Abstract
Polar and alpine microbial communities experience a variety of environmental stresses, including perennial cold and freezing; however, knowledge of genomic responses to such conditions is still rudimentary. We analyzed the metagenomes of cyanobacterial mats from Arctic and Antarctic ice shelves, using high-throughput pyrosequencing to test the hypotheses that consortia from these extreme polar habitats were similar in terms of major phyla and subphyla and consequently in their potential responses to environmental stresses. Statistical comparisons of the protein-coding genes showed similarities between the mats from the two poles, with the majority of genes derived from Proteobacteria and Cyanobacteria; however, the relative proportions differed, with cyanobacterial genes more prevalent in the Antarctic mat metagenome. Other differences included a higher representation of Actinobacteria and Alphaproteobacteria in the Arctic metagenomes, which may reflect the greater access to diasporas from both adjacent ice-free lands and the open ocean. Genes coding for functional responses to environmental stress (exopolysaccharides, cold shock proteins, and membrane modifications) were found in all of the metagenomes. However, in keeping with the greater exposure of the Arctic to long-range pollutants, sequences assigned to copper homeostasis genes were statistically (30%) more abundant in the Arctic samples. In contrast, more reads matching the sigma B genes were identified in the Antarctic mat, likely reflecting the more severe osmotic stress during freeze-up of the Antarctic ponds. This study underscores the presence of diverse mechanisms of adaptation to cold and other stresses in polar mats, consistent with the proportional representation of major bacterial groups.
INTRODUCTION
Microbial mats dominated by cyanobacteria are commonly found in extreme environments, such as geothermal springs, hypersaline basins, ultraoligotrophic ponds, and hot and cold desert soils (10, 14). Cyanobacterial mats are also a dominant feature of polar lake, pond, and river ecosystems, with some of the most luxuriant communities growing on the thick ice shelves that float on Arctic and Antarctic seas (55). The stresses encountered by organisms on polar ice shelves include sparse nutrients, freeze-thaw cycles, bright sunlight exposure during summer, prolonged darkness during winter, salinity fluctuations, desiccation, and persistent low temperatures (29, 56). Extreme cold is an overarching stress in the polar regions because it drastically modifies the physical-chemical environment of living cells, with effects on biochemical reaction rates, substrate transport, membrane fluidity, and conformation of macromolecules, such as DNA and proteins (45, 61). Once the freezing point is crossed, there are additional physical and chemical stresses imposed by ice crystal formation, water loss, and increasing solute concentrations.
Although polar ice shelf mats are visually dominated by cyanobacteria, other microorganisms, including Bacteria, Archaea, and protists, live within these mats, supporting microinvertebrates, such as nematodes, rotifers, and tardigrades (4). Previous studies on the mats have shown that they contain much higher concentrations of nutrients than the overlying ultraoligotrophic waters (54). Furthermore, proteins involved in diverse scavenging and recycling processes are coded for within the mat metagenome (53), suggesting that the cyanobacteria profit, in terms of recycled nutrient supply, from the close proximity to other microorganisms.
Heterotrophic Bacteria and Archaea isolated from polar environments appear to be true psychrophiles in that they show evidence of cold adaptation strategies, synthesizing common stress proteins (cold shock proteins, chaperone proteins, and antifreeze proteins) and producing cryoprotection substances, including exopolysaccharides (EPS), all of which may enable their optimal growth at low temperatures (40, 60). However, cyanobacteria isolated from both the Arctic and Antarctica are psychrotolerant rather than psychrophilic, with growth optima at temperatures that are well above those of the ambient environment (51). Consistent with these observations, in situ measurements of Arctic ice shelf mats have shown that photosynthesis increases with increasing temperatures up to the limit tested (20°C, well above the maximum ambient water temperature of 1.7°C), while bacterial production showed no such trend, with rates at 2.6°C that were as high as or higher than those at warmer temperatures (29). These differences led us to hypothesize that mat communities dominated by different major phyla could have differences in their potential responses to stress at a genetic level. We investigated this hypothesis by way of metagenomic analyses of ice shelf mats from both the Arctic and Antarctica. In addition, we tested the notion that consortia occupying similar extreme habitats, but on opposite sides of the planet, were genetically similar in terms of potential responses to environmental stress, irrespective of geographic origin.
MATERIALS AND METHODS
Study site and sample collection.
Sampling was undertaken from 12 to 14 July 2007 on the Ward Hunt Ice Shelf (WHI) and the Markham Ice Shelf (MIS), located along the northern coastline of Ellesmere Island in the Canadian High Arctic (29, 30), and on 8 March 2008 on the McMurdo Ice Shelf (MCM), located in the Ross Sea sector of Antarctica (Table 1). Mats sampled from the respective regions were visually representative of the mats in their area. The Arctic samples were collected from shallow (<25-cm-deep) unnamed ponds on the ice shelves. The mats were 1-cm-thick, loosely cohesive aggregates that were olive green in color with a thin (≤100-μm), more cohesive orange layer at the surface, as reported earlier (29). The Arctic mats were collected from three 10- to 20-cm-deep meltwater ponds on each ice shelf and combined to produce one composite sample for MIS and another for WHI. The samples were placed directly into sterile 50-ml Falcon tubes and stored in the dark at 0 to 4°C for a day before being transported to the field laboratory, where they were frozen at −20°C until further processing. The Antarctic sample (MCM) was collected from Fresh Pond, a 1-m-deep, perennial meltwater pond on the McMurdo Ice Shelf, which was above freezing in summer and had a higher conductivity and pH than the Arctic ponds (Table 1). The Antarctic mats were cohesive, 3- to 4-mm-thick biofilms that were green-gray at the surface and gray at depth (54). Several samples of the MCM mat from Fresh Pond were combined in a single tube and stored in the dark at 0°C for 3 days before being transferred to −20°C. Physical characteristics of the source ponds, pH, conductivity, and temperature (Table 1), were determined at each site during the maximum growth period in the summer using a portable pH/Con 10 Series instrument (Oakton Instruments, Vernon Hills, IL). Chlorophyll a (Chl a) concentrations were determined by high-performance liquid chromatography (HPLC) (methods as reported by Hawes et al. [13] and Jungblut et al. [18]).
Table 1.
Environmental and metagenomic comparisons for the three sampling sitesa
| Environmental and mat data | Value for indicated sampling site |
||
|---|---|---|---|
| MIS | WHI | MCMb | |
| Environmental data | |||
| Latitude | 83°02′N | 83°05′N | 78°01′S |
| Longitude | 71°31′W | 74°26′W | 165°33′E |
| Temp (°C) | 1.8 (0.9) | 0.9 (0.6) | 7.4 |
| pH | 6.5 (0.3) | 6.5 (0.5) | 9.6 |
| Conductivity (μS cm−1) | 637 (144) | 385 (345) | 1568 |
| Mat Chl a (μg cm−2) | 9 | 40 | 24 |
| Classes found in mat (% of total sequences) | |||
| Alphaproteobacteria | 26 | 20 | 9 |
| Betaproteobacteria | 17 | 20 | 25 |
| Other Proteobacteria | 12 | 11 | 9 |
| Cyanobacteria | 17 | 25 | 38 |
| Planctomycetes | 2.5 | 2 | 4 |
| Actinobacteria | 10.5 | 10 | 3 |
| Archaea | 0.28 | 0.26 | 0.25 |
| Eukaryota | 0.56 | 0.55 | 0.67 |
| Virus | 0.02 | 0.01 | 0.02 |
For MIS and WHI, the environmental data are the means (standard deviations [SDs]) of three meltwater ponds from which the mat samples were pooled for metagenomics analysis. The percentages are the percentages of significant matches to taxonomic groups for all assigned genes. The comparisons among the metagenomes from the Markham Ice Shelf (MIS), Ward Hunt Ice Shelf (WHI), and McMurdo Ice Shelf (MCM) used BLASTX against the SEED protein-coding gene database (E value of ≤10−5; alignment length of ≥50 bp; percentage of identity of >65%).
Data are for the closest date of sampling of Fresh Pond (24 Jan 2008) (I. Hawes, personal communication).
DNA extraction and sequencing.
The DNA extraction and sequencing protocols were as described by Varin et al. (53). Briefly, mat samples were freeze-dried to avoid interference from exopolymeric substances during subsequent steps and extracted in XS buffer (52). DNA was purified with four phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma-Aldrich) wash steps, precipitated in isopropanol (Sigma-Aldrich) with 1/10 volume of ammonium acetate (4 mM; Sigma-Aldrich), and rinsed with 70% ethanol. RNA was removed from the extracts by addition of 2 μl of RNase A (10 mg ml−1; Roche Lifesciences). DNA was then washed with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated, and resuspended in 1× Tris-EDTA (TE) buffer. For each site in the Arctic (MIS and WHI) and Antarctic (MCM), ca. 5 μg of total DNA was used for each pyrosequencing run (26) using a 454 Sequencing System (Roche 454 Life Sciences) at the McGill University and Genome Québec Innovation Centre (Montreal, Quebec, Canada).
Bioinformatics and statistical analyses.
The 454 replicate filter proposed by Gomez-Alvarez et al. (9) (http://microbiomes.msu.edu/replicates/) was used to screen for potential artificial pyrosequencing replicates. Short or low-quality sequences with ambiguous bases (multiple internal N′s) were not included in our analysis. All metagenomic sequences were compared to protein-coding gene databases using the MetaGenome Rapid Annotation with Subsystem Technology (MG-RAST) server, version 2.0 (http://metagenomics.nmpdr.org) (28, 37). MG-RAST used Basic Local Alignment Search Tool X (BLASTX) (2) algorithms for comparisons with protein-coding gene databases. Taxonomic analyses in MG-RAST consisted of comparing our metagenomic sequences with those in the SEED protein-coding gene database (http://www.theseed.org/wiki/index.php/Home_of_the_SEED), where we have deposited the three metagenomes (MIS, WHI, MCM). Only matches of >50 nucleotides and >65% similarity to a taxonomic group or a subsystem (subsets of sequences showing similarities to each major metabolic process) and with an E value of ≤10−5 were included. The best match for each sequence was automatically selected and classified as “known” if its match against the relevant database was significant, or the sequence was classified as “unknown” if no significant match was found in the database. We used the metagenomic SEED viewer of the MG-RAST server to identify sequences matching functions of interest. The metabolic comparisons performed within MG-RAST were conducted on the SEED subsystems. The tabular view filter option was used to narrow searches within subsystems and retrieve the number of matches for specific genes. Percentages of matches for a given gene were calculated according to the total number of significant sequences found with MG-RAST for each metagenome. BLAST output files were parsed by our custom scripts written in Ruby (www.ruby-lang.org/) as needed. We employed the Statistical Analysis of Metagenomic Profiles (STAMP) (version 1.08; Faculty of Computer Science, Dalhousie University) statistical probability model to identify biologically relevant differences between metagenomic communities (http://kiwi.cs.dal.ca/Software/STAMP) (38). This model takes into account the sampling effort, defined here by the total number of reads per metagenome, to evaluate differences in the proportions of gene groups (subsystems) found with MG-RAST. In order to determine biologically significant differences between the Arctic and Antarctic ice shelves using STAMP, which is valid only for two-way comparisons, we initially compared the two Arctic metagenomes separately with the Antarctic metagenome. Since results were similar in the two-way comparisons, we then combined the two Arctic samples (MIS-WHI) and all subsequent analyses were Arctic and Antarctic comparisons for both taxonomic and functional distributions. Statistically significant differences between subsystems of metagenomes were identified by Fisher's exact test combined with the Newcombe-Wilson method for calculating confidence intervals (nominal coverage of 95%). As a multiple-hypothesis test correction, a false-discovery-rate (FDR) method was applied (either the Storey or Benjamini-Hochberg FDR approach) to indicate the percentages of false positives (reported by q values) that should be expected among all significant subsystems. A filter was applied to remove features with a q value of >0.05.
In order to identify the likely taxonomic source of cold and stress gene content in metagenomes from both poles, an additional statistical analysis of Arctic and Antarctic metabolic profiles was performed with STAMP that considered only sequences related to cold adaptation-specific genes and not all sequences of each metagenome (45, 50). Exceptionally for this analysis, all matches obtained using MG-RAST (BLASTX) with an E value of ≤10−5 were selected, regardless of alignment length and percentage of similarity thresholds.
Metagenome sequence accession.
The sequence data are available under “Public Metagenomes” at http://metagenomics.nmpdr.org/. The individual files are named mis_wdu for the Markham Ice Shelf, whi_wdu for the Ward Hunt Ice Shelf, and McMurdo for the McMurdo Ice Shelf sequences.
RESULTS
Mat metagenomes.
The average guanine-cytosine (GC) contents of the polar metagenomes were similar, with 55% for MIS, 53% for WHI, and 52% for MCM. The pyrosequencing yields were 256,849 sequences from MIS, 335,705 sequences from WHI, and 83,271 sequences from MCM. The average lengths were 208 bp, 184 bp, and 372 bp, respectively, giving an overall average of 255 bp and a total of 146,080,402 bp. The metagenomes represent a sampling of hundreds of thousands of bacteria, belonging to many genera. The low number of significant matches reflects the fact that these are short reads and most do not contain sufficient taxonomic information to be of use. Around 15% of the total retained sequences from Arctic data sets had matches against the SEED phylogenetic profile database (alignment length of >50 bp; percent identity of >65%; E value of ≤10−5; 37,521 matches for the MIS samples and 47,423 matches for the WHI samples), whereas about 25% of the total MCM sequences were significantly assigned, with 18,607 matches. There were 23,184 significant matches for MIS, 29,505 for WHI, and 11,753 for MCM against the SEED metabolic profile subsystems database (alignment length of >50 bp; percent identity of >65%; E value of ≤10−5).
Taxonomic and functional comparisons of polar microbial mats.
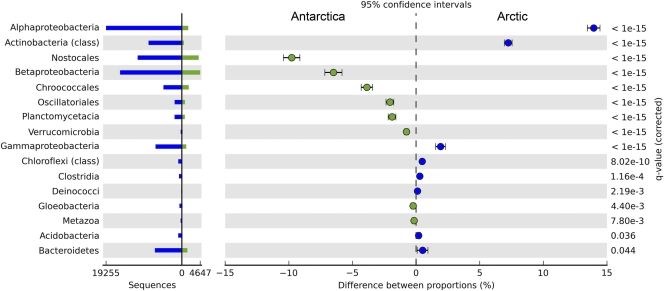
The protein-coding gene sequences indicated that there were diverse microbial taxa in all three polar mat communities, with similarities but also significant differences between the Arctic and Antarctic metagenomes (Fig. 1). In all three metagenomes, the most dominant sequences were Proteobacteria (Table 1). Cyanobacteria contributed the second-most dominant sequences but made a greater contribution to the Antarctic metagenome, with 38% of MCM sequences being from Cyanobacteria compared to 17% of MIS and 25% of WHI sequences. The Planctomycetes contributed a 2-fold-higher percentage of the total Antarctic sequences than of the Arctic sequences (Table 1). Conversely, Actinobacteria were over 3-fold more common in the Arctic, with ca. 10% of the total Arctic reads versus 3% of the total Antarctic reads. Within the Proteobacteria, there were also differences between the two polar regions. Betaproteobacteria contributed to a much greater percentage of Proteobacteria in the Antarctic mat (59%) than in the Arctic mats (31 to 39%), while Alphaproteobacteria were proportionately more important in the Arctic (40 to 48% of total Proteobacteria) than in Antarctica (20%). Archaea accounted for 0.61% (MIS), 0.58% (WHI), and 0.80% (MCM) of the sequences. Similarly, less than 1% of the total gene sequences were assigned as eukaryotes and viruses in the three samples. In this taxonomic analysis, 85% (219,328) of MIS, 86% (288,282) of WHI, and 78% (64,664) of MCM total sequences were not assigned to any taxon.
Fig 1.
Statistical analyses of taxonomic profiles for the Arctic (combined MIS and WHI samples) and Antarctic (MCM sample) metagenomes. Orders or classes overrepresented in the Antarctic have a negative difference between proportions (green dots); those overrepresented in the Arctic community have a positive-value difference between proportions (blue dots). Features (orders or classes) with a q value of >0.05 were considered significant.
Protein-coding sequences of polar bacterial isolates with available complete genomes had matches with the metagenomes (see Table S1 in the supplemental material). The match with the highest frequency in all metagenomes was to the marine Actinobacterium strain PHSC20C1, isolated from the surface waters of the western Antarctic Peninsula (32). Other high-frequency matches in all metagenomes were to Desulfotalea psychrophila LSv54 from cold marine Arctic sediments (43), Polaribacter irgensii from sea ice, and Flavobacterium psychrophilum from cold-water fish (7, 11). There were a small number of matches to protein-coding sequences from the psychrotolerant archaeal isolate Methanococcoides burtonii DSM 6242 from an Antarctic lake (8). Additionally, there were matches to Synechococcus strain WH5701 protein sequences associated with cold stress responses; although WH5701 is not a polar species, it falls in the same 16S rRNA gene clade as several isolates from Antarctic lakes (cluster 5.2 in reference 42).
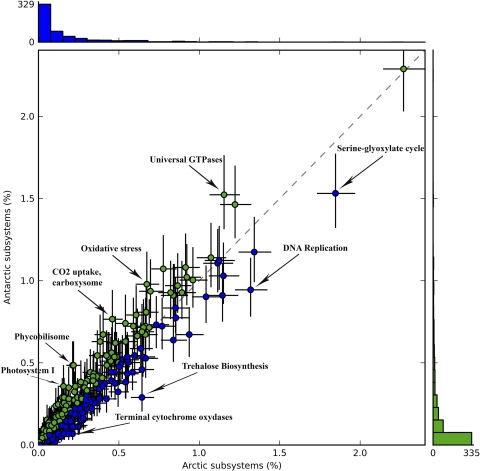
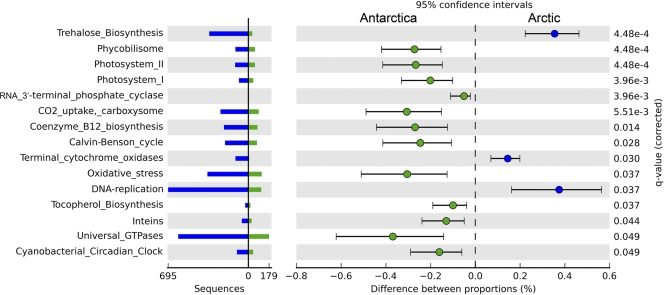
The functional analysis also revealed a high degree of similarity between the Arctic and Antarctic metagenomes (Fig. 2). However, again there were some differences, with significantly more reads in the Antarctic mat assigned to Photosystems I and II, phycobilisome proteins, and components of the cyanobacterial circadian clock (17). These results are consistent with the greater abundance of cyanobacterial genes in general in the Antarctic mat (Fig. 1 and 3). Other genes with significant overrepresentation in the Antarctic samples relative to Arctic samples included those for oxidative stress and universal GTPases (Fig. 3). In the Arctic mats, the trehalose biosynthesis coding genes were statistically more numerous than in the Antarctic, as were genes associated with DNA replication and terminal cytochrome oxidases (Fig. 3).
Fig 2.
Comparison of metabolic profiles for the Arctic (combined MIS-WHI samples) and Antarctic (MCM sample) metagenomes. Scatter plot of metabolic profiles of Arctic and Antarctic subsystems. Blue dots represent Arctic subsystems, and green dots represent Antarctic subsystems. Dots on either side of the dashed trend line are enriched in one of the two samples. Labeled dots indicate subsystems with the greatest distances from the dashed trend line; these subsystems had the greatest proportional differences (%) between Arctic and Antarctic metagenomes. Significant differences are shown in Fig. 3. The green bar graph (right) indicates the numbers of Antarctic subsystems as proportions of the total number of sequences in the Antarctic metagenome. The blue bar graph (top) indicates the numbers of Arctic subsystems as proportions of the Arctic metagenome, indicating that most subsystems represent a low proportion of the total number of sequences. The point in the upper right-hand corner is the tRNA aminoacylation subsystem, which accounted for a relatively high proportion of identified subsystems; however, since this was the same in both metagenomes, it was not labeled.
Fig 3.
Statistical analyses of metabolic profiles for the Arctic microbial mats (combined MIS-WHI samples) and the Antarctic metagenome (MCM sample). Total numbers of sequences in the different categories are shown in the left bar graph; the left side (blue) represents the Arctic mats, while the right side (green) represents the Antarctic mat. Subsystems in the Antarctic microbial mat community have negative differences between proportions (green dots). Subsystems overrepresented in the Arctic microbial mat samples have positive differences between proportions (blue dots). Features (orders or classes) with a q value of >0.05 were considered significant.
Taxonomy of genes involved in cold and other stresses.
Numbers presented in tables were normalized to the metagenome with the highest total number of sequences (WHI). The STAMP comparison adjusts for sampling coverage and hence reports only on the biological relevance of a feature between samples and, despite 5 times more Arctic sequences than Antarctic sequences, compensates for sampling artifacts. Genes implicated in adaptation to cold stress (45) were present in all three polar metagenomes, as shown in Table 2. These included the genes encoding DNA transcription and replication regulators (DnaA), recombination factor A (RecA), and topoisomerases (GyrA). RNA chaperones (Csp proteins) were rare and only sporadically recovered. Trehalose phosphate synthesis proteins (OtsA and OtsB) and fatty acid desaturases were present. Other sequences related to molecular chaperones identified as adaptations to psychrophilic lifestyles (6), such as DnaK, DnaJ, and peptidyl prolyl cis-trans isomerase, involved in protein folding, were more common. Translation initiation factor 1 (IF1) and ribosome-binding factor A (RbfA), both involved in protein biosynthesis, were present. Sequences associated with exopolysaccharide biosynthesis were also abundant in polar metagenomes (Table 2), as were fragments of genes coding for known cryo- and osmoprotectants, such as glutamate, glycine, betaine, and choline (50)(Table 2). Sequences matching other genes implicated in adaptation to cold stress (45), such as those encoding purine nucleoside phosphorylase (PNP), implicated in RNA degradation, and the AceE and AceF proteins (pyruvate dehydrogenases), were present in all three metagenomes. Sequences with nearest matches to several enzymes from cold-adapted Archaea were also recovered, including sequences coding for the archaeal tRNA modification process, such as the tRNA dihydrouridine synthase (Table 2).
Table 2.
Genes implicated in the adaptation to cold environments in the three metagenomesa
| Functional classification | Protein or subsystem name | No. of genes implicated in: |
Sigb | Phyla (no. of hits) found in indicated polar regionc |
||
|---|---|---|---|---|---|---|
| MIS-WHI | MCM | Arctic | Antarctic | |||
| DNA replication | GyrA (DNA gyrase A) | 97 | 90 | N | Alphaproteobacteria (28) | Betaproteobacteria (36) |
| Bacteroidetes (14) | Planctomycetes (18) | |||||
| Betaproteobacteria (9) | Undetermined phylum (14) | |||||
| Planctomycetes (9) | Alphaproteobacteria (9) | |||||
| Actinobacteria (8) | Bacteroidetes (5) | |||||
| RecA (recombination factor A) | 40 | 41 | N | Alphaproteobacteria (8) | Betaproteobacteria (23) | |
| Betaproteobacteria (4) | Undetermined phylum (9) | |||||
| Deltaproteobacteria (1) | ||||||
| Undetermined phylum (1) | ||||||
| DnaA (replication initiator protein) | 134 | 117 | N | Alphaproteobacteria (24) | Cyanobacteria (27) | |
| Cyanobacteria (19) | Gammaproteobacteria (14) | |||||
| Bacteroidetes (17) | Betaproteobacteria (14) | |||||
| Betaproteobacteria (14) | Bacteroidetes (9) | |||||
| Actinobacteria (11) | Verrucomicrobia (9) | |||||
| DNA metabolism | HU-β (DNA supercoiling) | 8 | 18 | — | ||
| Di- and oligosaccharides | OstA (trehalose phosphate synthase) | 84 | 50 | Arc | Alphaproteobacteria (17) | Deltaproteobacteria (14) |
| Actinobacteria (15) | Cyanobacteria (9) | |||||
| Betaproteobacteria (10) | Actinobacteria (5) | |||||
| Gammaproteobacteria (10) | Bacteroidetes (5) | |||||
| Deltaproteobacteria (3) | Planctomycetes (5) | |||||
| OstB (trehalose phosphatase) | 54 | 9 | Arc | Alphaproteobacteria (27) | Bacteroidetes (5) | |
| Actinobacteria (5) | ||||||
| Betaproteobacteria (4) | ||||||
| Deltaproteobacteria (2) | ||||||
| Bacteroidetes (2) | ||||||
| Unsaturated fatty acids | Fatty acid desaturases | 18 | 54 | Ant | Actinobacteria (7) | Cyanobacteria (41) |
| Cyanobacteria (4) | Undetermined phylum (5) | |||||
| Bacteroidetes (2) | Actinobacteria (5) | |||||
| Protein folding | Chaperone DnaK and DnaJ | 363 | 432 | N | Cyanobacteria (121) | Cyanobacteria (198) |
| Actinobacteria (29) | Betaproteobacteria (41) | |||||
| Alphaproteobacteria (29) | Alphaproteobacteria (23) | |||||
| Betaproteobacteria (26) | Actinobacteria (18) | |||||
| Bacteroidetes (25) | Planctomycetes (9) | |||||
| Peptidyl-prolyl cis-trans isomerase | 149 | 198 | N | Betaproteobacteria (32) | Betaproteobacteria (50) | |
| Bacteroidetes (13) | Gammaproteobacteria (18) | |||||
| Gammaproteobacteria (12) | Cyanobacteria (18) | |||||
| Alphaproteobacteria (12) | Verrucomicrobia (18) | |||||
| Cyanobacteria (10) | Bacteroidetes (9) | |||||
| Protein biosynthesis | Translation initiation factor 1 (IF1) | 33 | 27 | N | Bacteroidetes (7) | Betaproteobacteria (9) |
| Alphaproteobacteria (6) | Cyanobacteria (5) | |||||
| Actinobacteria (5) | Verrucomicrobia (5) | |||||
| Cyanobacteria (4) | Actinobacteria (5) | |||||
| Planctomycetes (2) | Bacteroidetes (5) | |||||
| Ribosome-binding factor A (RbfA) | 24 | 18 | — | |||
| Clustering-based subsystems | Exopolysaccharide biosynthesis | 57 | 113 | Ant | Cyanobacteria (10) | Cyanobacteria (27) |
| Betaproteobacteria (6) | Betaproteobacteria (23) | |||||
| Alphaproteobacteria (4) | Verrucomicrobia (14) | |||||
| Gammaproteobacteria (3) | Deltaproteobacteria (5) | |||||
| Verrucomicrobia (2) | Alphaproteobacteria (5) | |||||
| Capsular and extracellular polysaccharides | Glycine biosynthesis | 107 | 108 | N | Bacteroidetes (25) | Cyanobacteria (41) |
| Alphaproteobacteria (16) | Bacteroidetes (18) | |||||
| Gammaproteobacteria (11) | Gammaproteobacteria (9) | |||||
| Actinobacteria (10) | Alphaproteobacteria (9) | |||||
| Cyanobacteria (10) | Betaproteobacteria (5) | |||||
| Amino acids and derivatives | Glutamate biosynthesis | 20 | 5 | — | ||
| Choline and betaine uptake, betaine biosynthesis | 316 | 252 | N | Alphaproteobacteria (103) | Alphaproteobacteria (63) | |
| Actinobacteria (43) | Betaproteobacteria (45) | |||||
| Betaproteobacteria (37) | Cyanobacteria (45) | |||||
| Cyanobacteria (18) | Actinobacteria (14) | |||||
| Bacteroidetes (17) | Bacteroidetes (9) | |||||
| Nucleosides and nucleotides | Purine nucleoside phosphorylase (PNP) | 20 | 41 | — | ||
| Pyruvate metabolism II | AceE (pyruvate dehydrogenase E1 component) | 170 | 162 | N | Actinobacteria (41) | Alphaproteobacteria (41) |
| Alphaproteobacteria (27) | Cyanobacteria (32) | |||||
| Cyanobacteria (27) | Betaproteobacteria (27) | |||||
| Betaproteobacteria (24) | Verrucomicrobia (14) | |||||
| Gammaproteobacteria (14) | Actinobacteria (9) | |||||
| AceF (dihydrolipoamide acetyltransferase) | 65 | 81 | N | Actinobacteria (15) | Cyanobacteria (36) | |
| Cyanobacteria (11) | Planctomycetes (9) | |||||
| Alphaproteobacteria (9) | Betaproteobacteria (9) | |||||
| Betaproteobacteria (8) | Gammaproteobacteria (9) | |||||
| Bacteroidetes (4) | Bacteroidetes (5) | |||||
| tRNA modification Archaea | tRNA dihydrouridine synthase | 78 | 81 | N | Cyanobacteria (14) | Cyanobacteria (23) |
| Bacteroidetes (14) | Verrucomicrobia (14) | |||||
| Actinobacteria (13) | Bacteroidetes (14) | |||||
| Alphaproteobacteria (10) | Undetermined phylum (9) | |||||
| Undetermined phylum (8) | Alphaproteobacteria (5) | |||||
Number of genes in the mat metagenomes implicated in adaptation to cold environments (40, 45), number of other genes assumed to be adaptive to polar conditions, and number of genes in exopolysaccharide pathways. Using BLASTX with an E value of ≤10−5, metagenomic sequences were compared to those of genes present in the SEED database. Numbers were normalized to the combined Arctic metagenome with the highest number of BLASTX hits.
The biological significance (Sig) of differences between proportions using the STAMP protocols is indicated as nonsignificant (N) or significant, with Arc indicating overrepresentation in the Arctic and Ant indicating overrepresentation in the Antarctic. —, too few data to test.
Phyla or subphyla related to the gene representatives are indicated for the Arctic and Antarctica, and the number of hits belonging to those taxa, normalized to the Arctic metagenomes, are given in parentheses. The cold shock proteins CspA, CspE, CspG, and CspI had <20 matches per metagenome and are not listed in the table; CspB was not detected. Only the top five phyla, ranked by numbers of hits, are given; when fewer than five were present, all are listed.
Among non-cold-related stress genes, copper homeostasis and the sigma B stress response subsystems differed significantly. The Arctic metagenome was overrepresented in genes belonging to the copper homeostasis group (Table 3). Other non-cold-related stress genes were recovered from the metagenomes, most notably in subsystems from the heat shock dnaK gene cluster, flavohemoglobin, periplasmic stress, and acid resistance mechanisms, none of which were significantly more abundant in the metagenomes from either pole (Table 3).
Table 3.
Comparison of the numbers of genes implicated in non-cold stress responses in polar metagenomesa
| Subsystem name | No. of genes implicated in: |
Sigb | Phyla (no. of hits) found in indicated polar regionc |
||
|---|---|---|---|---|---|
| MIS-WHI | MCM | Arctic | Antarctic | ||
| Acid resistance mechanism | 196 | 108 | N | Cyanobacteria (59) | Cyanobacteria (54) |
| Alphaproteobacteria (28) | Verrucomicrobia (14) | ||||
| Bacteroidetes (25) | Undetermined phylum (14) | ||||
| Gammaproteobacteria (14) | Alphaproteobacteria (9) | ||||
| Verrucomicrobia (10) | Bacteroidetes (5) | ||||
| Detoxification | 19 | 14 | — | ||
| Periplasmic stress | 139 | 162 | N | Cyanobacteria (23) | Cyanobacteria (36) |
| Alphaproteobacteria (20) | Alphaproteobacteria (14) | ||||
| Betaproteobacteria (16) | Betaproteobacteria (9) | ||||
| Bacteroidetes (15) | Gammaproteobacteria (9) | ||||
| Planctomycetes (6) | Planctomycetes (5) | ||||
| Bacitracin stress response | 20 | 27 | — | ||
| Copper homeostasis | 1422 | 941 | Arc | Betaproteobacteria (235) | Cyanobacteria (180) |
| Alphaproteobacteria (202) | Betaproteobacteria (153) | ||||
| Bacteroidetes (201) | Bacteroidetes (104) | ||||
| Cyanobacteria (147) | Alphaproteobacteria (45) | ||||
| Actinobacteria (84) | Planctomycetes (36) | ||||
| Sigma B stress response regulation | 145 | 279 | Ant | Cyanobacteria (63) | Cyanobacteria (108) |
| Alphaproteobacteria (8) | Bacteroidetes (23) | ||||
| Bacteroidetes (7) | Verrucomicrobia (9) | ||||
| Actinobacteria (6) | Alphaproteobacteria (9) | ||||
| Gammaproteobacteria (5) | Planctomycetes (5) | ||||
| Universal stress protein family | 28 | 41 | — | ||
| Phage shock protein (psp) operon | 33 | 14 | — | ||
| Flavohemoglobin | 114 | 122 | N | Alphaproteobacteria (27) | Betaproteobacteria (27) |
| Betaproteobacteria (21) | Cyanobacteria (18) | ||||
| Actinobacteria (10) | Actinobacteria (14) | ||||
| Gammaproteobacteria (8) | Planctomycetes (9) | ||||
| Bacteroidetes (8) | Verrucomicrobia (9) | ||||
| Bacterial hemoglobin | 25 | 27 | — | ||
| Heat shock dnaK gene cluster extended | 367 | 342 | N | Alphaproteobacteria (63) | Bacteroidetes (59) |
| Bacteroidetes (57) | Betaproteobacteria (45) | ||||
| Betaproteobacteria (48) | Cyanobacteria (45) | ||||
| Cyanobacteria (41) | Deltaproteobacteria (23) | ||||
| Actinobacteria (22) | Alphaproteobacteria (23) | ||||
| Hfl operon | 83 | 63 | N | Alphaproteobacteria (33) | Betaproteobacteria (45) |
| Betaproteobacteria (21) | Deltaproteobacteria (5) | ||||
| Gammaproteobacteria (7) | |||||
| Deltaproteobacteria (4) | |||||
| Fusobacteria (1) | |||||
| Carbon starvation | 41 | 54 | N | Betaproteobacteria (14) | Planctomycetes (18) |
| Verrucomicrobia (4) | Betaproteobacteria (14) | ||||
| Gammaproteobacteria (4) | Gammaproteobacteria (9) | ||||
| Actinobacteria (4) | Undetermined phylum (9) | ||||
| Deltaproteobacteria (1) | |||||
Using BLASTX with an E value of ≤10−5, metagenomic sequences were compared to those of genes present in the SEED database. Numbers were normalized to the combined Arctic metagenome with the highest number of BLASTX hits.
The biological significance (Sig) of differences between proportions using the STAMP protocols is indicated as nonsignificant (N) or significant, with Arc indicating overrepresentation in the Arctic and Ant indicating overrepresentation in the Antarctic. —, too few data to test.
Phyla or subphyla related to the gene representatives are indicated for the Arctic and Antarctic, and the number of hits belonging to those taxa, normalized to the Arctic metagenomes, are given in parentheses. Only the top five phyla, ranked by numbers of hits, are given; when fewer than five were present, all are listed.
Taxonomy of functional differences.
Among the assumed cold-adaptive genes, there were several biologically significant differences between mats from the two polar regions. The di- and oligosaccharide genes involved in trehalose biosynthesis, those encoding OstA and OstB, were overrepresented in the Arctic and contributed by different bacterial groups than in the Antarctic (Table 2), with the majority of the Arctic sequences from Alphaproteobacteria, whereas in the Antarctic, ostA was most often found within the Deltaproteobacteria and ostB was most often found in the Bacteroidetes. Genes assigned as those encoding fatty acid desaturases were statistically overrepresented in Antarctica, and both Cyanobacteria and Actinobacteria were identified as important sources of the genes (Table 2). Similarly, Cyanobacteria contributed to the EPS biosynthesis gene pool in both poles. Betaproteobacteria represented the second-most abundant source of EPS genes (Table 2).
Among non-cold-related stress genes, the Arctic metagenome was overrepresented in genes belonging to the copper homeostasis group. Cyanobacteria, Alphaproteobacteria, Betaproteobacteria, and Bacteroidetes contributed to the copper homeostasis pool at both poles, but Actinobacteria were also an important source in the Arctic. The alternative sigma factor (sigma B) stress response regulation genes were overrepresented in the Antarctic, and Cyanobacteria accounted for most of these genes in both the Arctic and Antarctic metagenomes (Table 3).
DISCUSSION
Ice shelves provide similar habitats for microbial colonization and growth in the North and South polar regions, with liquid water conditions that persist for only a few weeks to months each year. Aqueous temperatures may occasionally rise above 5°C but are more typically around 0°C. During freeze-up, salts are excluded from the ice and the resultant brines may have temperatures that fall well below zero (13, 14, 29, 48). Calculations based on production-to-biomass ratios have shown that the ice shelf mats are perennial, with the standing stock representing many years of microbial biomass accumulation (29). The whole-mat metagenomes are therefore likely to reflect genetic responses to the ensemble of environmental conditions, including persistent cold, freeze-up, and variable salinities.
Consistent with the similar extreme conditions imposed by the ice shelf environments, the protein-coding genes indicated largely similar taxonomic compositions in the Arctic and Antarctica. Most of the matched sequences could be attributed to Proteobacteria, which likely profit from the organic carbon-rich environment within the mats, and Cyanobacteria, which provide the phototrophic energy source and structural biomass of the mats (55) while likely benefiting from the decomposition and nutrient recycling activities of the Proteobacteria (53) (Table 1). Archaea were a minor but detectable component of all three metagenomes, as were eukaryotes, including metazoans.
Cyanobacterial mats are generally found anywhere that larger metazoan grazers are absent or marginalized because of extremes in temperature, salinity, and UV (10, 16, 55). The vast range of temperatures where cyanobacterial mats are found, from continental polar regions to geothermal hot springs, suggests a diversity of strategies for coping with extremes in temperature (1, 59). Previous work has shown a relative absence in the High Arctic of cyanobacterial ribotypes from warmer latitudes and that many High Arctic 16S rRNA gene cyanobacterial sequences are >99% similar to sequences from Antarctica, including taxa previously assumed to be endemic to Antarctica (18). This apparent bipolar distribution might also imply similarities in the mechanisms of stress tolerance throughout the cold biosphere.
Despite the similarities, there were some conspicuous differences between the polar regions, both in the relative abundances of bacteria and in the proportions of the major classes of bacteria within the mats. The Antarctic mats had a higher representation of Cyanobacteria, and this was also reflected in the functional analysis, with a higher percentage of genes coding for photosynthetic functions found in MCM samples than in MIS-WHI samples. The greater proteobacterial and actinobacterial representation in the Arctic may reflect increased inputs of bacteria and terrigenous materials from the adjacent ice-free land. In addition to terrestrial inocula, the Arctic ice shelves are also exposed to diasporas from marine sources (12). This was reflected in the higher representation of bacteria, such as marine Alphaproteobacteria, in the Arctic mats than in the Antarctic. In addition, the shallower, more ephemeral ponds of the Arctic may be more prone to invasions by new groups (49) than the deeper, longer-persisting ponds in Antarctica.
The microbial mats showed broadly similar functional gene repertoires for acclimation to environmental stress. Among the detected genes were sequences coding for EPS production, cold shock proteins, and membrane modification. There were, however, some differences between the mats. The Arctic mats had higher representation of copper homeostasis genes, possibly indicating their greater exposure to long-range pollutants, including metals (for examples, see reference 31). Conversely, the Antarctic mat had a greater representation of the alternative sigma factor (sigma B) gene, which appears to be a general stress regulon that induces more than 100 genes in response to a great variety of stresses, including heat, acid, salt, and starvation (58). The greater frequency of this gene in Antarctica might reflect greater osmotic stresses at freeze-up in the more saline waters of the Antarctic ponds, which are more persistent and subject to salt accumulation over time.
Many studies have shown the important role of EPS in buffering and cryoprotection for diverse microorganisms against ice crystal damage and high salinity (25, 34, 35). Cyanobacteria produce large amounts of EPS (24, 36) and were the primary source of EPS genes in the mats from both Antarctica and the Arctic. EPS allow bacterial aggregate formation, which in turn provides opportunities for close biogeochemical interactions (34). In the cryophilic gammaproteobacterium Psychromonas ingrahamii, production of EPS may sequester water from the ambient saltwater, lowering the freezing point (44). Junge et al. (19) demonstrated a significant correlation between concentrations of local bacteria and EPS in Arctic winter sea ice. In harsh environments, such as the polar regions, it is likely that EPS contributes to the physical stability of microbial communities (40). Sulfate-reducing bacteria in the Deltaproteobacteria may also produce large amounts of EPS (5, 57). Nichols et al. (34) showed that Proteobacteria (especially Gammaproteobacteria) and Bacteroidetes, which were common phyla in all three polar metagenomes, are able to synthesize EPS in response to low temperatures, implying that this is a cold-adaptation process.
In all three polar samples, genes coding for xylose, mannose, rhamnose, and fucose synthesis were among the most abundant monosaccharide synthesis genes. These sugars are typically found in bacterial EPS (21), but the exact monosaccharide composition of EPS varies largely among bacterial strains (35). EPS produced by marine bacteria generally contains 20 to 50% of the total polysaccharide as uronic acid (22). Sequences assigned to uronic acid synthesis were rare in all three microbial mat samples. This is consistent with the presence of taxa, such as Pseudoalteromonas and Flavobacterium, that are known to produce EPS rich in neutral sugars (especially mannose and fucose) but with little uronic acid (35).
All of the polar metagenomes contained genes encoding cold shock proteins, which are a common feature of prokaryotes growing in low temperatures (47). RNA chaperones (cold shock proteins CspA, CspB, CspE, CspI, and CspG) are essential for proper protein folding, especially at low temperatures, guiding nascent polypeptides into functional three-dimensional configurations (44). All three polar metagenomes contained cold shock protein sequences. Matches assigned to the genes of more-constitutive proteins associated with cold adaptation, such as DNA transcription regulators (DnaA), recombination factors (RecA), topoisomerases (GyrA), trehalose synthesis proteins (OstA and OstB), and chaperones DnaK and DnaJ, were all numerous in the three polar microbial communities. These genes are known to be induced in bacteria upon exposure to cold temperatures (20, 45, 46, 61), and DnaA and GyrA are involved in the maintenance of functional DNA topology at cold temperatures (45).
It has long been known that exposure of microorganisms to lower temperatures results in substantial alteration of their membrane compositions, with changes in the ratio of saturated to unsaturated fatty acids (for examples, see reference 27). Saturation of the membrane fatty acids decreases at low temperatures in the psychrophilic gammaproteobacterium Psychrobacter arcticus (41), a widespread cold-adapted species that can survive for long periods under harsh conditions, including deep permafrost (3, 41). Gammaproteobacteria sequences with close similarity to those of P. arcticus were identified in the MIS, WHI, and MCM metagenomes. In cold environments, maintenance of cell membrane integrity requires an increased proportion of unsaturated and branched fatty acids (15, 23, 33). In cyanobacteria, this membrane composition adjustment occurred via desaturases (39), and the genes coding for these enzymes were prevalent in the three polar mat metagenomes. In P. arcticus, the effects of low temperatures on enzyme activity are compensated for by structural modifications that increase the flexibility of at least 50% of its proteome, thereby reducing energetic requirements (3).
In summary, this metagenomic analysis of polar microbial mat consortia has revealed the presence of many cold stress genes that to date have mostly been known only from laboratory studies on isolated microorganisms. Consistent with our hypothesis, the analyses showed diverse mechanisms of potential responses to cold and other stresses, and this reflects the taxonomic diversity within the mats. In both polar regions, Proteobacteria and Cyanobacteria dominated the sequences, including the cold stress genes. However, there were distinct differences in terms of taxonomy and preferred biological functions between the Antarctic and Arctic mats. For example, the greater representation of Cyanobacteria in MCM was reflected by a significantly higher percentage of genes coding for photosynthetic functions. Factors such as habitat stability and the connectivity to marine and terrestrial sources of microbiota may account for the differences between the Arctic and Antarctic ice shelf mats noted here; however, additional data from a broader range of sites and habitats are required to evaluate whether these reflect fundamental, consistent differences between the two poles of the cold biosphere.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ken Dewar for expert guidance in the pyrosequencing and members of the Lovejoy and Vincent laboratory and field teams for scientific and technical assistance. We are grateful to Ian Hawes and Brian Sorrell (NIWA, New Zealand) for sampling on the McMurdo Ice Shelf. We also thank three anonymous reviewers for their time and constructive comments on an earlier version of the manuscript. Polar Shelf Canada provided logistical support.
Funding was from the Natural Sciences and Engineering Research Council of Canada (NSERC), with additional support from Québec Océan, the International Polar Year program Microbiological and Ecological Responses to Global Environmental Change in the Polar Regions, the Networks of Centres of Excellence program ArcticNet, Genome Canada, and Genome Québec. We thank the staff of Quttinirpaaq National Park. J.C. and W.F.V. acknowledge support from the Canada Research Chair program, and T.V. was supported by a doctoral fellowship from the Canadian Institute of Health Research.
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allewalt JP, Bateson MM, Revsbech NP, Slack K, Ward DM. 2006. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the Octopus Spring microbial mat community of Yellowstone National Park. Appl. Environ. Microbiol. 72:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Ayala-del-Rio HL, et al. 2010. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl. Environ. Microbiol. 76:2304–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bottos EM, Vincent WF, Greer CW, Whyte LG. 2008. Prokaryotic diversity of arctic ice shelf microbial mats. Environ. Microbiol. 10:950–966 [DOI] [PubMed] [Google Scholar]
- 5. Braissant O, et al. 2007. Exopolymeric substances of sulfate-reducing bacteria: interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology 5:401–411 [Google Scholar]
- 6. Cavicchioli R. 2006. Cold-adapted Archaea. Nat. Rev. Microbiol. 4:331–343 [DOI] [PubMed] [Google Scholar]
- 7. Crump EM, Perry MB, Clouthier SC, Kay WW. 2001. Antigenic characterization of the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 67:750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franzmann PD, Springer N, Ludwig W, Conway de Macario E, Rohde M. 1992. A methanogenic archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst. Appl. Microbiol. 4:573–581 [Google Scholar]
- 9. Gomez-Alvarez V, Teal TK, Schmidt TM. 2009. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3:1314–1317 [DOI] [PubMed] [Google Scholar]
- 10. Gorbushina AA. 2007. Life on the rocks. Environ. Microbiol. 9:1613–1631 [DOI] [PubMed] [Google Scholar]
- 11. Gosink JJ, Woese CR, Staley JT. 1998. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus ’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Bacteriol. 48:223–235 [DOI] [PubMed] [Google Scholar]
- 12. Harding T, Jungblut AD, Lovejoy C, Vincent WF. 2011. Microbes in High Arctic snow and implications for the cold biosphere. Appl. Environ. Microbiol. 77:3234–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hawes I, Safi K, Sorrell B, Webster-Brown J, Arscott D. 2011. Summer-winter transitions in Antarctic ponds. I. The physical environment. Antarct. Sci. 23:235–242 [Google Scholar]
- 14. Hawes I, Safi K, Webster-Brown J, Sorrell B, Arscott D. 2011. Summer-winter transitions in Antarctic ponds. II. Biological responses. Antarct. Sci. 23:243–254 [Google Scholar]
- 15. Hazel J, Williams EE. 1990. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 29:167–227 [DOI] [PubMed] [Google Scholar]
- 16. Hoffman L. 1999. Marine cyanobacteria in tropical regions: diversity and ecology. Eur. J. Phycol. 34:371–379 [Google Scholar]
- 17. Ito H, et al. 2009. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A. 106:14168–14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jungblut AD, Lovejoy C, Vincent WF. 2010. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 4:191–202 [DOI] [PubMed] [Google Scholar]
- 19. Junge K, Imhoff F, Staley T, Deming JW. 2002. Phylogenetic diversity of numerically important Arctic sea-ice bacteria cultured at subzero temperature. Microb. Ecol. 43:315–328 [DOI] [PubMed] [Google Scholar]
- 20. Kandror O, DeLeon A, Goldberg AL. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U. S. A. 99:9727–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenne L, Lindberg B. 1983. Bacterial polysaccharides, p 287–363 In Aspinall GO. (ed), The polysaccharides. Academic Press, New York, NY [Google Scholar]
- 22. Kennedy AF, Sutherland IW. 1987. Analysis of bacterial exopolysaccharides. Biotechnol. Appl. Biochem. 9:12–19 [PubMed] [Google Scholar]
- 23. Klein W, Weber MHW, Marahiel MA. 1999. Cold shock response of Bacillus subtilis: isoleucine-dependent switch in the fatty acid branching pattern for membrane adaptation to low temperatures. J. Bacteriol. 181:5341–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klock J-H, Wieland A, Seifert R, Michaelis W. 2007. Extracellular polymeric substances (EPS) from cyanobacterial mats: characterisation and isolation method optimisation. Mar. Biol. 152:1077–1085 [Google Scholar]
- 25. Krembs C, Eicken H, Junge K, Deming JW. 2002. High concentrations of exopolymeric substances in Arctic winter sea ice: implications for the polar ocean carbon cycle and cryoprotection of diatoms. Deep Sea Res. Part I Oceanogr. Res. Pap. 49:2163–2181 [Google Scholar]
- 26. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maslova IP, Mouradyan EA, Lapina SS, Klyachko-Gurvich GL, Los DA. 2004. Lipid fatty acid composition and thermophilicity of Cyanobacteria. Russ. J. Plant Physiol. 51:353–360 [Google Scholar]
- 28. Meyer F, et al. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mueller DR, Vincent WF, Bonilla S, Laurion I. 2005. Extremotrophs, extremophiles and broadband pigmentation strategies in a High Arctic ice shelf ecosystem. FEMS Microbiol. Ecol. 53:73–87 [DOI] [PubMed] [Google Scholar]
- 30. Mueller DR, Vincent WF, Jeffries MO. 2006. Environmental gradients, fragmented habitats, and microbiota of a northern ice shelf cryoecosystem, Ellesmere Island, Canada. Arct. Antarct. Alp. Res. 38:593–607 [Google Scholar]
- 31. Muir DCG, et al. 2009. Spatial trends and historical deposition of mercury in eastern and northern Canada inferred from lake sediment cores. Environ. Sci. Technol. 43:4802–4809 [DOI] [PubMed] [Google Scholar]
- 32. Murray AE, Grzymski JJ. 2007. Diversity and genomics of Antarctic marine micro-organisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mykytczuk NCS, Trevors JT, Twine SM, Ferroni GD, Leduc LG. 2010. Membrane fluidity and fatty acid comparisons in psychrotrophic and mesophilic strains of Acidithiobacillus ferrooxidans under cold growth temperatures. Arch. Microbiol. 192:1005–1018 [DOI] [PubMed] [Google Scholar]
- 34. Nichols CM, Bowman JP, Guezennec J. 2005. Effects of incubation temperature on growth and production of exopolysaccharides by an Antarctic sea ice bacterium grown in batch culture. Appl. Environ. Microbiol. 71:3519–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nichols CM, et al. 2005. Chemical characterization of exopolysaccharides from Antarctic marine bacteria. Microb. Ecol. 49:578–589 [DOI] [PubMed] [Google Scholar]
- 36. Nicolaus B, Kambourova M, Oner ET. 2010. Exopolysaccharides from extremophiles: from fundamentals to biotechnology. Environ. Technol. 31:1145–1158 [DOI] [PubMed] [Google Scholar]
- 37. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parks D, Beiko R. 2010. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26:715–721 [DOI] [PubMed] [Google Scholar]
- 39. Phadtare S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125–136 [PubMed] [Google Scholar]
- 40. Poli A, Anzelmo G, Nicolaus B. 2010. Bacterial exopolysaccharides from extreme marine habitats: production, characterization and biological activities. Mar. Drugs 8:1779–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ponder M. 2005. Ph.D. thesis Characterization of physiological and transcriptome changes in the ancient Siberian permafrost bacterium Psychrobacter arcticum 273-4 with low temperature and increased osmotica. Michigan State University, East Lansing, MI [Google Scholar]
- 42. Powell LM, Bowman JP, Franzmann PD, Burton HR. 2005. Ecology of a novel Synechococcus clade occurring in dense populations in saline Antarctic lakes. Mar. Ecol. Prog. Ser. 291:65–80 [Google Scholar]
- 43. Rabus R, et al. 2004. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6:887–902 [DOI] [PubMed] [Google Scholar]
- 44. Riley M, et al. 2008. Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics 9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodrigues DF, Tiedje JM. 2008. Coping with our cold planet. Appl. Environ. Microbiol. 74:1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosen R, Ron EZ. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 21:244–265 [DOI] [PubMed] [Google Scholar]
- 47. Scherer S, Neuhaus K. 2006. Life at low temperatures, p 210–262 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes, 3rd ed Springer, New York, NY [Google Scholar]
- 48. Schmidt S, Moskall W, de Mora SJ, Howard-Williams C, Vincent WF. 1991. Limnological properties of Antarctic ponds during winter freezing. Antarct. Sci. 3:379–388 [Google Scholar]
- 49. Schneider DW, Frost TM. 1996. Habitat duration and community structure in temporary ponds. J. North Am. Benthol. Soc. 15:64–86 [Google Scholar]
- 50. Simon C, Wiezer A, Strittmatter AW, Daniel R. 2009. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 75:7519–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang EPY, Tremblay R, Vincent WF. 1997. Cyanobacterial dominance of polar freshwater ecosystems: are high-latitude mat-formers adapted to low temperatures? J. Phycol. 33:171–181 [Google Scholar]
- 52. Tillett D, Neilan BA. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251–258 [Google Scholar]
- 53. Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeil J. 2010. Metagenomic profiling of Arctic microbial mat communities as nutrient scavenging and recycling systems. Limnol. Oceanogr. 55:1901–1911 [Google Scholar]
- 54. Vincent WF, Castenholz RW, Downes MT, Howard-Williams C. 1993. Antarctic cyanobacteria: light, nutrients, and photosynthesis in the microbial mat environment. J. Phycol. 29:745–755 [Google Scholar]
- 55. Vincent WF. 2000. Cyanobacterial dominance in the polar regions, p 321–340 In Whitton BA, Potts M. (ed), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 56. Vincent WF. 2007. Cold tolerance in cyanobacteria and life in the cryosphere, p 287–301 In Seckbach J. (ed), Algae and cyanobacteria in extreme environments. Springer, Heidelberg, Germany [Google Scholar]
- 57. Visscher PT, Reid RP, Bebout BM. 2000. Microscale observations of sulfate reduction: correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology 28:919–922 [Google Scholar]
- 58. Volker U, Maul B, Hecker M. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ward DM, et al. 2006. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yergeau E, Hogues H, Whyte LG, Greer CW. 2010. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 4:1206–1214 [DOI] [PubMed] [Google Scholar]
- 61. Zhao D, Chen X, He H, Shi M, Zhang Y. 2007. Gene cloning and sequence analysis of the cold-adapted chaperones DnaK and DnaJ from deep-sea psychrotrophic bacterium Pseudoalteromonas sp. SM9913. Acta Oceanol. Sin. 26:91–100 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.