Abstract
Isolation, cultivation, and characterization of the intestinal microorganisms are important for understanding the comprehensive physiology of the human gastrointestinal (GI) tract microbiota. Here, we isolated two novel bacterial strains, YIT 12067T and YIT 12068, from the feces of healthy human adults. Phylogenetic analysis indicated that they belonged to the same species and were most closely related to Phascolarctobacterium faecium ACM 3679T, with 91.4% to 91.5% 16S rRNA gene sequence similarities, respectively. Substrate availability tests revealed that the isolates used only succinate; they did not ferment any other short-chain fatty acids or carbohydrates tested. When these strains were cocultured with the xylan-utilizing and succinate-producing bacterium Paraprevotella xylaniphila YIT 11841T, in medium supplemented with xylan but not succinate, their cell numbers became 2 to 3 orders of magnitude higher than those of the monoculture; succinate became undetectable, and propionate was formed. Database analysis revealed that over 200 uncultured bacterial clones from the feces of humans and other mammals showed high sequence identity (>98.7%) to YIT 12067T. Real-time PCR analysis also revealed that YIT 12067T-like bacteria were present in 21% of human fecal samples, at an average level of 3.34 × 108 cells/g feces. These results indicate that YIT 12067T-like bacteria are distributed broadly in the GI tract as subdominant members that may adapt to the intestinal environment by specializing to utilize the succinate generated by other bacterial species. The phylogenetic and physiological properties of YIT 12067T and YIT 12068 suggest that these strains represent a novel species, which we have designated Phascolarctobacterium succinatutens sp. nov.
INTRODUCTION
Advances in culture-independent molecular techniques have allowed a more complete and accurate assessment of the biodiversity of the ecosystem of the human gastrointestinal (GI) tract microbiota (10, 52) and have revealed that most of the phylotypes detected are uncultured (42). For a better understanding of the physiology of the human GI tract microbiota, it is important to isolate, cultivate, and characterize the intestinal microorganisms. Such undertakings provide information on the ecology and physiology of the GI tract microbiota that cannot be acquired from gene sequence information alone. For example, a study that used an isolated strain in the gnotobiotic mouse model revealed that one of the bacteria most frequently detected from human feces, Bacteroides thetaiotaomicron, modulates the expression of host genes involved in nutrient absorption, mucosal barrier fortification, xenobiotic metabolism, angiogenesis, and postnatal intestinal maturation (19). Therefore, cultivation of previously uncultured bacteria will also contribute to a more comprehensive understanding of the human GI tract microbiota through not only the phenotypic characterization of these species but also the sequencing of their whole genomes as a reference for metagenomic studies (53).
For these reasons, we have attempted to isolate uncultured bacteria from human feces. To date, we have successfully isolated 17 species, including 4 new genera (33–40, 43, 56). Here, we isolated another novel species. Many uncultured bacterial clones found in the feces of humans and other mammals show high levels of identity in their 16S rRNA gene sequences to those of our isolates, implying that these isolates are common among GI tract microbiota. A substrate availability test (described below) revealed that these isolates did not use any of the carbohydrates we tested. On the other hand, for human resident gut microbiota, dietary carbohydrates that resist degradation in the upper intestinal tract are important energy sources (7), and most of the phylotypes frequently detected in the human GI tract seem to be closely related to species that have functions in carbohydrate catabolism (48). Therefore, it was not clear what kind of substrate was used by these novel isolates and why they were prevalent in the GI tract. To address these questions, we analyzed the biochemical properties and energy source acquisition of these isolates. We also investigated the distribution of isolate-like bacteria in human feces by using real-time PCR analysis with species-specific primers, in addition to carrying out a database search.
MATERIALS AND METHODS
Isolation of strains YIT 12067T and YIT 12068.
Fecal samples were collected from two healthy Japanese males (subjects S and O, 37 and 40 years old, respectively) and immediately transferred into an anaerobic glove box that contained 88% N2, 7% H2, and 5% CO2. Each sample was weighed and diluted with prereduced 0.1 M phosphate-buffered saline (0.145 M NaCl, 0.15 M sodium phosphate; pH 7.0), and 100 μl of each was spread on modified Gifu anaerobic medium (GAM) agar plates (Nissui Pharmaceutical Co., Tokyo, Japan) supplemented with bile (2%, 4%, 6%, and 8% Bacto oxgall; Difco Laboratories, Detroit, MI), NaCl (1%, 3%, 6%, or 10%), or antibiotics (1 of 12 different antibiotics at three concentrations) in an attempt to suppress species that grew well and may therefore have inhibited our ability to isolate objective species. The composition of the modified GAM was as follows (per liter): peptone, 5.0 g; soy peptone, 3.0 g; proteose peptone, 5.0 g; serum powder, 10.0 g; yeast extract, 2.5 g; beef extract powder, 2.2 g; liver extract powder, 1.2 g; glucose, 0.5 g; soluble starch, 5.0 g; l-cysteine monohydrochloride, 0.3 g; sodium thioglycolate, 0.3 g; potassium dihydrogen phosphate, 2.5 g; sodium chloride, 3.0 g; l-tryptophan, 0.2 g; l-arginine, 1.0 g; vitamin K1, 5 mg; and hemin, 10 mg. Plates were incubated at 37°C for 3 days in an anaerobic glove box, as described above. Single colonies showing different morphologies were picked out and grown on modified GAM agar plates. This step was repeated until pure cultures were obtained. Strain YIT 12067T was isolated from a modified GAM agar plate supplemented with vancomycin (20 μg ml−1) and inoculated with a 10−5 serially diluted fecal sample from subject S. Strain YIT 12068 was isolated from a modified GAM agar plate supplemented with oxacillin (4 μg ml−1) and inoculated with a 10−5 serially diluted fecal sample from subject O. Both strains formed small pinpoint colonies on modified GAM agar plates.
Phylogenetic analysis.
DNA extraction, PCR, and sequencing of the 16S rRNA genes were performed as described previously (43). Closely related sequences were retrieved from the EMBL-EBI database (http://www.ebi.ac.uk/tools/sss/fasta/nucleotide.html) by using the FASTA program. Pairwise sequence similarity values for strains YIT 12067T and YIT 12068 and for the other species derived from the database search were calculated by using EzTaxon (5). Sequences were aligned by using ClustalX software (version 2.0) (50). Alignments were corrected manually by using the BioEdit program (16), and a phylogenetic tree was then produced by using the neighbor-joining (NJ) method with ClustalX. The topology of the tree was evaluated by a bootstrap analysis with 1,000 replicates. The tree was visualized by using the TreeView program (version 1.6.6) (41). Maximum-parsimony (MP) and maximum-likelihood (ML) methods were used to confirm the phylogenetic placement of the aligned sequences. MP analysis was performed by using the MEGA4 software package (47). The ML tree was calculated with the SEQBOOT and DNAML programs in the PHYLIP package (13). Fluorometric DNA-DNA hybridization in microdilution wells was performed as described previously (11). PCR-based random amplified polymorphic DNA (RAPD) fingerprinting was performed with the primers 5′-CCGCAGCCAA-3′ and 5′-AACGCGCAAC-3′ (2). Each 50-μl reaction mixture contained each deoxynucleoside triphosphate at a concentration of 200 μM, 80 pmol of primer, 3 mM MgCl2, 1× final concentration of Taq polymerase reaction buffer, 2 U of TaKaRa ExTaq polymerase Hot Start version (Takara Bio, Otsu, Japan), and genomic DNA (50 to 100 ng). PCR conditions were as follows: (i) 94°C for 2 min; (ii) 5 cycles consisting of 94°C for 30 s, 36°C for 60 s, and 72°C for 90 s; (iii) 29 cycles consisting of 94°C for 20 s, 36°C for 30 s, and 72°C for 90 s; (iv) 72°C for 3 min; and (v) hold at 4°C. The PCR products were subjected to agarose gel electrophoresis.
Substrate availability test.
Substrate availability for strains YIT 12067T and YIT 12068 was examined by using prereduced, anaerobically sterilized peptone-yeast extract (PY) (18) broth tubes with the addition of different carbohydrates and short-chain fatty acids (SCFA). The carbohydrates and SCFA tested were esculin, l-arabinose, d-cellobiose, glycerol, lactose, d-maltose, d-mannitol, d-mannose, d-raffinose, rhamnose, salicin, d-sorbitol, sucrose, d-trehalose, d-xylose, glucose, xylan, acetate, citrate, lactate, pyruvate, and succinate. Each carbohydrate was added at 0.5% or 1.0% (wt/wt), and the SCFA were supplemented to a final concentration of 20 mM. Growth was determined by observing visible turbidity.
Other phenotypic characterization.
Strains YIT 12067T and YIT 12068 underwent further phenotypic characterization. Phascolarctobacterium faecium ACM3679T, purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, served as a reference strain throughout. Growth in anaerobic and microaerophilic conditions was tested on PY agar plates supplemented with 80 mM sodium succinate (PYS). Addition of 80 mM succinate to PY medium was found to be better than 20 mM on the basis of a growth test using PY medium supplemented with 10, 20, 80, or 160 mM succinate. Anaerobic and microaerophilic atmospheres were created by using Anaeropack (Mitsubishi Gas Chemical, Tokyo, Japan) and CampyPak Plus microaerophilic system envelopes with a palladium catalyst (BBL, Sparks, MD), respectively. The morphology of the cells after a 2-day culture in the PYS medium was observed by using phase-contrast microscopy (BX51 system; Olympus, Tokyo, Japan) and scanning electron microscopy (SEM) (S-3400 system; Hitachi High-Technologies, Tokyo, Japan). For sample preparation for SEM, cells were placed on Sempore (JEOL Datum, Tokyo, Japan), fixed with glutaraldehyde and OsO4, critical point dried, and coated with osmium plasma. Spore formation by isolates grown on PYS agar plates for 2, 4, and 9 days was examined by using the spore-staining method (44). Sensitivity to bile was determined by comparing rates of growth of the strains on modified GAM agar with and without 2% Bacto oxgall after a 5-day incubation. The biochemical characteristics of strains YIT 12067T and YIT 12068 were determined by using the API 20A, API Rapid ID 32A, and APIZYM systems (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions, except that PYS agar plates were used for cell growth. All API tests were performed in duplicate. The end products from the isolates grown in prereduced PYS and PY medium supplemented with succinate were analyzed by using high-pressure liquid chromatography (HPLC), as described previously (4). Oxidase activity was examined by using oxidase test strips (Eiken Chemical Co., Ltd., Tokyo, Japan). Cellular fatty acid methyl esters (FAMEs) were obtained from lyophilized cells grown in PYS medium at 37°C for 2 days by saponification, methylation, and extraction, with minor modifications (24) of a method described previously (32). FAMEs were determined by using the MIDI system with MOORE5 5.00 of the MIS Standard Libraries. The DNA G+C contents were determined by hydrolyzing the DNA enzymatically and quantifying the nucleosides by using HPLC, as previously described (12).
Coculture experiments with succinate-producing bacteria.
To investigate the potential cross-feeding of succinate to the novel isolates from the succinate producer Paraprevotella xylaniphila YIT 11841T (34), we conducted coculture experiments in test tubes containing 3 ml of PY medium supplemented with 1% birchwood xylan (PYX). Strains YIT 12067T and YIT 12068 were grown at 37°C in PYS medium, and P. xylaniphila YIT 11841T was grown at 37°C in PYX medium. After incubation for 2 days under anaerobic conditions, strains YIT 12067T and YIT 12068 were cocultured with P. xylaniphila YIT 11841T in PYX medium. The inoculum size was 2% (vol/vol). The inoculated tubes were incubated at 37°C for 72 h. Growth was followed by plating the diluted inocula on the appropriate selective agar plates. Strains YIT 12067T and YIT 12068 were enumerated on PYS agar plates and P. xylaniphila YIT 11841T was enumerated on PY agar plates with 1% glucose and 25 μg/ml streptomycin. The plates were then incubated for 48 h at 37°C under anaerobic conditions, as described above. Triplicate experiments were performed using the same inocula. The concentrations of SCFA (succinate, propionate, acetate, formate, lactate, butyrate, isobutylate, valerate, and isovalerate) were determined by using HPLC, as described above. The coculture experiments were performed in triplicate.
Quantification of YIT 12067T-like bacteria in human feces by using real-time PCR.
Fecal numbers of strain YIT 12067T-like bacteria were quantified by using real-time PCR with 16S rRNA gene-targeted oligonucleotide primers. To design the species-specific primers, we obtained the sequences of the 16S rRNA genes of YIT 12067T, YIT 12068, and other closely related bacteria from the Ribosomal Database Project (6) and aligned them by using the ClustalX program; specific regions for both strains YIT 12067T and YIT 12068 were detected. Primers were designed by using the NCBI Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers, PhasF (5′-AGCAATCTCGCATGAGGATGCTGT-3′) and PhasR (5′-GCCGTGGCTTATTCGTTTACTACCG-3′), provided a 341-bp amplicon. Primer specificity was verified by using the Primer-BLAST and ProbeCheck programs (http://131.130.66.200/cgi-bin/probecheck/content.pl?id=home) (28). Specificity was further confirmed by use of DNA extracted from bacterial cells at values corresponding to 5 × 106 cells of the YIT 12067T strain and 96 different species, comprising five phyla: Actinobacteria (including 7 species of Bifidobacterium, 3 each of Atopobium and Collinsella, and 1 of Eggerthella), Bacteroidetes (including 8 species of Bacteroides and 3 of Prevotella), Firmicutes (including 18 species of Clostridium, 10 of Lactobacillus, 8 of Eubacterium, 5 of Ruminococcus, 4 of Roseburia, 3 of Veillonella, 2 of Peptostreptococcus, and 1 each of Anaerostipes, Coprococcus, Faecalibacterium, Megasphaera, Phascolarctobacterium, and Pseudobutyrivibrio), Fusobacteria (including 5 species of Fusobacterium), and Proteobacteria (including 2 species each of Desulfovibrio, Enterobacter, and Enterococcus and 1 each of Citrobacter, Escherichia, and Proteus). Bacterial cell counts were determined by using the 4′,6-diamidino-2-phenylindole (DAPI) staining method (20). Real-time PCR using each DNA extract of the 96 reference strains revealed that no amplified signal was detected under the conditions described below.
Real-time PCR amplification and detection were performed in a 10-μl volume with the ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA) and SYBR Premix EX Taq II (Takara Bio, Otsu, Japan), according to the manufacturers' instructions, with a final concentration of 0.4 μM for each primer. The amplification program consisted of 1 cycle at 95°C for 30 s, then 40 cycles at 95°C for 5 s and 60°C for 34 s. The fluorescent product was detected in the last step of each cycle. To distinguish the target PCR product from nontarget product, melting curve analysis was performed after amplification using the default settings of the ABI software from 60°C to 95°C.
A standard curve was created by using serial 10-fold dilutions of strain YIT 12067T pure culture DNA corresponding to 106 to 101 cells per reaction, as determined microscopically by using the DAPI staining method. The bacterial concentration of each sample was calculated by comparing the threshold cycle (CT) values obtained from the standard curve. All samples were analyzed in triplicate. A linear relationship between the cell counts and the CT values (r2 = 0.9975) was observed when the number of cells per reaction mixture was between 106 and 101.
Thirty-three healthy volunteers (30 males and 3 females; ages, 20 to 64 years [average, 39.4 ± 10.3 years]) provided fresh fecal samples. DNA extraction from fecal samples was performed according to the method using glass beads and phenol described previously (30).
Nucleotide sequence accession numbers.
Sequences for the 16S rRNA gene of strains YIT 12067T and YIT 12068 have been deposited in the GenBank/EMBL/DDBJ database under accession no. AB490811 and AB490812, respectively. Accession numbers for sequences used as references are indicated in the phylogenetic tree.
RESULTS
Phylogenetic analysis of strains YIT 12067T and YIT 12068.
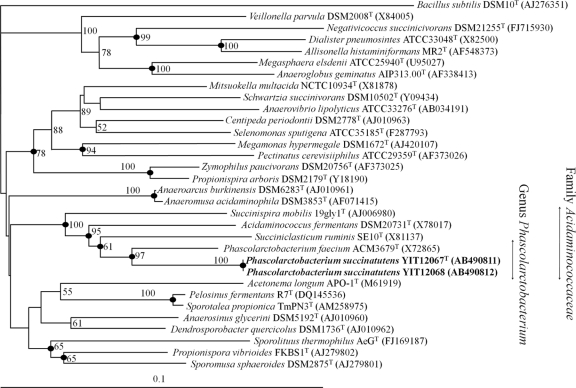
We assessed 1,610 bp and 1,611 bp of the 16S rRNA genes of strains YIT 12067T and YIT 12068, respectively, and found that they had 99.9% sequence identity. Although the isolates could be discriminated by RAPD fingerprinting (data not shown), they had 93% DNA-DNA relatedness. Database searches revealed that the strain with the highest 16S rRNA gene sequence relatedness to strains YIT 12067T and YIT 12068 was Phascolarctobacterium faecium ACM 3679T, with 91.4% and 91.5% identity, respectively. The 16S rRNA gene sequences of strains YIT 12067T and YIT 12068 and those of related species of the family Acidaminococcaceae in the order Selenomonadales were aligned, and a phylogenetic tree was constructed by using Bacillus subtilis as an outgroup (Fig. 1). The phylogenetic tree shows that the new isolates belonged to the family Acidaminococcaceae. The positions of the strains were phylogenetically distinct, and the specific lineages were robust and supported by high bootstrap values. These results were supported by those obtained with the MP and ML methods (data not shown).
Fig 1.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic position of strains YIT 12067T and YIT 12068 among related taxa. Bootstrap values (>50%) are given at the branch nodes. Filled circles indicate that the corresponding nodes were also recovered in the tree generated with the maximum-parsimony and maximum-likelihood algorithms. Bar, 0.1 substitutions per nucleotide position.
Carbohydrate and SCFA utilization.
Dietary carbohydrates that escape digestion in the upper gastrointestinal tract are the most important energy sources for microbial growth in the large intestine (7). Carbohydrate utilization tests using PY broth tubes showed that strains YIT 12067T and YIT 12068 did not utilize any carbohydrate tested. We then tested whether these strains used SCFA, which are major end products of the bacterial breakdown of carbohydrates and also serve as important energy sources for GI tract microbiota (51). PY broth tube tests indicated that strains YIT 12067T and YIT 12068 grew well only in PY medium supplemented with succinate. HPLC analysis showed that succinate was quantitatively converted to propionate. These results indicate that succinate decarboxylation stimulated the growth of strains YIT 12067T and YIT 12068. Addition of succinate to modified GAM or PY agar plates at a final concentration of 80 mM resulted in the formation of larger colonies (up to 1.2 mm in diameter) than the pinpoint colonies on these media without succinate.
Phenotypic characteristics.
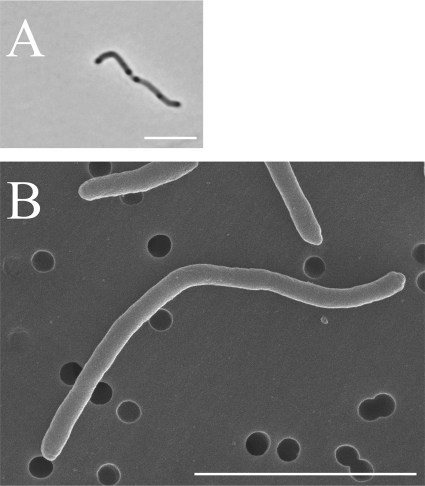
Strains YIT 12067T and YIT 12068 grew under strict anaerobic conditions, but not in aerobic or microaerobic atmospheres. The cells of both strains were Gram negative, nonmotile, non-spore-forming, and long rod shaped. The cells had several darker regions, like nodes, after 2 days of incubation in PYS medium (Fig. 2A). Strain YIT 12067T cells were 0.4 to 0.5 μm wide and 2.4 to 22.3 μm long (Fig. 2B); strain YIT 12068 cells were nearly identical in size to those of strain YIT 12067T. Both strains were oxidase and catalase negative. Growth of the isolates was completely suppressed by the addition of bile to the PYS agar. In contrast, P. faecium formed colonies on PYS agar supplemented with bile, although there were fewer colonies than on normal PYS agar. The cellular fatty acid compositions of strains YIT 12067T and YIT 12068 and that of P. faecium ACM3679T as the closest related reference strain are shown in Table 1. The major (>10%) cellular fatty acids of strains YIT 12067T and YIT 12068 were C14:0 dimethyl acetal and summed feature 4, comprising C15:2 and/or C15:1ω8c and/or an unknown fatty acid of ECL 14.762. The DNA G+C contents of strains YIT 12067T and YIT 12068 and P. faecium ACM3679T, as determined by HPLC, were 46.5, 47.3, and 45.4 mol%, respectively. The value for strain YIT 12067T was also determined by genome sequence analysis and found to be 47 mol%. In the phenotypic analyses with the API systems (API 20A, API Rapid ID32A, and API ZYM), the novel isolates and P. faecium ACM3679T showed no positive reactions.
Fig 2.
Phase-contrast (A) and scanning electron (B) micrographs of strain YIT 12067T grown in PYS broth for 2 days. Bars, 5 μm.
Table 1.
Cellular fatty acid compositions of strains YIT 12067T and YIT 12068 and Phascolarctobacterium faecium ACM3679T
| Fatty acid | % total fatty acidsa for: |
||
|---|---|---|---|
| YIT 12067T | YIT 12068 | P. faecium | |
| Saturated straight chain | |||
| C11:0 | 8.23 | 8.38 | 5.11 |
| C12:0 | 1.54 | 4.76 | 5.47 |
| C13:0 | 9.92 | 7.03 | 2.18 |
| C14:0 | 1.47 | ||
| C15:0 | 8.99 | 7.54 | 14.21 |
| C16:0 | 2.64 | 2.50 | 5.23 |
| C18:0 | 1.12 | 1.26 | |
| Unsaturated straight chain | |||
| C16:1ω9c | 3.12 | 12.90 | |
| C16:1ω7c | 5.31 | 3.83 | 5.09 |
| C18:1ω9c | 4.21 | 3.98 | 4.51 |
| C18:2ω6,9c | 1.42 | 1.36 | 1.46 |
| Hydroxy acid | |||
| C15:0 3-OH | 1.92 | 1.86 | 4.05 |
| Dimethyl acetal | |||
| C14:0 DMA | 22.43 | 23.99 | 15.53 |
| C16:0 DMA | 1.33 | 2.02 | |
| C16:1ω7c DMA | 2.77 | 3.20 | 1.07 |
| Summed featuresb | |||
| 4 | 18.27 | 11.89 | 4.22 |
| 5 | 3.53 | 5.28 | 8.60 |
| 6 | 1.18 | 2.21 | |
| 7 | 1.75 | ||
Only those fatty acids that make up >1% of the total are shown.
Summed feature compositions are as follows: 4, C15:2 and/or C15:1ω8c and/or an unknown fatty acid of ECL 14.762; 5, C15:0 DMA and/or C14:0 3-OH; 6, anteiso-C15:0 3-OH and/or C16:1ω9cDMA; 7, C17:1ω7c and/or an unknown fatty acid of ECL 16.760.
Effect of coculture with a succinate producer.
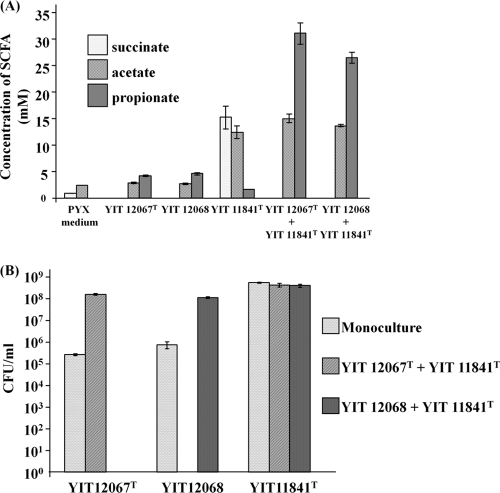
Succinate is not a major fermentation product in human feces (51); however, it is produced by many of the microorganisms that colonize the human GI tract, such as members of the Bacteroides and Prevotella genera (45). Therefore, to create a nutritional situation analogous to what takes place in vivo, coculture experiments with the succinate producer P. xylaniphila YIT 11841T were performed. Paraprevotella xylaniphila YIT 11841T can use xylan as a growth substrate, producing mainly succinate (34), whereas strains YIT 12067T and YIT 12068 do not ferment xylan. Xylan is one of the most abundant components of plant fibers, which are important sources of substrates for the resident gut microorganisms in humans. After 72 h of coculture with P. xylaniphila YIT 11841T, the cell numbers of the strains YIT 12067T and YIT 12068 became approximately 2 to 3 orders of magnitude higher than those of the monoculture; succinate became undetectable, and propionate was formed (Fig. 3). Growth of P. xylaniphila YIT 11841T was not affected by the presence of strains YIT 12067T and YIT 12068.
Fig 3.
Coculture of the succinate-producing bacterium P. xylaniphila YIT 11841T and strain YIT 12067T or YIT 12068 after 72 h of growth in PY medium supplemented with 1% birchwood xylan. (A) Concentrations of succinate, acetate, and propionate, as determined by using HPLC. Triplicate experiments were performed. (B) CFU of strains YIT 12067T and YIT 12068 and P. xylaniphila YIT 11841T. Diluted inocula were plated on the appropriate selective agar plates. Strains YIT 12067T and YIT 12068 were enumerated on PYS agar plates, and P. xylaniphila YIT 11841T was enumerated on PY agar plates with 1% glucose and 25 μg/ml of streptomycin. Plates were incubated for 48 h at 37°C under anaerobic conditions. Triplicate experiments were performed. The initial cell numbers of strains YIT 12067Tand 12068 and of P. xylaniphila YIT 11841T were 3.85 ×103, 8.44 × 104, and 7.67 × 105 CFU ml−1, respectively.
Database search for YIT 12067T-like uncultured bacterial clones.
Recently improved sequencing technology has led to the deposition of a large number of uncultured bacterial clones in the GenBank/EMBL/DDBJ public databases. To obtain a broader estimate of the redundancy of these isolates, we searched for uncultured bacterial clones that were closely related to the sequence of strain YIT 12067T. With 98.7% identity as the threshold (this is the % identity proposed for distinguishing species 46), we found 210 published uncultured bacterial clones by using the FASTA program (Table 2). These clones were isolated from fecal samples from humans and other mammals, especially primates. Similar sequences were also detected from the feces of other mammals, namely, rats, beef cattle, and swine. These results indicate that YIT 12067T-like bacteria are broadly distributed in the intestines of humans and other mammals, especially primates.
Table 2.
Sources of uncultured bacterial clones that showed ≥98.7% 16S rRNA gene sequence identity to that of strain YIT 12067T listed in the EMBL database
| Source | Diet | No. of sequences retrieveda | Reference(s) of study(ies) in which 16S rRNA gene sequences were detected |
|---|---|---|---|
| Primates | |||
| Human | Omnivore | 132 | 10, 14, 15, 23, 26, 27, 29, 31, 52 |
| Chimpanzee | Omnivore | 12 | 26 |
| Bonobo | Omnivore | 4 | 26 |
| East Angolan colobus | Herbivore | 4 | 26 |
| Francois' langur | Herbivore | 4 | 26 |
| Mongoose lemur | Omnivore | 4 | 26 |
| Eastern black and white colobus | Herbivore | 2 | 26 |
| Black lemur | Omnivore | 1 | 26 |
| Sumatran orangutan | Herbivore | 1 | 26 |
| Other mammals | |||
| Rat | Omnivore | 36 | 1, 3 |
| Beef cattle | Herbivore | 9 | 9 |
| Swine | Omnivore | 1 | 25 |
Retrieved from published data.
Quantification of strain YIT 12067T-like bacteria in human feces.
To determine the actual prevalence and proportion of strain YIT 12067T-like bacteria in human feces, we performed real-time PCR with species-specific primers. Application of real-time PCR to fecal samples from 33 volunteers showed that strain YIT 12067T-like bacteria were detected in 7 of the 33 fecal samples (21%), at an average level of 3.34 × 108 cells/g of feces (Table 3). The detection ratio and concentration of strain YIT 12067T-like bacteria were similar to those of members of subdominant species of human GI tract microbiota, such as Bifidobacterium bifidum (detection ratio, 28%; concentration, 2.00 × 108 cells/g) and Bifidobacterium breve (17%, 2.00 × 107 cells/g) (30), indicating that strain YIT 12067T-like bacteria are also subdominant members of the human GI microbiota.
Table 3.
Cell counts of strain YIT 12067T-like bacteria in fecal samples from Japanese volunteers determined by using real-time PCR
| Subjecta | Count (cells/g feces) of: |
% relative to the total no. of cells | |
|---|---|---|---|
| Strain YIT 12067T-like bacteria | Total cellsb | ||
| E036 | 3.70 × 108 | 7.94 × 1010 | 0.45 |
| E038 | 2.20 × 107 | 1.26 × 1011 | 0.02 |
| E041 | 2.44 × 107 | 2.51 × 1010 | 0.10 |
| E046 | 1.19 × 107 | 1.58 × 1011 | 0.01 |
| E049 | 1.28 × 109 | 2.51 × 1011 | 0.53 |
| E059 | 4.74 × 108 | 1.26 × 1012 | 0.41 |
| E061 | 1.54 × 108 | 1.00 × 1011 | 0.14 |
| Mean | 3.34 × 108 | 2.86 × 1011 | 0.24 |
Subjects from which strain YIT 12067T-like bacteria were detected, among 33 volunteers.
Total number of bacteria as determined by DAPI count.
DISCUSSION
The use of molecularly based methods has revealed that many of the phylotypes found in the human GI tract are detected only through the use of culture-independent methods (42). In this uncultured phylogenetic group, however, some species were isolated from other mammals or environmental samples. The genus Phascolarctobacterium had consisted of only one species, P. faecium, isolated from koala feces (8). Although uncultured clones that were closely related to P. faecium were frequently detected in samples from the human GI tract, there had been no isolates belonging to the genus Phascolarctobacterium from the human GI tract.
On the basis of the findings described here, we propose that strains YIT 12067T and YIT 12068 belong to the genus Phascolarctobacterium within the family Acidaminococcaceae; these strains are strictly anaerobic, nonmotile, non-spore-forming, Gram negative, and asaccharolytic. They utilize succinate and produce propionate. In the API systems, they were unreactive. They had cellular fatty acid compositions and DNA G+C contents similar to those of P. faecium ACM3679T. However, these novel isolates could be differentiated from P. faecium ACM3679T on the basis of bile sensitivity and 16S rRNA gene sequence identity (91.4%). Although the isolates and P. faecium shared low sequence identity, these two species had quite similar phenotypic characteristics. Accordingly, we propose that strains YIT 12067T and YIT 12068 represent a new species of the genus Phascolarctobacterium. To our knowledge, this is the first report of the isolation of members of the genus Phascolarctobacterium from the human GI microbiota.
Isolation of members of the genus Phascolarctobacterium might have been difficult because of the very narrow range of energy sources available. In our experiments, strains YIT 12067T and YIT 12068 used only succinate as an energy source, as has been shown for P. faecium ACM3679T. We isolated these novel strains by using modified GAM-based medium to which succinate had not been added. HPLC analysis revealed that the modified GAM contained a small quantity of succinate (0.7 mM), which may have allowed these strains to form very small pinpoint colonies. The isolation of bacteria highly specific for using succinate might have been caused by selective pressure that suppressed the growth of other bacterial strains forming large colonies on the basal agar. To this end, we have isolated 17 new species from human GI tract microbiota by using this strategy (33–40, 43, 56); the use of nutrient-rich basal agar plates supplemented with various selective pressures thus appears to be effective for isolating various uncultured species from the GI microbiota.
In the substrate utility test, strains YIT 12067T and YIT 12068 required succinate as an energy source. Although the free energy change of succinate decarboxylation is small compared with that of carbohydrates, amino acids, and other major SCFA (49), succinate decarboxylation to propionate is coupled with ATP synthesis (21), and the ability to decarboxylate succinate is found mainly among members of Clostridium cluster IX (e.g., P. faecium [8], Succiniclasticum ruminis [54], and Schwartzia succinivorans [55] as exclusively succinate utilizers and Succinispira mobilis [22] as a succinate and amino acid utilizer). Succinate is seldom detected as a major fermentation product in human feces (51), yet succinate-producing saccharolytic bacteria are abundant among the GI microbiota. Therefore, the specific fermentation of succinate may result in not only the acquisition of a constant energy source but also the ability to coexist with bacteria that use other high-energy-yielding substrates, such as resistant carbohydrates. In fact, our coculture experiments showed that the growth of strains YIT 12067T and YIT 12068 was stimulated by the presence of succinate-producing bacteria, as succinate was converted to propionate (Fig. 3A and B). It seems likely that, to adapt to the GI tract environment with its variety of bacterial species, strains YIT 12067T and YIT 12068 might have evolved to specialize in the utilization of succinate produced by other microorganisms. Analysis of the EMBL database library (Table 2) and real-time PCR analysis (Table 3) support the concept that strain YIT 12067T-like bacteria have adapted to the GI tract and are prevalent in humans and other mammals as subdominant members. Harmsen et al. (17) reported that the detection ratio for the Phascolarctobacterium group, including the genera Succiniclasticum and Acidaminococcus, was 81% (8 of 11 samples) at an average level of 9.0 × 108 cells/g feces (dry weight). Thus, other members of this group with characteristics similar to those of strains YIT 12067T and YIT 12068 are also likely to be persistent in the human GI tract.
Interestingly, the database search for uncultured bacterial clones with high levels of similarity in their 16S rRNA gene sequences to those of the novel isolates also revealed many clones in the feces of several kinds of primates and other mammals (Table 2). These host organisms were omnivorous or herbivorous. Ley et al. (26), through a network-based analysis of bacterial diversity from fecal samples of humans and 59 other mammalian species, suggested that both diet and phylogeny of the host influence GI bacterial diversity. Presumably, strain YIT 12067T-like bacteria might have undergone widespread distribution in the GI tracts of herbivorous or omnivorous mammals—mainly primates, including humans.
Strain YIT 12067T has been selected for inclusion in the catalog of reference genomes by the human microbiome project (53), and a high-quality draft sequence has been completed. The assembled and annotated genomic sequences of this bacterium have been submitted to the GenBank/EMBL/DDBJ databases (http://www.ncbi.nlm.nih.gov/genome/2893). These sequences contain 2,203 predicted genes in 2,122,261 bp, as read by the Genome Sequencing Center, Washington University School of Medicine. Further genomic research, such as exploring the genes involved in energy metabolism or comparing the genomic structure with those of other members of the human GI tract, may provide additional insights into the role of this succinate-specific utilizer in the GI tract or into its evolutionary strategy of adaptation for competition and survival in this complex microbial ecosystem.
The phylogenetic and phenotypic analysis indicated that strains YIT 12067T and YIT 12068 belonged to the same species. Although these isolates could be differentiated from their nearest relative, P. faecium, in terms of 16S rRNA gene sequence similarity and bile resistance, they shared similar phenotypic characteristics. We therefore propose that strains YIT 12067T and YIT 12068 represent a novel species of the genus Phascolarctobacterium, for which the name Phascolarctobacterium succinatutens sp. nov. is proposed.
Description of Phascolarctobacterium succinatutens sp. nov.
Phascolarctobacterium succinatutens (suc.ci.nat.u′tens; N.L. n. succinas -atis, succinate; L. v. utor, make use of; N.L. part. adj. sucinatutens, using succinate). Cells are Gram negative, nonmotile, non-spore-forming, and long rod shaped. They are strictly anaerobic. Cells are 0.4 to 0.5 μm wide and 2.4 to 22.3 μm long. Colonies after 2 days of growth at 37°C on PY agar supplemented with 80 mM succinate are 0.8 to 1.2 mm in diameter, circular, and beige. Oxidase, catalase, and bile resistance are negative. API 20A tests are negative for acid production from l-arabinose, d-cellobiose, glucose, glycerol, lactose, maltose, d-mannitol, d-mannose, d-melezitose, d-raffinose, l-rhamnose, salicin, sucrose, d-sorbitol, d-trehalose, and d-xylose. API Rapid ID32A and API ZYM results are negative for nitrate reduction, urease, and hydrolysis of esculin and gelatin. Indole is not produced. Negative reactions are obtained for alanine, arginine, cystine, glutamyl glutamic acid, glycine, histidine, leucine, phenylalanine, proline, pyroglutamic acid, serine, tyrosine, valine, and leucyl glycine arylamidase and for naphthol-AS-BI-phosphohydrolase, acid phosphatase, N-acetyl-β-glucosaminidase, alkaline phosphatase, arginine dihydrolase, α-arabinosidase, α-fucosidase, α- and β-galactosidase, 6-phospho-β-galactosidase, α- and β-glucosidase, β-glucuronidase, α-mannosidase, chymotrypsin, esterase (C4), esterase lipase (C8), glutamic acid decarboxylase, lipase (C14), and trypsin. The major cellular fatty acids are C14:0 dimethyl acetal and summed feature 4, comprising C15:2 and/or C15:1ω8c and/or an unknown fatty acid of ECL 14.762. The DNA G+C content is 46.5% to 47.3%. The type strain, YIT 12067T (DSM 22533T or JCM 16074T), was isolated from human feces. Strain YIT 12068 (DSM 22521 or JCM 16075) is also assigned to this species.
ACKNOWLEDGMENTS
We are grateful to Jean P. Euzéby of the Ecole Nationale Vétérinaire in Toulouse for his suggestions regarding the etymology of the species epithet. We thank M. Ando and C. Hata for helping with the electron micrographs. We thank T. Sato and T. Matsuda for their advice and help with the real-time PCR assay. We also thank R. Tanaka and H. Sawada for their understanding and encouragement through our research activities.
Footnotes
Published ahead of print 11 November 2011
REFERENCES
- 1. Abnous K, et al. 2009. Diets enriched in oat bran or wheat bran temporally and differentially alter the composition of the fecal community of rats. J. Nutr. 139:2024–2031 [DOI] [PubMed] [Google Scholar]
- 2. Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brooks SPJ, et al. 2009. In-feed administered sub-therapeutic chlortetracycline alters community composition and structure but not the abundance of community resistance determinants in the fecal flora of the rat. Anaerobe 15:145–154 [DOI] [PubMed] [Google Scholar]
- 4. Chonan O, Matsumoto K, Watanuki M. 1995. Effect of galactooligosaccharides on calcium absorption and preventing bone loss in ovariectomized rats. Biosci. Biotechnol. Biochem. 59:236–239 [DOI] [PubMed] [Google Scholar]
- 5. Chun J, et al. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2259–2261 [DOI] [PubMed] [Google Scholar]
- 6. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cummings JH, Englyst HN. 1987. Fermentation in the human large intestine and the available substrates. Am. J. Clin. Nutr. 45:1243–1255 [DOI] [PubMed] [Google Scholar]
- 8. Del Dot T, Osawa R, Stackebrandt E. 1993. Phascolarctobacterium faecium gen. nov, spec. nov., a novel taxon of the Sporomusa group of Bacteria. Syst. Appl. Microbiol. 16:380–384 [Google Scholar]
- 9. Durso LM, et al. 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 76:4858–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ezaki T, Hashimoto Y, Yabuuchi E. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224–229 [Google Scholar]
- 12. Ezaki T, et al. 1990. Rapid procedure to determine the DNA base composition from small amounts of Gram-positive bacteria. FEMS Microbiol. Lett. 55:127–130 [DOI] [PubMed] [Google Scholar]
- 13. Felsenstein J. 2007. PHYLIP (Phylogeny Inference Package) version 3.67. Department of Genetics, University of Washington, Seattle, WA [Google Scholar]
- 14. Gill SR, et al. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. 2006. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44:4136–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 17. Harmsen HJM, Raangs GC, He T, Degener JE, Welling GW. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holdeman LV, Cato EP, Moore WEC. 1977. VPI anaerobe laboratory manual. Virginia Polytechnic Institute and State University, Blacksburg, VA [Google Scholar]
- 19. Hooper LV, et al. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884 [DOI] [PubMed] [Google Scholar]
- 20. Jansen GJ, Wildeboer-Veloo AC, Tonk RH, Franks AH, Welling GW. 1999. Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37:215–221 [DOI] [PubMed] [Google Scholar]
- 21. Janssen PH. 1992. Growth yield increase and ATP formation linked to succinate decarboxylation in Veillonella parvula. Arch. Microbiol. 157:442–445 [DOI] [PubMed] [Google Scholar]
- 22. Janssen PH, O'Farrell KA. 1999. Succinispira mobilis gen. nov., sp. nov., a succinate-decarboxylating anaerobic bacterium. Int. J. Syst. Bacteriol. 49:1009–1013 [DOI] [PubMed] [Google Scholar]
- 23. Khachatryan ZA, et al. 2008. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One 3:e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuykendall LD, Roy MA, O'Neill JJ, Devine TE. 1988. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Bacteriol. 38:358–361 [Google Scholar]
- 25. Leser TD, et al. 2002. Culture–independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 28. Loy A, et al. 2008. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol. 10:2894–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mai V, Greenwald B, Morris JG, Raufman J-P, Stine OC. 2006. Effect of bowel preparation and colonoscopy on post-procedure intestinal microbiota composition. Gut 55:1822–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuki T, et al. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McLaughlin SD, et al. 2010. The bacteriology of pouchitis: a molecular phylogenetic analysis using 16S rRNA gene cloning and sequencing. Ann. Surg. 252:90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller LT. 1982. Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J. Clin. Microbiol. 16:584–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morotomi M, Nagai F, Sakon H, Tanaka R. 2008. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 58:2716–2720 [DOI] [PubMed] [Google Scholar]
- 34. Morotomi M, Nagai F, Sakon H, Tanaka R. 2009. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family “Prevotellaceae” isolated from human faeces. Int. J. Syst. Evol. Microbiol. 59:1895–1900 [DOI] [PubMed] [Google Scholar]
- 35. Morotomi M, Nagai F, Watanabe Y. Christensenella minuta gen. nov., sp. nov., isolated from human faeces that forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 36. Morotomi M, Nagai F, Watanabe Y. 2011. Parasutterella secunda sp. nov., isolated from human faeces and proposal of Sutterellaceae fam. nov. in the order Burkholderiales. Int. J. Syst. Evol. Microbiol. 61:637–643 [DOI] [PubMed] [Google Scholar]
- 37. Morotomi M, Nagai F, Watanabe Y, Tanaka R. 2010. Succinatimonas hippei gen. nov., sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 60:1788–1793 [DOI] [PubMed] [Google Scholar]
- 38. Nagai F, Morotomi M, Sakon H, Tanaka R. 2009. Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces. Int. J. Syst. Evol. Microbiol. 59:1793–1797 [DOI] [PubMed] [Google Scholar]
- 39. Nagai F, Morotomi M, Watanabe Y, Sakon H, Tanaka R. 2010. Alistipes indistinctus sp. nov. and Odoribacter laneus sp. nov., common members of the human intestinal microbiota isolated from faeces. Int. J. Syst. Evol. Microbiol. 60:1296–1302 [DOI] [PubMed] [Google Scholar]
- 40. Nagai F, Watanabe Y, Morotomi M. 2010. Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int. J. Syst. Evol. Microbiol. 60:2639–2646 [DOI] [PubMed] [Google Scholar]
- 41. Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 42. Rajilić-Stojanović M, Smidt H, de Vos WM. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9:2125–2136 [DOI] [PubMed] [Google Scholar]
- 43. Sakon H, Nagai F, Morotomi M, Tanaka R. 2008. Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 58:970–975 [DOI] [PubMed] [Google Scholar]
- 44. Schaeffer AB, Fulton MD. 1933. A simplified method of staining endospores. Science 77:194. [DOI] [PubMed] [Google Scholar]
- 45. Shah HN, et al. 2009. Approaches to the study of the systematics of anaerobic, Gram-negative, non-sporeforming rods: current status and perspectives. Anaerobe 15:179–194 [DOI] [PubMed] [Google Scholar]
- 46. Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152–155 [Google Scholar]
- 47. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 48. Tap J, et al. 2009. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11:2574–2584 [DOI] [PubMed] [Google Scholar]
- 49. Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Topping DL, Clifton PM. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031–1064 [DOI] [PubMed] [Google Scholar]
- 52. Turnbaugh PJ, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Turnbaugh PJ, et al. 2007. The human microbiome project. Nature 449:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Gylswyk NO. 1995. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 45:297–300 [DOI] [PubMed] [Google Scholar]
- 55. van Gylswyk NO, Hippe H, Rainey FA. 1997. Schwartzia succinivorans gen. nov., sp. nov., another ruminal bacterium utilizing succinate as the sole energy source. Int. J. Syst. Bacteriol. 47:155–159 [DOI] [PubMed] [Google Scholar]
- 56. Watanabe Y, Nagai F, Morotomi M, Sakon H, Tanaka R. 2010. Bacteroides clarus sp. nov., Bacteroides fluxus sp. nov. and Bacteroides oleiciplenus sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 60:1864–1869 [DOI] [PubMed] [Google Scholar]