cassette located between the 3′ terminus of tnpISAba11 and the 13-bp terminal IRR. The tagged ISAba11 (ISAba11-aacC1) sequence was introduced into the suicide vector pDS132, generating pDSISAba11Gm. This plasmid was then transferred from the E. coli S17.1 λ pir-derived donor into A. baumannii strain A424 by conjugation. A. baumannii strain A424 was chosen as the recipient because it lacked ISAba11 and carried a comM-borne TnAbaR element, thus providing the opportunity to investigate whether ISAba11 is inserted preferentially into TnAbaR. Colonies were selected by plating the overnight conjugation mixture on Simmons citrate medium supplemented with gentamicin (80 μg/ml). Plating of donor-donor and recipient-recipient mock conjugations, as expected, led to no colonies on this medium. In contrast, each donor-recipient conjugation experiment yielded more than 300 gentamicin-resistant colonies, suggesting de novo ISAba11-aacC1 transposition or homologous recombination between the suicide plasmid and the recipient genome. Seventy randomly selected gentamicin-resistant colonies from two independent conjugation experiments were screened by PCR for aacC1 and tnpISAba11 and found to be positive for both targets. However, 67 of these colonies yielded a positive band with primers aacC1R/orf1-F that amplified a 829-bp fragment spanning the orf1-ISAba11-aacC1 junction present in pDSISAba11Gm (see Fig. S1B in the supplemental material). Subsequent analysis of a further 28 independently derived gentamicin-resistant colonies suggested that few, if any, of these original 67 colonies were the result of targeted transposition of ISAba11-aacC1 into the A424-borne TnAbaR orf1 gene. Instead, the vast majority almost certainly resulted from chromosomal integration of pDSISAba11Gm following orf1-targeted homologous recombination between the suicide plasmid and the A424-borne orf1 gene (see Fig. S1 in the supplemental material for details). The remaining three original colonies, representative of likely ISAba11-aacC1 transposition, were further characterized by genomic walking and confirmed as bearing newly transposed copies of ISAba11-aacC1 flanked by characteristic 5-bp DRs (Fig. 1C).
A single copy of ISAba11-aacC1 was mapped to a distinct genomic location in each of the three insertion mutants. In clone A424-BR1, ISAba11-aacC1 was inserted into a TonB-dependent receptor gene (Fig. 1C; GenBank accession no. ZP_06795313). Homologs of this gene exhibiting upwards of 98% BLASTN identity were found on eight different A. baumannii plasmids, including pAB0057 (8.7 kb) (1), pABVA01 (9.0 kb) (3), and pMMCU3 (9.0 kb) (GenBank accession no. GQ904227). An inverse PCR on A424-BR1 genomic DNA performed with outwardly facing ISAba11-specific primers (tnp-R/DFISAba11-2) amplified an ∼9.0-kb band (Fig. 1C), thus confirming that the gene targeted in A424-BR1 also lay on a similarly sized plasmid. In clone A424-BR2, ISAba11-aacC1 was inserted upstream of a typically chromosomally located ABC-type multidrug efflux pump gene (GenBank accession no. YP_001713963; 100% BLASTP identity) conserved in all of the A. baumannii strains sequenced to date (Fig. 1C). The third de novo ISAba11-aacC1 transposition event observed in A424-BR3 mapped to an entirely novel sequence that was shown to be plasmid borne by inverse PCR. The closest match, of questionable significance, was to a sequence upstream of a gene coding for a hypothetical protein in the fungus Gibberella zeae PH-1 (GenBank accession no. FG00266; 26% amino acid identity) (Fig. 1C). Hence, two of the three de novo ISAba11-aacC1 transposition events identified targeted plasmids rather than the chromosome.
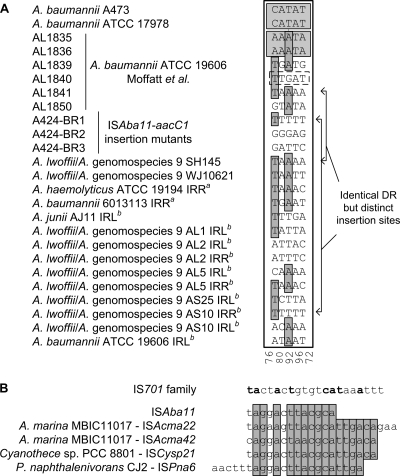
All three characterized transposition events generated distinct but perfect 5-bp DR sequences (Fig. 1C), consistent with the IS element having targeted a different local sequence in each case and suggesting a lack of strong target sequence preference. Furthermore, examination of all of the 5-bp DR sequences associated with mapped intact iso-ISAba11 elements revealed insertions into at least 11 distinct target sequences (Fig. 2A). The exceptions were the previously observed targeting of a site with the same sequence within the TnAbaR orf1 gene and the recently reported targeting of the same site within lpxC in ATCC 19606 (15). If data restricted to only one end of an ISAba11 element were also considered, a further 12 unique insertion sites would also be recognized. The sequences TTTTT and TAAAA occur twice but in these cases correspond to ISAba11 insertions into distinct loci (Fig. 2A).
Fig 2.
Repeat sequences associated with ISAba11 and its homologs. (A) Alignment of identified and putative 5-bp DRs flanking mapped ISAba11 elements. Data generated by Moffatt et al. (15) and available within the genome database are also included. The suffix IRL or IRR indicates that the data are not definitive, as flanking sequences are only available for the indicated end. The superscript letters a and b indicate a terminally truncated ISAba11 element and limited sequence data, respectively. Nucleotides conserved in the majority of sequences are shaded dark gray. Pairs of DRs in lighter boxes highlight identified instances of insertion at identical sites in independent strains. The DR shown in the dashed-outline box corresponds to an insertion site in lpxC shared by a second ISAba11 insertion mutant that harbors a 34-bp DR instead (15). The numbers at the bottom are the A+T percentages of the columns. (B) Alignment of IRL sequences from ISAba11 and its four homologs. For comparison, the consensus IS701 family IRL, as defined by De Palmenaer et al. (6), is shown at the top (shared residues are in bold). ISAba11 family IRL residues that are strictly conserved are shaded gray.
ISAba11 copy number estimation in Acinetobacter species.
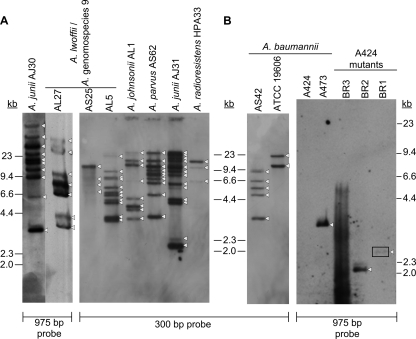
Southern hybridization with a DIG-labeled probe specific for ISAba11 was performed on EcoRI-digested genomic DNA from four wild-type A. baumannii strains, eight non-A. baumannii strains, and the three newly generated ISAba11 insertion mutants. All were ISAba11 positive, except for A. baumannii A424. EcoRI does not cut within ISAba11 or aacC1 and thus allowed the determination of minimum ISAba11 copy number values. This analysis showed that the ISAba11 copy number varied from 1 to 5 in the three natural ISAba11-positive A. baumannii strains studied to ≥7 copies in six of the eight non-A. baumannii strains investigated. A. junii J31 possessed an estimated 13 copies (Fig. 3). As expected, no ISAba11-specific band was detected for A. baumannii A424. Single bands were observed for A424-BR2 and A424-BR1, while the intense and smeared A424-BR3 hybridization pattern was attributed to the ∼2.5-kb ISAba11-aacC1-bearing plasmid that had been identified by inverse PCR. Nevertheless, the possibility of additional ISAba11-aacC1 copies in A424-BR3 could not be ruled out (Fig. 3B).
Fig 3.
Southern blot-based estimation of ISAba11 copy numbers in selected Acinetobacter strains. EcoRI-digested and resolved genomic DNA from eight non-A. baumannii strains (A) and seven A. baumannii strains (B) was hybridized against a 300-bp or 975-bp ISAba11-specific, DIG-labeled probe amplified from A. baumannii A473 genomic DNA with primers tnp-F/tnp-R and ISAba11-F/tnp-F, respectively. The positions of DNA size markers are shown. Arrowheads highlight individual detected bands, while the weak BR1-associated band is boxed.
In silico analysis of the ISAba11 transposase.
BLASTP analysis of TnpISAba11 against the NCBI genome database revealed the presence of highly similar homologs (98 to 100% sequence identity) in nine Acinetobacter strains belonging to four Acinetobacter species, and in Enhydrobacter aerosaccus SK60 (Table 2). However, intact copies of iso-ISAba11 elements could be identified in only six strains, while variably truncated or indeterminate copies were found in the others (Table 2). Four proteins, encoded by phylogenetically distant organisms, were found to exhibit 36 to 49% identity to TnpISAba11 (Fig. 4 and 5; Table 2). Genes coding for two of these resided on plasmids found in Cyanothece sp. strain PCC 8801 (GenBank accession no. ACK683871) and Polaromonas naphthalenivorans CJ2 (GenBank accession no. CP000533). Examination of flanking sequences confirmed that both mapped to typical insertion sequences (Table 2). The remaining two distant homologues, found in Acaryochloris marina MBIC11017, were encoded by the chromosomally borne ISAcma22 and ISAcma42 elements, which had been deposited in ISfinder by Swingley et al. (34).
Table 2.
Characteristic features of members of the ISAba11 family identified in available bacterial sequence dataa
| Strain/IS designation | Insertion sequence |
Transposase |
||||
|---|---|---|---|---|---|---|
| Length (bp) | TIR length (bp)/no. of mismatchese | DR length (bp) | BLASTN hit (%)b | No. of aa | BLASTP hit (%)b | |
| Acinetobacter baumannii ATCC 17978/ISAba11 | 1,101 | 13/0 | 5 | 100 | 324 | 100 |
| Acinetobacter baumannii A473/ISAba11 | 1,101 | 13/0 | 5 | 100 | 324 | 100 |
| Acinetobacter baumannii ATCC 19606/iso-ISAba11f | 1,101 | 13/0 | NA | 99 | 324 | 98 |
| Acinetobacter lwoffii/Acinetobacter genomospecies 9 SH145/iso-ISAba11 | 1,101 | 13/0 | 5 | 95c | 324 | 100 |
| Acinetobacter lwoffii/Acinetobacter genomospecies 9 WJ10621/iso-ISAba11 | 1,101 | 13/0 | 5 | 99 | 324 | 100 |
| Enhydrobacter aerosaccus SK60/ISEnae1 | 1,101 | 13/0 | None | 98 | 324 | 100 |
| Acinetobacter haemolyticus ATCC 27244/iso-ISAba11c | ≥1,062 | 13/NA | NA | 98 | 324 | 99 |
| Acinetobacter johnsonii SH046/iso-ISAba1c | ≥833 | 13/NA | NA | 98 | ≥241 | 100 |
| Acinetobacter baumannii 6013113/iso-ISAba11d | 1,066 | 13/NA | NA | 98 | 324 | 98 |
| Acinetobacter haemolyticus ATCC 19194/iso-ISAba11d | 557 | 13/NA | NA | 98 | 185 | 100 |
| Acaryochloris marina MBIC11017/ISAcma22 | 1,251 | 22/2 | 5 | NS | 350 | 45 |
| Acaryochloris marina MBIC11017/ISAcma42 | 1,140 | 18/1 | 5 | NS | 308 | 36 |
| Polaromonas naphthalenivorans CJ2(pPNAP04)/ISPna6 | 1,134 | 22/1 | None | NS | 326 | 49 |
| Cyanothece sp. strain PCC 8801(pP880101)/ISCysp21 | 1,139 | 19/0 | None | NS | 321 | 44 |
Abbreviations: aa, amino acids; NA, not available (due to incomplete sequence data and/or IS truncation); NS, not significant.
BLASTN and BLASTP results are shown as percentages of identity to the ISAba11 and TnpISAba11 sequences, respectively.
Incomplete and/or poor-quality sequence data.
Truncated copy of iso-ISAba11.
Number of mismatches between the left and right TIRs.
The sequence of an intact iso-ISAba11 element in A. baumannii ATCC 19606 was recently reported by Moffatt et al. (15).
Fig 4.
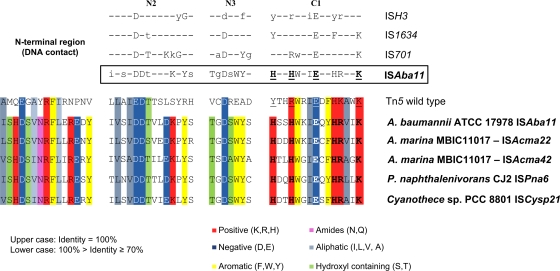
Alignment of key regions of TnpISAba11, the four HHEK-bearing TnpISAba11 homologs, and TnpTn5. The N2, N3, and C1 regions bear the archetypal DDE residues common to many transposases. The N-terminal region is also shown, as it is highly conserved and known to partake in DNA contact during Tn5 transposition. TnpTn5 is included as a representative IS4 family transposase. The YREK motif of TnpTn5 (underlined residues), found in the C1 region, characterizes all of the IS4 family transposases. The glutamic acid residue is shared between the DDE and YREK motifs. Residues within the N2, N3, and C1 regions shared by members of the ISH3, IS1634, and IS701 families, as identified by De Palmenaer et al. (6), are shown in uppercase (strictly conserved) or lowercase (highly conserved) letters (upper panel), as appropriate. Residues shared by members of the ISAba11 family are similarly highlighted. The HHEK motif, which is characteristic of the ISAba11 family, is shown in bold and underlined. Colors indicate conserved residues with shared physiochemical properties, as depicted in the key. This figure has been adapted from a schematic first presented by De Palmenaer et al. (6).
Fig 5.
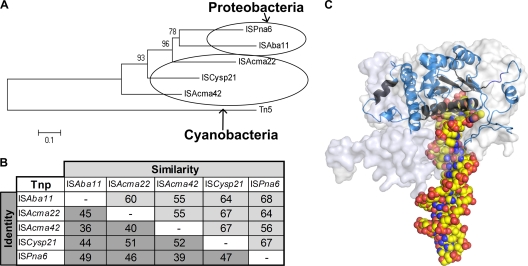
Phylogeny, predicted structure, and similarities of TnpISAba11. (A) Phylogenetic relationships of ISAba11 family transposases and TnpTn5. The phylogenetic tree was generated in Mega5 (35) using the maximum-likelihood method with default settings based on the underlying profile-profile alignment between the Tn5 and the TnpISAba11 homologs as described in Materials and Methods. Ovals indicate the two associated host phyla. (B) Pairwise amino acid sequence similarity/identity matrix for TnpISAba11 and its four homologs encoded by ISAcma22, ISAcma42, ISCysp21, and ISPna6. (C) Low-resolution homology model of the TnpISAba11 monomer shown in a cartoon representation (α-helices as helices, β-strands as arrows). Color is used to indicate conservation. The most-conserved regions used for anchoring the TnpISAba11-TnpTn5 alignment are highlighted in black in the TnpISAba11 model (TnpISAba11 residues: His20-Leu27, Ile51-Thr57, Phe135-Tyr142, Gly155-Asp163, and Phe224-Val247), while other regions of the TnpISAba11 model are shown in blue. For size comparison, the overall shape of the experimentally determined TnpTn5 monomer structure (PDB entry 1MUS) is shown as a transparent surface representation. A generic DNA substrate (sphere model) is overlaid.
All of the available TIR sequences for the iso-ISAba11 elements identified perfectly matched those of the ISAba11 element described in this study, while those of the four distant homologs ranged from 18 bp to 22 bp, with up to two mismatches between cognate IRL and IRR sequences. Remarkably, there was extensive identity among the IRL sequences of all five homologs (Fig. 2B). However, a characteristic flanking DR could be found for only the two elements in A. marina MBIC11017 and those in Acinetobacter spp. (Table 2), suggesting that DR generation was not necessarily a feature of these elements or that restructuring events had occurred following initial transposition.
TnpISAba11 modeling based on the crystal structure of Tn5 transposase.
An InterProScan search predicts that TnpISAba11 contains the “transposase DDE domain” (Pfam entry PF01609; E value, 1.4e-9), which belongs to the RNase H-like superfamily. The transposase DDE domain contains a motif of three conserved carboxylate residues (DDE), which are thought to be responsible for coordinating metal ions needed for catalysis. This has been shown conclusively in the E. coli TnpTn5 structure (33).
The homology model of TnpISAba11 is based on the E. coli TnpTn5 structure (PDB entry 1MUS [33]). TnpTn5 consists of 476 amino acids, whereas TnpISAba11 is only 324 amino acids long. This is attributed to significantly shorter N- and C-terminal regions and in part shorter loops between secondary structure elements in TnpISAba11. For example, there are no corresponding TnpISAba11 residues for the ∼35 residues of the N-terminal region of TnpTn5. Also, the 40 C-terminal TnpISAba11 residues could not be aligned with the ∼100 residues of the C terminus of TnpTn5 and hence were not modeled (Fig. 5C). However, a number of regions were found to be relatively conserved across the TnpTn5-TnpISAba11 profile-profile alignment. These regions are located mostly at the core of the DNA binding region, in particular, the functionally important N2, N3, and C1 regions of TnpTn5, which include the DDE motif (D97, D188, and E326 in TnpTn5). The DDE motif is fully conserved between Tn5 and TnpISAba11 homologs. The C1 region also contains the YREK motif (Y319, R322, E326, and K333 in TnpTn5). The residues of the latter motif are involved in metal coordination (E326) and DNA binding (Y319, R322, and K333) (11). A variation of the YREK motif, HHEK, was found to be conserved across all five TnpISAba11 homologs shown in Fig. 4. Furthermore, the N-terminal domain showed sequence conservation between TnpTn5 and the TnpISAba11 homologs in a region required for initial DNA recognition by TnpTn5 (4). This region includes R62 (corresponding to R25 in TnpISAba11), which is critical for TnpTn5-DNA interactions (37) and is fully conserved across the entire TnpISAba11 protein alignment (Fig. 4).
DISCUSSION
A. baumannii exhibited the highest TnAbaR carriage rate (56/99; 57%) but a much lower ISAba11 positivity ranking (7%; ranked 12/19) among the species studied. In contrast, 90% of the 31 A. lwoffii/Acinetobacter genomospecies 9 strains analyzed were ISAba11 positive but only 19% harbored TnAbaR elements. The limited Southern data also suggest that ISAba11 is typically found at a lower copy number in A. baumannii than in selected non-A. baumannii species. Species-specific niches may partly explain the seemingly distinct distributions of ISAba11 and TnAbaR. For example, the vast majority of Acinetobacter clinical isolates are A. baumannii (36), a species rarely isolated from other environments. In contrast, many other Acinetobacter species are found primarily in soil, water, and/or inanimate environments (7), potentially hampering horizontal gene transfer between environmental and human-resident Acinetobacter species (5).
The presence of ISAba11 at the same location within the TnAbaR-borne orf1 gene in two A. baumannii strains could reflect divergence from a common clone or independent ISAba11 insertion events. As TnAbaR has been hypothesized to transpose via a Tn7-analogous mechanism, Orf1 would be predicted to be essential for transposition. Hence, a cut-and-paste TnAbaR element carrying an ISAba11-disrupted orf1 gene would be stabilized, as with the role of IS26 in locking-in the blaCTX-M-laden ISEcp element (40). Moffatt et al. have recently shown that ISAba11 can target two lipid A biosynthesis pathway genes (lpxA, lpxC). Eight such mutants were identified in their study. Two each carried ISAba11 elements that had targeted distinct sites with the same sequences in lpxC, suggesting the presence of insertion hot spots within this gene (15). Nonetheless, available data suggest that ISAba11 displays target promiscuity, as we have identified 23 distinct definite or likely insertion sites, suggesting, at best, low-level targeting of the TnAbaR orf1 and lpxC genes and a possible slight bias toward A+T-rich loci (Fig. 2A).
The low pairwise identity values (36 to 52%) among the five HHEK-bearing TnpISAba11 homologs suggest that these proteins and, by extension, the associated IS elements have evolved from a common ancestor that existed in the distant past (Fig. 5A). Horizontal gene transfer between phylogenetically distant species is thought to be rare, and IS dissemination of this nature typically reflects ancient transposition and subsequent evolutionary divergence (38). Transphylum and even transdomain distribution of members of an IS family has been reported. Cassier-Chauvat et al. identified an IS element found in the cyanobacterium Synechocystis which had a significant homolog in alphaproteobacteria (2), while the Shigella dysenteriae IS1 transposase exhibits amino acid identity of up to 48% with IS-encoded transposases in archaea (20). In contrast, copies of IS elements in closely related species often approach identity, reflecting very recent dissemination. This is the case for the iso-ISAba11 elements identified in Acinetobacter spp. and the single E. aerosaccus representative, consistent with both Acinetobacter and Enhydrobacter belonging to the Moraxellaceae family of the class Gammaproteobacteria. The restricted distribution and low copy number of ISAba11 in A. baumannii, as opposed to its frequent and abundant presence in other selected Acinetobacter species, suggest that ISAba11 has only recently entered the A. baumannii gene pool, most probably via direct transfer from another Acinetobacter species. Assuming that ISAba11 behaves as a cut-and-paste element, like its IS4 relatives, copy number expansion would require repeated horizontal acquisition and/or entry on a multicopy plasmid (Table 1).
Three distinct variations of the archetypal IS4 transposase family YREK motif had been previously identified by De Palmenaer et al. (6), leading these authors to regroup IS elements with these motifs into the independent IS families, IS701, ISH3, and IS1634. ISAba11 had been posted on ISfinder as a member of the IS701 family (31). However, identification of the novel HHEK motif in TnpISAba11 and its homologs; high-level conservation of cognate TIRs; and recognition that insertion of ISAba11, ISAcma22, and ISAcma42 generates a 5-bp DR instead of the 4-bp DR typical of IS701 family members (6) led us to assign ISAba11 and its homologs to a new family designated the ISAba11 family. Indeed, based on differences in DR and TIR lengths and the extent of nucleotide conservation between ISAba11 and members of the IS701 and IS4 families, Moffatt et al. had also recently proposed that ISAba11 be assigned to a new IS family (15). Notably, the single representatives of ISCysp21 and ISPna6 and the only identified non-Acinetobacter-borne iso-ISAba11 element were not associated with flanking DRs (Table 2). Additionally, Moffatt et al. had reported a single instance of a newly transposed ISAba11 element being flanked by 34-bp perfect DRs (15), suggesting the possibility of low-frequency alternate cleavage sites during ISAba11 transposition (22). A further 22 IS elements present in seven bacterial phyla and one archaeal phylum encode transposases with marked similarity to TnpISAba11. Remarkably, these elements possess TIRs resembling those of the ISAba11 family, but none encodes a transposase with a complete HHEK motif, raising the possibility of broadening the ISAba11 family to include elements with the less stringent HEK or EK submotif as well (see Fig. S2 in the supplemental material).
The five HHEK-bearing ISAba11 family transposases share significant sequence similarity with the TnpTn5 DDE domain that is known to bind to DNA during transposition; in particular, the DDE motif is fully conserved (33). This suggests that ISAba11 family transposases could utilize a similar method of TIR sequence recognition to that of TnpTn5 (33). However, a variety of distinctive features between ISAba11 family and Tn5 transposases suggests divergence of the DNA sequence recognition process. Indeed, the contrasting lengths of the 19-bp Tn5 TIR and the 13-bp ISAba11 TIR appear to correspond to the different sizes of the DNA binding domains in TnpTn5 and TnpISAba11, the latter having been predicted by the three-dimensional modeling performed in this study. TnpISAba11 appears to be a “minimal” version of TnpTn5, consistent with the reduced protein-DNA interaction space associated with the shorter ISAba11 TIRs. Secondary structure predictions of TnpISAba11 also suggest the presence of a C-terminal α-helix, which is not included in the TnpISAba11 model, as this region could not be reliably aligned with TnpTn5. The equivalent region in TnpTn5 forms an α-helical subdomain which has been shown to mediate protein-protein interactions (33). Analogously, the predicted C-terminal α-helix of TnpISAba11 may well be involved in dimerization.
The precise mechanisms of ISAba11 transposition, factors that govern transposition efficiency, barriers to wider taxonomic dissemination, and the potential impact of this element on the downstream evolution of Acinetobacter warrant further study. The recent report of ISAba11-mediated colistin resistance is clearly of major concern (15). Indeed, should ISAba11 permeate A. baumannii as it has done in a number of other Acinetobacter species, widespread resistance to this last-resort agent may become a feature of endemic A. baumannii strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Owen Lancaster for help with bioinformatics, Donna Mathew for technical assistance, and Jon van Aartsen for helpful advice. Major thanks to Te-Li Chen (Division of Infectious Diseases, Taipei Veterans General Hospital, Taipei, Taiwan, Republic of China), Simon Hewson, Eva Horvath-Papp, and Kevin Towner for providing Acinetobacter strains.
This work was funded by a BSAC grant to K.R.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adams MD, et al. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cassier-Chauvat C, Poncelet M, Chauvat F. 1997. Three insertion sequences from the cyanobacterium Synechocystis PCC6803 support the occurrence of horizontal DNA transfer among bacteria. Gene 195:257–266 [DOI] [PubMed] [Google Scholar]
- 3. D'Andrea MM, et al. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3528–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davies DR, Goryshin IY, Reznikoff WS, Rayment I. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77–85 [DOI] [PubMed] [Google Scholar]
- 5. de la Cruz F, Davies J. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128–133 [DOI] [PubMed] [Google Scholar]
- 6. De Palmenaer D, Siguier P, Mahillon J. 2008. IS4 family goes genomic. BMC Evol. Biol. 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doughari HJ, Ndakidemi PA, Human IS, Benade S. 2011. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes Environ. 26:101–112 [DOI] [PubMed] [Google Scholar]
- 8. Fournier PE, et al. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hallet B, Rezsohazy R, Mahillon J, Delcour J. 1994. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol. 14:131–139 [DOI] [PubMed] [Google Scholar]
- 10. Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB. 1998. Tn10 transposition via a DNA hairpin intermediate. Cell 95:125–134 [DOI] [PubMed] [Google Scholar]
- 11. Klenchin VA, et al. 2008. Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif. Nucleic Acids Res. 36:5855–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krizova L, Dijkshoorn L, Nemec A. 2011. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob. Agents Chemother. 55:3201–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martí-Renom MA, et al. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291–325 [DOI] [PubMed] [Google Scholar]
- 15. Moffatt J, et al. 2011. The insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:3022–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mugnier PD, Poirel L, Nordmann P. 2009. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 191:2414–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mussi MA, Limansky AS, Viale AM. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagai T, Phan Tran LS, Inatsu Y, Itoh Y. 2000. A new IS4 family insertion sequence, IS4Bsu1, responsible for genetic instability of poly-gamma-glutamic acid production in Bacillus subtilis. J. Bacteriol. 182:2387–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ochman H, Gerber AS, Hartl DL. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohta S, et al. 2002. Presence of a characteristic D-D-E motif in IS1 transposase. J. Bacteriol. 184:6146–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pilhofer M, et al. 2007. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res. 35:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plikaytis BB, Crawford JT, Shinnick TM. 1998. IS1549 from Mycobacterium smegmatis forms long direct repeats upon insertion. J. Bacteriol. 180:1037–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poirel L, et al. 2005. OXA-58, a novel class D {beta}-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poirel L, Nordmann P. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reznikoff WS. 2008. Transposon Tn5. Annu. Rev. Genet. 42:269–286 [DOI] [PubMed] [Google Scholar]
- 27. Rose A. 2010. TnAbaR1: a novel Tn7-related transposon in Acinetobacter baumannii that contributes to the accumulation and dissemination of large repertoires of resistance genes. Biosci. Horiz. 3:40–48 [Google Scholar]
- 28. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1989 [Google Scholar]
- 29. Siguier P, Filee J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9:526–531 [DOI] [PubMed] [Google Scholar]
- 30. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith MG, et al. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960 [DOI] [PubMed] [Google Scholar]
- 33. Steiniger-White M, Rayment I, Reznikoff WS. 2004. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 14:50–57 [DOI] [PubMed] [Google Scholar]
- 34. Swingley WD, et al. 2008. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl. Acad. Sci. U. S. A. 105:2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Turton JF, Shah J, Ozongwu C, Pike R. 2010. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48:1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Twining SS, Goryshin IY, Bhasin A, Reznikoff WS. 2001. Functional characterization of arginine 30, lysine 40, and arginine 62 in Tn5 transposase. J. Biol. Chem. 276:23135–23143 [DOI] [PubMed] [Google Scholar]
- 38. Wagner A, de la Chaux N. 2008. Distant horizontal gene transfer is rare for multiple families of prokaryotic insertion sequences. Mol. Genet. Genomics 280:397–408 [DOI] [PubMed] [Google Scholar]
- 39. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple-sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zong Z, Partridge SR, Iredell JR. 2010. ISEcp1-mediated transposition and homologous recombination can explain the context of bla(CTX-M-62) linked to qnrB2. Antimicrob. Agents Chemother. 54:3039–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.